Abstract
Three-dimensional (3D) cell culture is regarded as a more physiologically relevant method of growing cells in the laboratory compared to traditional monolayer cultures. Recently, the application of polystyrene-based scaffolds produced using polyHIPE technology (porous polymers derived from high internal phase emulsions) for routine 3D cell culture applications has generated very promising results in terms of improved replication of native cellular function in the laboratory. These materials, which are now available as commercial scaffolds, are superior to many other 3D cell substrates due to their high porosity, controllable morphology, and suitable mechanical strength. However, until now there have been no reports describing the surface-modification of these materials for enhanced cell adhesion and function. This study, therefore, describes the surface functionalization of these materials with galactose, a carbohydrate known to specifically bind to hepatocytes via the asialoglycoprotein receptor (ASGPR), to further improve hepatocyte adhesion and function when growing on the scaffold. We first modify a typical polystyrene-based polyHIPE to produce a cell culture scaffold carrying pendent activated-ester functionality. This was achieved via the incorporation of pentafluorophenyl acrylate (PFPA) into the initial styrene (STY) emulsion, which upon polymerization formed a polyHIPE with a porosity of 92% and an average void diameter of 33 μm. Histological analysis showed that this polyHIPE was a suitable 3D scaffold for hepatocyte cell culture. Galactose-functionalized scaffolds were then prepared by attaching 2′-aminoethyl-β-d-galactopyranoside to this PFPA functionalized polyHIPE via displacement of the labile pentafluorophenyl group, to yield scaffolds with approximately ca. 7–9% surface carbohydrate. Experiments with primary rat hepatocytes showed that cellular albumin synthesis was greatly enhanced during the initial adhesion/settlement period of cells on the galactose-functionalized material, suggesting that the surface carbohydrates are accessible and selective to cells entering the scaffold. This porous polymer scaffold could, therefore, have important application as a 3D scaffold that offers enhanced hepatocyte adhesion and functionality.
Introduction
Substantial evidence exists to support three-dimensional (3D) cell culture as a more physiologically relevant growth environment compared to that with conventional monolayer cultures.1−4 Cells cultured in 3D more closely mimic their native morphology, unlike monolayer cultures in which cells are often flattened into a two-dimensional (2D) shape. Cells in the 3D environment can also experience more interaction with their neighbors, in turn increasing cellular communication that is important in regulating normal cell function. Recognizing these advantages associated with a 3D growth environment, many researchers now require practical technologies that can enable routine 3D cell culture in the laboratory.5
Synthetic, nonbiodegradable, porous polymers are attractive materials as routine 3D scaffolds as they are inert, reproducible, and can be engineered into a versatile range of morphologies. Recently, several groups have employed porous polymers derived from high internal phase emulsions (polyHIPEs) as scaffolds for 3D cell culture.6−11 In particular, polystyrene-based polyHIPE scaffolds have shown very promising results with a range of different cell types due to their controllable morphology, high porosity and suitable mechanical properties.12−16 These materials are also now available as commercial 3D scaffolds (AlvetexScaffold by Reinnervate) and have already been adopted by a broad range of research groups.17,18
One potential limitation of polystyrene-based polyHIPEs as 3D cell scaffolds is surface chemistry. Cells in vivo are surrounded by a complex extracellular matrix that contributes to cell anchorage and function. They also receive a plethora of biochemical cues from molecules such as carbohydrates and proteins that serve to regulate normal cell behavior. Being able to mimic some of these biological interactions on the surface of synthetic 3D scaffolds is therefore an attractive prospect.19 However, achieving this for polystyrene-based polyHIPEs is challenging. Postpolymerization modifications of polystyrene are possible but often require harsh reaction conditions due to the inert nature of the polymer.20,21 Similarly, including a new functional comonomer into the prepolymerized emulsion can often disrupt emulsion stability and thus distort polyHIPE morphology. Recently, several groups have successfully overcome the latter issue by employing functional comonomers that are sufficiently soluble in the styrene (STY) external emulsion phase. For example, Krajnc et al. demonstrated that methacrylic acid could be incorporated.22 Heise et al. reported that 4-vinylbenzylphthalimide could be incorporated for subsequent polypeptide attachment.23,24 Other groups have also previously included 4-vinylbenzyl chloride.25,26 All of these functional comonomers potentially offer a useful functional ‘hook’ on the polyHIPE surface as a route to facile attachment of various (bio)molecules, such as proteins or carbohydrate residues.
Our work therefore describes the inclusion of an ester comonomer into a styrene-based emulsion as a facile route to polyHIPE surface modification with aminoethyl glycosides. We show that a polystyrene-based polyHIPE containing pentafluorophenyl acrylate (PFPA) can be produced with a suitable morphology to support 3D cell growth (photopolymerised polyacrylate polyHIPEs incorporating PFPA have been described previously27). A straightforward postpolymerization functionalization of this material was then employed to attach pendent galactose residues via aminoethyl glycoside coupling with PFPA, similar to the strategy described by Boyer and Davis for solution-based glycopolymer synthesis.28−30 The selectivity and accessibility of the pendent galactose residues was demonstrated by the culture of primary rat hepatocytes, which contain a cell-surface asialoglycoprotein receptor (ASGPR31) that can specifically bind to galactose for enhanced cell adhesion and function.32,33 Glucose-functionalized polyHIPEs were also prepared as a selectivity control for galactose during the primary rat cell adhesion experiments.
Materials and Methods
Materials
The monomers styrene (STY), divinylbenzene (DVB), and 2-ethylhexylacrylate (EHA) were obtained from Sigma Aldrich and used without further purification. Span80 and anhydrous dimethylformamide (DMF) were also obtained from Sigma Aldrich. The polymerization initiator 2,2′-azobisisobutyronitrile (AIBN) was supplied by Acros Organics and recrystallized from methanol before use. Pentafluorophenyl acrylate (PFPA),34 2′-aminoethyl-β-d-glucopyranoside, and 2′-aminoethyl-β-d-galactopyranoside35 were synthesized as described in the literature. Characterization data were in accord with published values.
