Abstract
In Caulobacter crescentus, the PopZ polar scaffold protein supports asymmetric cell division by recruiting distinct sets of binding partners to opposite cell poles. To understand how polar organizing centers are established by PopZ, we investigated a set of mutated PopZ proteins for defects in sub-cellular localization and recruitment activity. We identified a domain within the C-terminal 76 amino acids that is necessary and sufficient for accumulation as a single subcellular focus, a domain within the N-terminal 23 amino acids that is necessary for bipolar targeting, and a linker domain between these localization determinants that tolerates large variation. Mutations that inhibited dynamic PopZ localization inhibited the recruitment of other factors to cell poles. Mutations in the C-terminal domain also blocked discrete steps in the assembly of higher-order structures. Biophysical analysis of purified wildtype and assembly-defective mutant proteins indicates that PopZ self-associates into an elongated trimer, which readily forms a dimer of trimers through lateral contact. The final six amino acids of PopZ are necessary for connecting the hexamers into filaments, and these structures are important for sub-cellular localization. Thus, PopZ undergoes multiple orders of self-assembly, and the formation of an interconnected superstructure is a key feature of polar organization in Caulobacter.
INTRODUCTION
In asymmetric cell divisions, unequal partitioning of regulatory factors results in daughter cells with different characteristics and activities. This is a fundamental aspect of cell type differentiation and stem cell maintenance in eukaryotes (Yamashita and Fuller 2008; Kusek et al., 2012), and it is also a key aspect in the cell cycle of many bacterial species, which upon division produce morphologically and behaviorally distinct cell types. In all cases, a set of behavioral determinants is specifically targeted to one side of the division plane and held in place until septation is complete. This is a particularly remarkable feat in prokaryotes given that it occurs in the absence of extracellular cues, without directed vesicular transport, and in a small contiguous cytoplasm, across which molecules can readily diffuse. Bacterial asymmetry typically involves unequal distribution of proteins between the two cell poles, though the underlying processes of pole recognition and protein complex assembly are not fully understood (Bowman et al., 2011).
The gram negative bacterium Caulobacter crescentus divides asymmetrically during each cell cycle, producing daughter cells with different morphologies and transcriptional programs (Fig 1A). In predivisional cells, one of the poles has a membranous stalk and the other a flagellum. Each pole accumulates a different set of cytoplasmic regulatory proteins that govern the behavior of the nascent daughter cells. Differentially localized polar proteins include components of the proteolytic machinery, kinases, phosphatases, di-guanylate cyclases, and phosphodiesterases that control the timing of DNA replication, global transcriptional activity, and also regulate the production and activity of stalks and flagella (Kirkpatrick and Viollier 2012; Abel et al., 2011). Additionally, the pole is used to tether the chromosomal centromere, which facilitates the distribution of newly replicated DNA between daughter cells (Ptacin et al., 2010; Schofield et al., 2010) and regulates the timing and localization of the division plane (Thanbichler 2009).
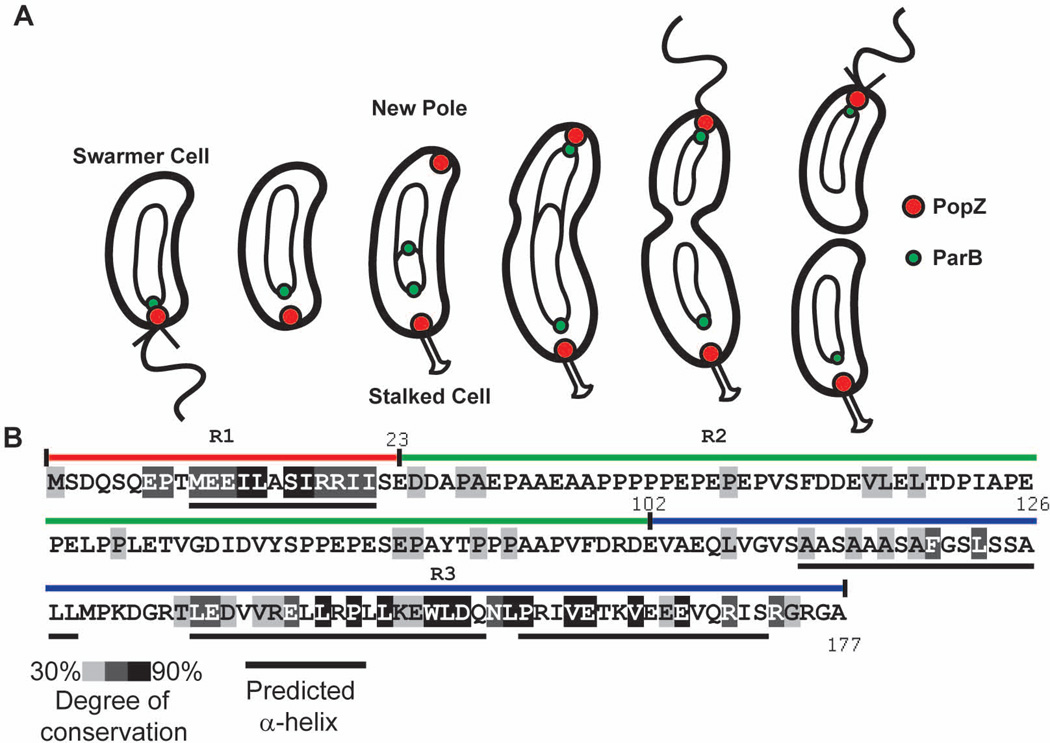
Figure 1.