The hepatocellular carcinoma HepG2 cell line was supplied by the American Type Culture Collection (ATCC). Rat Sprague–Dawley pooled cryopreserved primary hepatocytes (Grade P) were obtained from Biopredic International along with the thawing medium. Culture medium materials were used according to ATCC and Biopredic recommendations for HepG2 and primary rat hepatocytes, respectively. The Albumin ELISA kit was obtained from Universal Biologicals Cambridge. The AlvetexScaffold polyHIPE and associated plastic clips and well inserts used to house the polyHIPE membranes during cell-culture were supplied by Reinnervate.
PolyHIPE Nomenclature
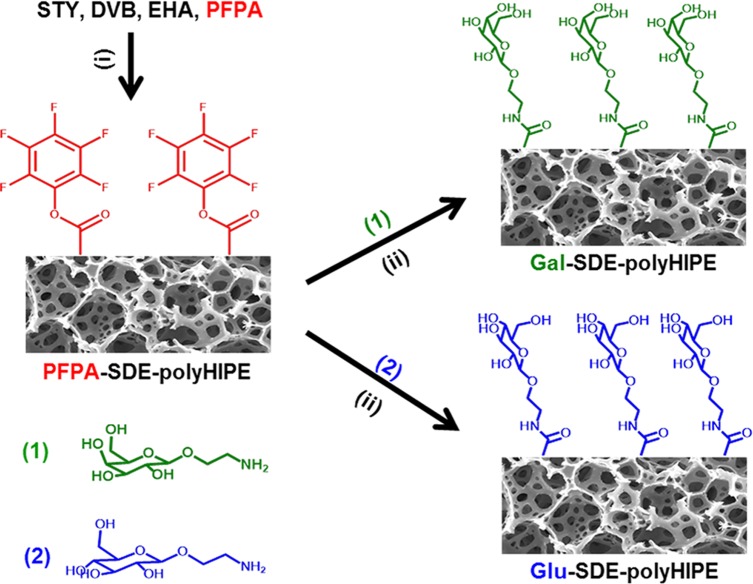
The SDE-polyHIPE corresponds to the parent unfunctionalised polyHIPE derived from STY, DVB, and EHA. The xPFPA-SDE-polyHIPEs correspond to the different PFPA-functionalized SDE-polyHIPEs, where x denotes the % PFPA in the initial HIPE monomer mixture. The Gal-SDE-polyHIPE refers to the resulting material after coupling the 26PFPA-SDE-polyHIPE with 2′-aminoethyl-β-d-galactopyranoside. The Glu-SDE-polyHIPE refers to the resulting material after coupling the 26PFPA-SDE-polyHIPE with 2′-aminoethyl-β-d-glucopyranoside.
PFPA-SDE-polyHIPE Synthesis and Morphological Characterization
The preparation of polystyrene-based porous polymers by emulsion templating has already been well documented.8,14,15 Specific HIPE formulations for this study are shown in Table S1 in the Supporting Information. Briefly, an oil phase consisting of the organic monomers, AIBN initiator, and surfactant Span80 were placed in a 250 mL 3-necked round-bottomed flask and stirred continually at 350 rpm using a PTFE paddle connected to an overhead stirrer. To this organic phase, deionized water was slowly added at room temperature via a dropping funnel. Stirring continued for an additional 2 min after the last water droplet was added, before transferring the high internal phase emulsion (HIPE) to a 50 mL centrifuge tube. The tube was then placed in a 60 °C oven for 24 h to polymerize. The resulting polyHIPE monolith was then Soxhlet washed in acetone for 24 h and left to air-dry overnight in a fume hood.
The morphology of the polyHIPEs was investigated using a Phillips XL30 ESEM operating in SEM mode between 10 kV and 25 kV. Samples were first mounted on carbon fiber pads preadhered to aluminum stubs and then gold coated using an Edwards Pirani 501 sputter coater before imaging. ImageJ software was used to measure void diameters on each micrograph. Mercury intrusion porosimetry was performed using a Micromeritics AutoPore IV using penetrometers with a stem volume of 1.836 mL and a bulb volume of 5 mL. Analysis was performed from 0.5 to 1600 psi.
HepG2 Culture and Histological Analysis on the 26PFPA-SDE-polyHIPE
HepG2 cells were precultured in T75 flasks before use. The 26PFPA-SDE-polyHIPE monolith was sectioned into 200 μm membranes using a Leica VT1000S vibrotome and then cut into circles of 15 mm diameter using a bore-cutter. These discs, along with AlvetexScaffold discs (commercial SDE-polyHIPE control), were quickly submerged in ethanol and then washed extensively with PBS before being housed in plastic inserts in a 12-well plate. Then, 0.4 million cells were added to each polyHIPE via a 100 μL media suspension, and cells were cultured for 5 days at 37 °C and 5% CO2 in 4 mL of media. After this period, cells were fixed in Bouin’s fixative, dehydrated using ethanol, and embedded into paraffin wax. Ten micrometer sections were then used for hematoxylin and eosin staining.
PFPA-SDE-polyHIPE Functionalization with Aminoethyl Glycosides
The 26PFPA-SDE-polyHIPE monolith was sectioned into 200 μm membranes using a Leica VT1000S vibrotome and then cut into circles of 15 mm diameter using a bore-cutter. Six discs were then placed in a glass vial containing 12 mL of dimethylformamide and 60 mg of either 2′-aminoethyl-β-d-galactopyranoside or 2′-aminoethyl-β-d-glucopyranoside. The vials were then placed in a shaker oven at 40 °C for 48 h with the shaker operating at 150 rpm. Samples were then slowly rehydrated though a series of Milli-Q water-dimethylformamide gradients so as not to collapse the polyHIPE structure during deswelling. The functionalized polyHIPEs were then washed in Milli-Q water and then left to dry before analysis.
Detection of Surface Carbohydrates
Attenuated Total Reflection Fourier Transform Infra Red (ATR-FTIR) spectra were recorded on a Perkin-Elmer 1600 Series FTIR spectrometer fitted with a Golden Gate ATR element. Solid samples were pressed using a spatula before being placed on the crystal. Spectra were evaluated with Omnic, version 7.3. 13C solid state NMR spectra were recorded on a Varian VNMRS 400 spectrometer at a frequency of 100.56 MHz using direct excitation with proton decoupling. Spectra were obtained with total sideband suppression (TOSS). 19F solid state NMR spectra were recorded on a Varian Unity Inova 300 spectrometer at a frequency of 282.10 MHz, using direct polarization and no decoupling. Spectra were evaluated with MestReNova, version 8.1.1–11591.
X-ray photoelectron spectroscopy (XPS) was performed at the National EPSRC XPS User’s Service (NEXUS) at Newcastle University. A K-Alpha instrument equipped with a monochromated AlKa source (Thermo Scientific) was used. A pass energy of 40 eV and a step size of 0.1 eV was used for high resolution spectra of the elements of interest. Spectra were analyzed using Casa XPS licensed at Newcastle University.
Primary Rat Hepatocyte Culture and Albumin Analysis
Cyropreserved primary rat hepatocytes were brought up from frozen and diluted into prewarmed thawing medium. Cells were then resuspended into culture medium to give a final concentration of 0.2 million cells per 1 mL of media. PolyHIPE discs were quickly submerged in ethanol and then washed extensively with PBS before being clipped into the wells of a 24-well cell-culture plate. One milliliter of the cell suspension was added to each polyHIPE, and the cells were cultured at 37 °C and 5% CO2. Albumin assessments were carried out using a rat-specific albumin ELISA (Assaypro ERA3201-1) according to the protocol provided with the kit.
Results and Discussion
PolyHIPEs derived from styrene STY, DVB, and EHA have been previously reported as suitable scaffolds for routine 3D cell culture.12,16,36 The main component of these polyHIPEs is STY (∼60% of monomer mixture), with DVB present as a cross-linking comonomer to increase the mechanical strength and EHA present to increase polymer elasticity by reducing the Tg. This type of polyHIPE, termed SDE-polyHIPE for the purpose of this study, was therefore chosen as the parent material to be functionalized via a two-step strategy, as shown in Figure 1.
Figure 1.
Schematic showing the synthetic strategy for incorporating carbohydrate functionality onto the surface of a polystyrene-based polyHIPE (SDE-polyHIPE). Step 1 involves the incorporation of PFPA into the material to leave pendent ester functionality (PFPA-SDE-polyHIPE). Step 2 then involves a coupling reaction between the ester functional groups on the polyHIPE and the aminoethyl glycosides (1) and (2). Reaction (i) conditions: AIBN, 60 °C, and 24 h. Reaction (ii) conditions: DMF, 40 °C, and 48 h.
SDE-polyHIPEs are often fabricated using a nonionic surfactant with a low HLB, such as Span80, which is insoluble in the dispersed phase and so inhibits emulsion phase separation. Electrolytes such as calcium chloride (CaCl2) are also sometimes used in the fabrication process of these materials to promote emulsion stabilization by improving surfactant packing at the interface.37 However, in this study we chose to exclude CaCl2 because the additional organic phase component PFPA was expected to provide some extra emulsion stabilization and consequently a reduction in void diameter. SDE-polyHIPEs are also usually fabricated with potassium persulfate (KPS) as the free radical initiator, which being water-soluble, favors polymerization of those monomers found in excess at the interface. However, we chose AIBN as the initiator for this study since the interfacial activity of PFPA in comparison with that of the other monomers was unknown.
PFPA Functionalization of SDE-polyHIPE
The first step toward galactose-functionalized polystyrene-based polyHIPEs was to include PFPA as a functional comonomer into the parent HIPE to render pendent ester functionality in the polymerized monolith. PFPA was chosen as a suitable monomer as it has hydrophobicity similar to that of STY (log PPFPA = 2.5538 and log PSTY = 2.9539) and thus was not expected to significantly disrupt HIPE stability. PFPA has also been previously reported to undergo facile coupling reactions with amines when incorporated into photopolymerized polyacrylate polyHIPEs,27 as well as when polymerized as a poly(PFPA) homopolymer.40 Furthermore, PFPA contains 5 flourine atoms that are conveniently detectable by NMR and X-ray photoelectron spectroscopy (XPS).
An oil phase was prepared containing the organic monomers (STY, DVB, EHA, and PFPA), AIBN and Span80. To this, an aqueous water phase was added and the mixture stirred vigorously to form HIPE. Increasing PFPA monomer concentrations were attempted, namely, 10 wt %, 20 wt %, 26 wt %, 33 wt %, and 43 wt % relative to the total monomer mixture (see Table S1 in Supporting Information for complete HIPE compositions). All formulations formed a stable HIPE with no apparent signs of phase separation. The HIPE mixtures were then thermally polymerized at 60 °C for 24 h to form PFPA-SDE-polyHIPEs.