Subcellular localization of PopZ as a function of the cell cycle and PopZ sequence.
A. A schematic of the Caulobacter cell cycle. Circles and theta structures within the cell indicate the replicating chromosome. Following the initiation of DNA replication, a new focus of PopZ assembles at the pole opposite the stalk, where it is required for tethering the newly duplicated ParB/parS centromere.
B. PopZ primary sequence, with shaded regions indicating the degree of conservation in Alphaproteobacteria. Black bars under the sequence represent predicted α-helices. The R1, R2, and R3 regions of the PopZ protein are indicated above the sequence as red, green, and blue bars, respectively.
Many polar proteins are de-localized in the absence of a protein called PopZ (Polar Organizing Protein Z), which is hypothesized to act as a scaffold for polar assembly. PopZ itself is a polar protein, and its activity is regulated as a function of the cell cycle. Prior to the initiation of DNA replication, it is localized only to the flagellar pole of the cell (Fig 1A). Replication initiation is a cue that triggers PopZ accumulation at the opposite cell pole (Bowman et al., 2010), and at this location it tethers the newly replicated chromosome by interacting directly with the centromere binding protein ParB (Bowman et al., 2008, Ebersbach et al., 2008). At the old pole, ParB anchoring is relaxed, a set of polar regulatory proteins is recruited, and the flagellum is replaced by a stalk (Fig 1A). At least seven different stalked pole proteins are known to be delocalized in the absence of PopZ (Bowman et al., 2010; Ebersbach et al., 2008), and no other stalked pole proteins are known to be correctly localized in this mutant. PopZ tethering of the ParB/parS centromere to the cell poles is critical for the placement of the FtsZ division ring and the assembly of the divisome. The MipZ ATPase binds to ParB and functions to inhibit FtsZ polymerization (Thanbichler and Shapiro 2006). A gradient of MipZ emanating from each polar focus of ParB restricts the assembly of FtsZ to mid-cell where the concentration of MipZ is at its lowest (Kiekebusch et al., 2012). Thus, deletion of PopZ results in filamentous cells where cell division is often delayed or incorrectly positioned at a site near the cell pole (Ebersbach et al. 2008).
The mechanism by which PopZ itself is localized within the cell is not fully understood. Previous work has shown that PopZ accumulates in polar foci when expressed heterologously in E. coli (Bowman et al., 2008; Ebersbach et al., 2008). Since E. coli is evolutionarily separated from C. crescentus by a large distance and do not produce any proteins that are homologous to PopZ, these observations suggest that sub-cellular accumulation of PopZ occurs by a mechanism that is, to some extent, intrinsic to PopZ itself. One model proposes that localized PopZ accumulation is driven by the self-assembly of PopZ sub-units into large macromolecular structures (Ebersbach et al., 2008). Evidence for self-assembly comes from the observations that purified PopZ behaves like a large multiprotein complex during native gel electrophoresis, and that it can undergo further assembly into filament networks, as observed by transmission electron microscopy (Bowman et al., 2008).
A related question is how the assembling PopZ complexes become localized to the cell poles. In E. coli, this could be explained by entropic differences between the area of condensed chromosome that fills the bulk of the cell and relatively open areas of cytoplasm at the poles (Ebersbach et al., 2008). Biophysical models predict that large macromolecular complexes, such as those that may be formed by PopZ, are more stable and more likely to accumulate in areas that offer more free space. Such a mechanism has also been described for the polar accumulation of misfolded protein aggregates in E. coli (Winkler et al., 2010; Rokney et al., 2009), but it is a novel concept when applied to a natively expressed, fully functional protein such as PopZ.