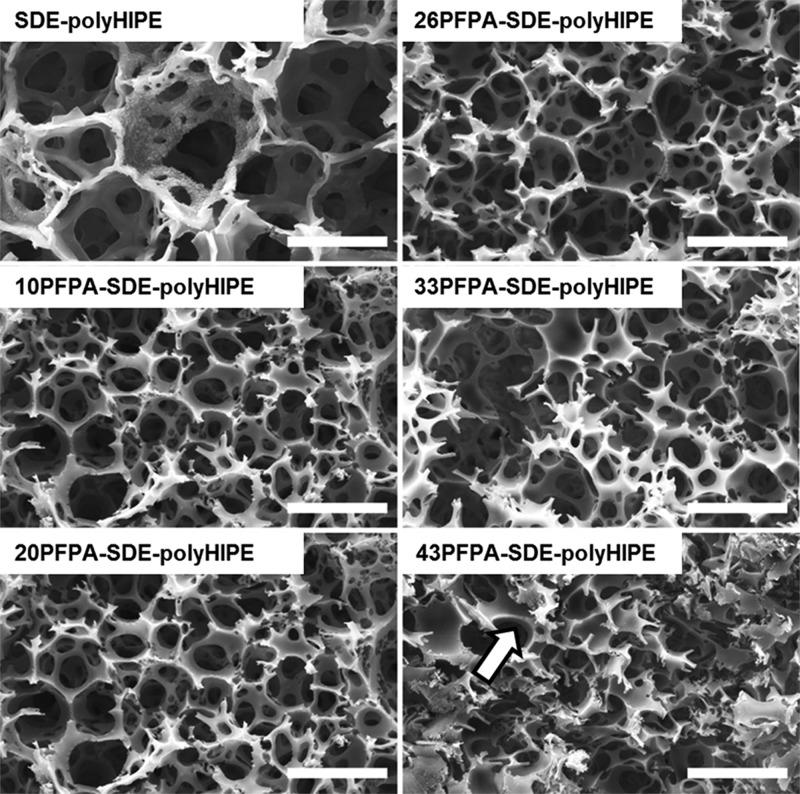
The morphologies of the polymerized PFPA-SDE-polyHIPEs are shown in Figure 2. A typical SDE-polyHIPE morphology was observed for all PFPA-SDE-polyHIPE materials except for the 43PFPA-SDE-polyHIPE (where 43 corresponds to ∼43 wt % of the initial monomer mixture being attributed to PFPA). The morphology of this material was found to be collapsed and with unidentifiable voids. For those materials that resembled typical polyHIPE morphologies, an overall decrease in void diameter was observed with increasing PFPA concentration, from approximately 69 μm (control: 0 wt % PFPA) to 28 μm (33 wt % PFPA). Table S2 in the Supporting Information contains specific physical characteristics for each polyHIPE. This trend toward smaller void sizes suggests an increase in emulsion stability with higher PFPA incorporation, which is likely the result of the higher organic content helping to form wider and more robust continuous phase films around the internal phase droplets. This hypothesis is supported in the case of the 43PFPA-SDE-polyHIPE, where the strut thickness of the material is visibly larger than those polyHIPEs with lower PFPA content (Figure 2, white arrow).
Figure 2.
Scanning electron micrographs showing the morphologies of the different PFPA-SDE-polyHIPEs. The white arrow indicates thicker struts in the 43PFPA-SDE-polyHIPE material. Scale bars = 50 μm.
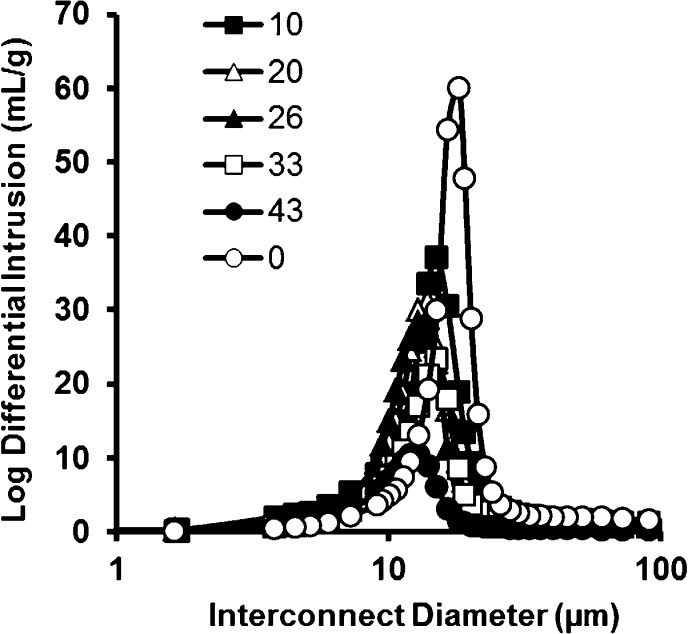
The average interconnect diameter also decreases with increasing PFPA content, which is to be expected given that void diameter decreases (again refer to Table S2, Supporting Information). Figure 3 shows the interconnect diameter distribution for PFPA-SDE-polyHIPEs. Generally, a wider distribution in interconnect diameter was observed with higher levels of PFPA incorporation.
Figure 3.
Interconnect diameter distribution for the different PFPA-SDE-polyHIPEs. The numbers in the legend correspond to the different PFPA concentrations added to the SDE-HIPE formulations (wt % in total monomer mixture).
The porosities of all materials remained high (∼90%), although a general decreasing trend was observed with increasing PFPA concentration (Table S2, Supporting Information), which is to be expected with an increasing organic phase content and therefore a lowered internal phase volume fraction.
Overall, 26PFPA-SDE-polyHIPE appeared to display optimum physical characteristics for the maximum theoretical PFPA loading. The material has an average void diameter of 33 μm and an average interconnect diameter of 10 μm, compared to 69 and 15 μm, respectively, in the parent control SDE-polyHIPE. This difference was deemed acceptable, given that many cells have diameters in the region of 15 to 25 μm and therefore sufficiently smaller than the 33 μm voids in 26PFPA-SDE-polyHIPE. Furthermore, the commercial AlvetexScaffold has an average void diameter of 42 μm. To check the compatibility of the 26PFPA-SDE-polyHIPE morphology for 3D cell growth, we cultured HepG2 cells (a hepatocyte-derived cell line) on the material. Figure 4 shows a histological cross-section of the cells after 5 days of growth. Cells anchored onto the PFPA-functionalized polyHIPE in a manner similar to that with AlvetexScaffold and formed a tissue-like layer in the top portion of the membrane. All cells appeared healthy and viable with no signs of necrosis. The penetration of cells into the material was slightly less compared to that with AlvetexScaffold. This would be expected given that AlvetexScaffold has a slightly larger average void diameter. Nonetheless, cells still managed to enter the PFPA-functionalized material after only 5 days of culture.
Figure 4.
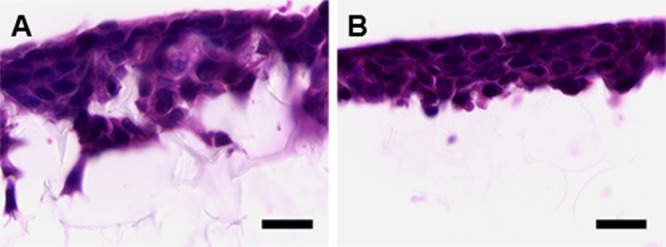
Haematoxylin and Eosin staining of HepG2 cells cultured on commercial SDE-polyHIPE (AlvetexScaffold) (A) and 26PFPA-SDE-polyHIPE (B) after 5 days. Scale bar = 20 μm.
Carbohydrate Functionalization of 26PFPA-SDE-polyHIPE
2′-Aminoethyl-β-d-galactopyranoside (galactose-amine (1)) and 2′-aminoethyl-β-d-glucopyranoside (glucose-amine (2)) were chosen for reaction with the 26PFPA-SDE-polyHIPE. These aminoethyl glycosides were chosen over galactosamine and glucosamine in order to lock the carbohydrate in the beta conformation after coupling with the PFPA ester. With galactosamine and glucosamine, the amine group is attached to the 2-carbon leaving the 1-carbon free. This can therefore lead to ring-opening of the carbohydrate into the open-chair form and thus the subsequent cyclic rearrangement into alpha, beta, and furanoside forms that may jeopardize binding with the ASGPR.
PolyHIPE was first sectioned into 200 μm membranes and cut into discs of 15 mm in diameter. These discs were then mixed with a solution of aminoethyl glycoside in DMF for 48 h at 40 °C under constant agitation. The resulting materials, termed either Gal-SDE-polyHIPE or Glu-SDE-polyHIPE due to galactose or glucose coupling, respectively, were then slowly rehydrated and washed extensively with Milli-Q water before characterization.
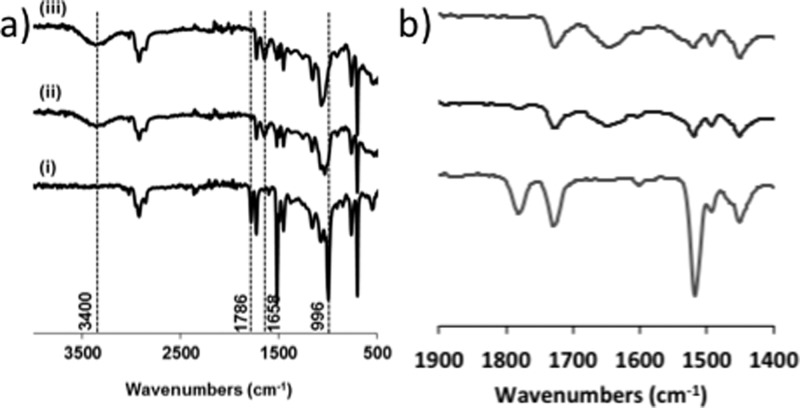
Figure 5 shows the ATR-FTIR spectra for 26PFPA-SDE-polyHIPE along with Gal-SDE-polyHIPE and Glu-SDE-polyHIPE. All materials contain a peak at 1732 cm–1, which corresponds to the EHA carbonyl group from the parent SDE-polyHIPE formulation (EHA ester C=O stretching). The presence of the PFPA in the starting material is confirmed by the peaks at 996 cm–1 (C–F stretching), 1520 cm–1 (Ar C=C stretching), and 1786 cm–1 (PFPA ester C=O stretching). For the carbohydrate-functionalized materials, these peaks almost disappear, which is consistent with the loss of pentafluorophenol during nucleophilic substitution. The spectra of the carbohydrate-functionalized materials have additional peaks at 1658 cm–1 (amide C=O stretching) and 3400 cm–1 (O–H stretching).
Figure 5.
(a) ATR-FTIR spectra of 26PFPA-SDE-polyHIPE (i), Gal-SDE-polyHIPE (ii), and Glu-SDE-polyHIPE (iii). (b) Expansion of the range 1400–1900 cm–1 (order of spectra is the same as that in panel a).
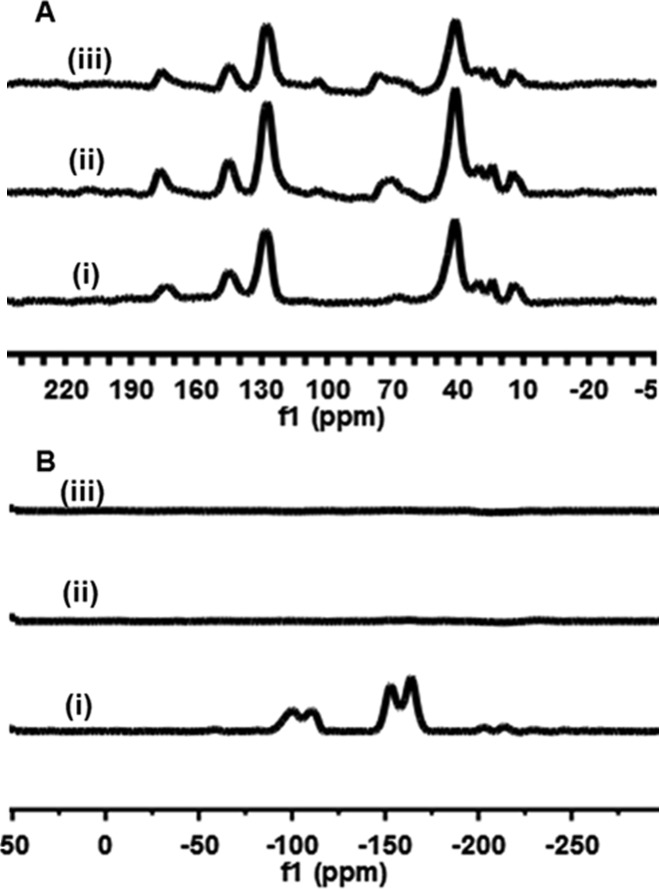
Figure 6 shows the solid state NMR spectra (13C and 19F) for 26PFPA-SDE-polyHIPE along with Gal-SDE-polyHIPE and Glu-SDE-polyHIPE. A new carbon peak occurs at ∼70 ppm in the 13C spectra of the carbohydrate-functionalized materials (C–OH) that is not present in the starting 26PFPA-SDE-polyHIPE. Similarly, a complete disappearance of fluorine peaks is observed in the 19F spectra for both carbohydrate-functionalized materials, suggesting a near complete conversion to the amide-carbohydrate.
Figure 6.
13C (A) and 19F (B) solid state NMR spectra of 26PFPA-SDE-polyHIPE (i), Gal-SDE-polyHIPE (ii), and Glu-SDE-polyHIPE (iii).
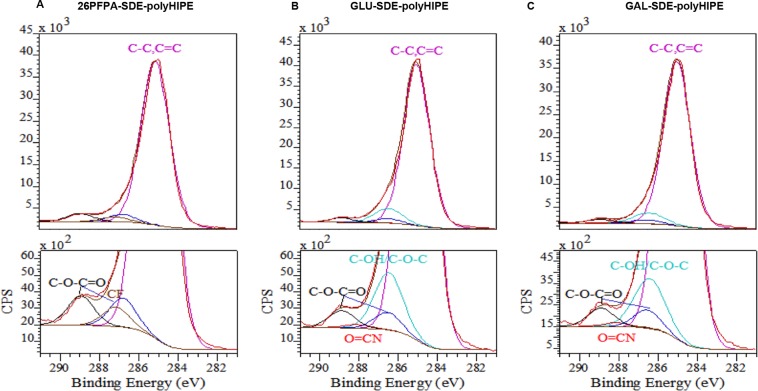
XPS was used to quantify the amount of PFPA and carbohydrate on the surface of 26PFPA-SDE-polyHIPE, Gal-SDE-polyHIPE, and Glu-SDE-polyHIPE materials. Figure 7 shows the peak-fitted high resolution C1s spectra for the three materials. F1s and N1s high resolution spectra can also be found in the Supporting Information, along with surface atomic concentrations from survey spectra (Table S3, Supporting Information). Peaks were fitted for (a) C–C, C=C at a binding energy (BE) of 285.11 eV, (b) C–OH, C-OC at a BE of 286.00 eV, (c) C–O–C=O at BE’s of 286.81 and 288.93 eV, (d) C–F at BE of 287.12 eV, and (e) O=C–N at BE of 288.13 eV. Noticeably larger peak areas for C–F and C–O–C=O are observed for 26PFPA-SDE-polyHIPE compared to the carbohydrate-functionalized materials. Quantification shows that the 26PFPA-SDE-polyHIPE contains ca. 9% PFPA, whereas Gal-SDE-polyHIPE and Glu-SDE-polyHIPE contain ca. 0%. This loss of PFPA compared to the original 26 wt % in the initial emulsion has also been observed in other polyHIPE systems27 and may be attributed to either partial PFPA solubility in the aqueous phase, hydrolysis of the pentafluorophenyl ester, or incomplete PFPA polymerization. Both carbohydrate-functionalized materials display new peak areas for C–OH/C-OC and O=CN that are absent in 26PFPA-SDE-polyHIPE. Quantification shows that the materials contain between ca. 7% and 9% carbohydrate, suggesting a near complete conversion from ester to amide under the reaction conditions employed.
Figure 7.
High resolution peak-fitted C1s spectra for 26PFPA-SDE-polyHIPE (A), Glu-SDE-polyHIPE (B), and Gal-SDE-polyHIPE (C).
Primary Rat Hepatocyte Culture on Galactose-Functionalized SDE-polyHIPEs
Hepatocytes are the main functional cells of the liver and are used extensively as in vitro models for drug toxicity screening.41 However, primary hepatocytes taken directly from a patient or animal are notoriously difficult to culture, with a rapid loss of differentiated phenotype occurring almost instantly after removal from the native tissue. Consequently most functional experiments with primary rat hepatocytes occur soon after seeding onto the substrate and rarely progress beyond 2–3 days.
Galactose is a carbohydrate that is known to specifically bind to hepatocytes to promote cell adhesion and function via the ASGPR. We therefore wanted to assess if the pendent galactose residues on our Gal-SDE-polyHIPE were accessible and selective to primary rat hepatocytes as a means of improving cell adhesion onto the scaffold. As glucose is not recognized by the ASGPR, it was chosen as a selective control.
Cryopreserved primary rat hepatocytes were cultured in the presence of serum proteins for up to 24 h and assessed for albumin production, a typical marker for hepatic function (Figure 8). We chose to culture in the presence of serum as this is typical laboratory practice for hepatocyte culture, even though this may increase nonselective cell binding onto the scaffold via protein deposition.
Figure 8.
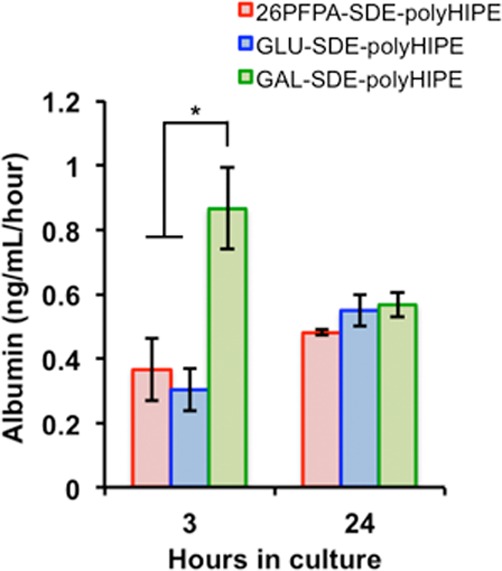
Primary rat hepatocyte albumin synthesis after culture on 26PFPA-SDE-polyHIPE, Gal-SDE-polyHIPE, and Glu-SDE-polyHIPE. Data represent the mean ± SEM, n = 3. * denotes p < 0.05 as determined by Student’s t test.
After 3 h (cell adhesion period), hepatocytes produced a significantly higher amount of albumin on Gal-SDE-polyHIPE compared to that in Glu-SDE-polyHIPE and PFPA-SDE-polyHIPE, suggesting that galactose is accessible and can maintain selectivity with ASGPR. These data are consistent with other 2D and 3D substrates functionalized with galactose residues.42−44 However, as the culture period progressed, this enhanced albumin synthesis was diminished, potentially due to nonselective adherence onto the scaffold via serum protein coating.
Conclusions
We have developed a polystyrene-based porous polymer using polyHIPE technology that carries pendent-activated ester functionality. The morphology of this material is comparable to that of typical polystyrene-based polyHIPE scaffolds and supports the 3D growth of hepatocyte-based cells. This material can also undergo facile surface coupling reactions with aminoethyl glycosides to render pendent carbohydrate functionality on the material surface. Preliminary studies with primary rat hepatocytes on a galactose-functionalized polyHIPE show that the carbohydrates are accessible and selective to cells entering the scaffold, seen by an enhanced functional activity of the cells as they initially adapt to the culture environment. This is the first report of a surface-functional polystyrene-based polyHIPE for 3D hepatocyte applications. We therefore believe this material could be a cornerstone for the development of tailored polyHIPE scaffolds that promote the survival and differentiation of primary hepatocytes used in drug discovery models.
Acknowledgments
Funding from the EPSRC, CIKTN, Reinnervate and the Leverhulme Trust is greatly acknowledged. We thank Dr. David Apperley from Durham University Solid State NMR Service. X-ray photoelectron spectra were obtained at the National EPSRC XPS User’s Service (NEXUS) at Newcastle University, an EPSRC Mid-Range Facility. N.R.C. acknowledges the P2M RNP programme of the European Science Foundation.
Glossary
Abbreviations
- STY
styrene
- DVB
divinylbenzene
- EHA
2-ethylhexylacrylate
- PFPA
pentafluorophenyl acrylate
- HIPE
high internal phase emulsion
- polyHIPE
porous polymer fabricated from a high internal phase emulsion
- ASGPR
asialoglycoprotein receptor
Supporting Information Available
Details of HIPE compositions, polyHIPE physical characteristics, and additional XPS data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare the following competing financial interest(s): Professor Stefan Przyborski is Chief Scientific Officer and Director of Reinnervate Limited. The work was funded in part by Reinnervate Limited through the EPSRC Industrial CASE scheme.
Supplementary Material
References
- Kimlin L. C.; Casagrande G.; Virador V. M. Mol. Carcinog. 2013, 52, 167–182. [DOI] [PubMed] [Google Scholar]
- Pampaloni F.; Reynaud E. G.; Stelzer E. H. K. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [DOI] [PubMed] [Google Scholar]
- Abbott A. Nature 2003, 424, 870–872. [DOI] [PubMed] [Google Scholar]
- Yamada K. M.; Cukierman E. Cell 2007, 130, 601–610. [DOI] [PubMed] [Google Scholar]
- Maltman D. J.; Przyborski S. A. Biochem. Soc. Trans. 2010, 38, 1072–1075. [DOI] [PubMed] [Google Scholar]
- Barbetta A.; Dentini M.; De Vecchis M. S.; Filippini P.; Formisano G.; Caiazza S. Adv. Funct. Mater. 2005, 15, 118–124. [Google Scholar]
- Barbetta A.; Massimi M.; Devirgiliis L. C.; Dentini M. Biomacromolecules 2006, 7, 3059–3068. [DOI] [PubMed] [Google Scholar]
- Akay G.; Birch M. A.; Bokhari M. A. Biomaterials 2004, 25, 3991–4000. [DOI] [PubMed] [Google Scholar]
- Bokhari M. A.; Akay G.; Zhang S. G.; Birch M. A. Biomaterials 2005, 26, 5198–5208. [DOI] [PubMed] [Google Scholar]
- Lumelsky Y.; Lalush-Michael I.; Levenberg S.; Silverstein M. S. J. Polym. Sci., Part A: Polym. Chem. 2009, 47, 7043–7053. [Google Scholar]
- Lumelsky Y.; Zoldan J.; Levenberg S.; Silverstein M. S. Macromolecules 2008, 41, 1469–1474. [Google Scholar]
- Bokhari M.; Carnachan R. J.; Cameron N. R.; Przyborski S. A. Biochem. Biophys. Res. Commun. 2007, 354, 1095–1100. [DOI] [PubMed] [Google Scholar]
- Bokhari M.; Carnachan R. J.; Cameron N. R.; Przyborski S. A. J. Anat. 2007, 211, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayman M. W.; Smith K. H.; Cameron N. R.; Przyborski S. A. Biochem. Biophys. Res. Commun. 2004, 314, 483–488. [DOI] [PubMed] [Google Scholar]
- Hayman M. W.; Smith K. H.; Cameron N. R.; Przyborski S. A. J. Biochem. Biophys. Methods 2005, 62, 231–240. [DOI] [PubMed] [Google Scholar]
- Schutte M.; Fox B.; Baradez M.-O.; Devonshire A.; Minguez J.; Bokhari M.; Przyborski S.; Marshall D. Assay Drug Dev. Technol. 2011, 9, 475–486. [DOI] [PubMed] [Google Scholar]
- Neofytou E. A.; Chang E.; Patlola B.; Joubert L. M.; Rajadas J.; Gambhir S. S.; Cheng Z.; Robbins R. C.; Beygui R. E. J. Biomed. Mater. Res., Part A 2011, 98, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan N.; Elliott R.; Clewes O.; Mackay A.; Reis-Filho J. S.; Burn J.; Langtry J.; Sieber-Blum M.; Lord C. J.; Ashworth A. Oncogene 2011, 30, 4243–4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens M. M.; George J. H. Science 2005, 310, 1135–1138. [DOI] [PubMed] [Google Scholar]
- Mercier A.; Deleuze H.; Mondain-Monval O. React. Funct. Polym. 2000, 46, 67–79. [Google Scholar]
- Cameron N. R.; Sherrington D. C.; Ando I.; Kurosu H. J. Mater. Chem. 1996, 6, 719–726. [Google Scholar]
- Sevsek U.; Krajnc P. React. Funct. Polym. 2012, 72, 221–226. [Google Scholar]
- Audouin F.; Larragy R.; Fox M.; O’Connor B.; Heise A. Biomacromolecules 2012, 13, 3787–3794. [DOI] [PubMed] [Google Scholar]
- Audouin F.; Fox M.; Larragy R.; Clarke P.; Huang J.; O’Connor B.; Heise A. Macromolecules 2012, 45, 6127–6135. [Google Scholar]
- Alexandratos S. D.; Beauvais R.; Duke J. R.; Jorgensen B. S. J. Appl. Polym. Sci. 1998, 68, 1911–1916. [Google Scholar]
- Benicewicz B. C.; Jarvinen G. D.; Kathios D. J.; Jorgensen B. S. J. Radioanal. Nucl. Chem. 1998, 235, 31–35. [Google Scholar]
- Kircher L.; Theato P.; Cameron N. R. Polymer 2013, 54, 1755–1761. [Google Scholar]
- Boyer C.; Davis T. P. Chem. Commun. 2009, 6029–6031. [DOI] [PubMed] [Google Scholar]
- Beija M.; Li Y.; Lowe A. B.; Davis T. P.; Boyer C. Eur. Polym. J. 2013, 49, 3060–3071. [Google Scholar]
- Li Y.; Duong H. T. T.; Jones M. W.; Basuki J. S.; Hu J.; Boyer C.; Davis T. P. ACS Macro Lett. 2013, 2, 912–917. [DOI] [PubMed] [Google Scholar]
- Ashwell G.; Morell A. G. Adv. Enzymol. Relat. Areas Mol. Biol. 1974, 41, 99–128. [DOI] [PubMed] [Google Scholar]
- Cho C. S.; Hoshiba T.; Harada I.; Akaike T. React. Funct. Polym. 2007, 67, 1301–1310. [Google Scholar]
- Cho C. S.; Seo S. J.; Park I. K.; Kim S. H.; Kim T. H.; Hoshiba T.; Harada I.; Akaike T. Biomaterials 2006, 27, 576–585. [DOI] [PubMed] [Google Scholar]
- Jochum F. D.; Theato P. Macromolecules 2009, 42, 5941–5945. [Google Scholar]
- Chernyak A. Y.; Sharma G. V. M; Kononov L. O.; Krishna P. R.; Levinsky A. B.; Kochetkov N. K.; Rao A. V. R. Carbohydr. Res. 1992, 223, 303–309. [Google Scholar]
- Bokhari M.; Carnachan R. J.; Cameron N. R.; Przyborski S. A. J. Anat. 2007, 211, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan V.; Solans C.; Kunieda H. Colloid Polym. Sci. 1994, 272, 1166–1173. [Google Scholar]
- ChemSpider ID: 2056299. http://www.chemspider.com/Chemical-Structure.2056299.html (accessed May 28, 2013).
- ChemSpider ID: 7220. http://www.chemspider.com/Chemical-Structure.7220.html (accessed May 28, 2013).
- Jochum F. D.; Theato P. Polymer 2009, 50, 3079–3085. [Google Scholar]
- Hewitt N. J.; Lechon M. J. G.; Houston J. B.; Hallifax D.; Brown H. S.; Maurel P.; Kenna J. G.; Gustavsson L.; Lohmann C.; Skonberg C.; Guillouzo A.; Tuschl G.; Li A. P.; LeCluyse E.; Groothuis G. M. M.; Hengstler J. G. Drug Metab. Rev. 2007, 39, 159–234. [DOI] [PubMed] [Google Scholar]
- Park T. G. J. Biomed. Mater. Res. 2002, 59, 127–135. [DOI] [PubMed] [Google Scholar]
- Ying L.; Yin C.; Zhuo R. X.; Leong K. W.; Mao H. Q.; Kang E. T.; Neoh K. G. Biomacromolecules 2003, 4, 157–165. [DOI] [PubMed] [Google Scholar]
- Yin C.; Ying L.; Zhang P. C.; Zhuo R. X.; Kang E. T.; Leong K. W.; Mao H. Q. J. Biomed. Mater. Res., Part A 2003, 67A, 1093–1104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.