While this passive mechanism of self-assembly and preferential accumulation in nucleoid-free regions may be sufficient to explain sub-cellular localization patterns in E. coli, it is not sufficient to explain how PopZ becomes localized in C. crescentus, where PopZ localization is dynamic and under cell cycle control. Recent work (Laloux and Jacobs-Wagner 2013) has shown that ParA, which is active in chromosome segregation and accumulates at the new pole during this process, is localized at the correct time and place for stimulating the assembly of a new macromolecular complex of PopZ at this location. Additionally, there is evidence that ParA interacts with the N-terminal region of PopZ and that perturbations in ParA distribution affect the dynamics of PopZ localization. From a structural standpoint, the relationship between the self-assembly of PopZ into homotypic complexes and additional effects that could come from the ParA interaction is unclear.
Some additional clues about PopZ assembly and localization may come from comparing it to a functionally analogous protein, DivIVA, which is found in gram positive bacteria. Like PopZ, DivIVA organizes the cytoplasm by acting as a polar recruitment factor for chromosome centromeres and other regulatory proteins (Kaval and Halbedel 2012; Donovan et al., 2012). PopZ and DivIVA are also similar in that they form oligomers of defined size, which undergo further assembly into a branched filamentous superstructure in vitro (Stahlberg et al., 2004; Bowman et al., 2008). It is significant that DivIVA and PopZ are both capable of forming oligomers and accumulating at cell poles when expressed heterologously, indicating that these proteins are self-sufficient for assembly and localization activity even in a non-native cytoplasmic environment (Bowman et al., 2008; Ebersbach et al., 2008; Edwards et al., 2000). DivIVA localizes to cell poles because it has specific affinity for negatively curved membrane surfaces (Lenarcic et al., 2009; Ramamurthi and Losick 2009). Membrane insertion is achieved by a small hydrophobic loop near the N-terminus, and the C-terminus links DivIVA into a tetramer through interactions along a long coiled coil domain (Oliva et al., 2010). A model for pole specificity posits that higher-order assembly between oligomers is important for the recognition of cell poles, as such superstructures are likely to be more sensitive to the geometry of a gently curved membrane. Consistent with this, a DivIVA mutant protein that lacks the final 38 amino acids of the C-terminus is capable of localizing to the membrane, but does not accumulate at the pole (Ramamurthi and Losick 2009), and a protein that lacks the final 20 amino acids exhibits reduced affinity for curved liposomes (Lenarcic et al., 2009). However, a link between higher-order assembly and localization to curved membranes has not been established.
As monomers, PopZ and DivIVA have nearly the same molecular weight and both are predominantly alpha helical. However, their primary sequences share no regions of homology, and although coiled coil prediction programs correctly identify those regions in DivIVA, none are found in PopZ. Here, we use mutational analysis to divide the PopZ primary sequence into distinct functional regions, and then combine these results with biophysical analysis of purified wildtype PopZ and mutated PopZ complexes to elucidate the relationship between PopZ structure and function. We demonstrate that the C-terminal 76 amino acids of PopZ are necessary and sufficient for accumulation as a single localized focus, and that this same region is also necessary and sufficient for oligomer formation. We also find that the central 76 amino acids of PopZ is a flexibly sized linker that connects the C-terminal domain with a 26 amino acid region in the N-terminus that is required for cell cycle-controlled bi-polar distribution of PopZ. Overall, these results suggest that PopZ assembly and sub-cellular accumulation are mechanistically linked, yet not sufficient to achieve controlled bi-polar localization. Biophysical analysis of purified PopZ revealed that the oligomeric form of PopZ is comprised of paired three-helix bundles, with a quaternary structure unlike DivIVA. Through direct visualization, we find that PopZ oligomers undergo higher-order assembly into elongated filaments, and demonstrate that filaments are the active, localization competent form of PopZ.
RESULTS
Homologs of popZ are present in nearly all genera of Alphaproteobacteria (Supplementary Figure S1). The most strongly conserved parts of the protein are found within an 18 amino acid region at the N-terminus and a 65 amino acid region at the C-terminus (Fig1B). Structural prediction algorithms suggest that both of these regions contain alpha helices. The intervening sequence, which has less conservation identity and is somewhat variable in length, is negatively charged and proline rich in many homologs. The coincidence of areas of sequence conservation and alpha helical content within C. crescentus PopZ revealed three distinct regions in the 178 amino acid protein, henceforth referred to as R1 (amino acids 1–23), R2 (amino acids 24–101), and R3 (amino acids 102–177). Because PopZ performs multiple functions in the cell, we reasoned that these distinct regions may be responsible for different aspects of PopZ activity. The distinct nature of these regions is supported by our study of phylogenetic sequence conservation, which suggests that N and C termini are evolving under different processes in some lineages (Supplementary Figure S2).
The C-terminal region of PopZ is necessary and sufficient for sub-cellular accumulation
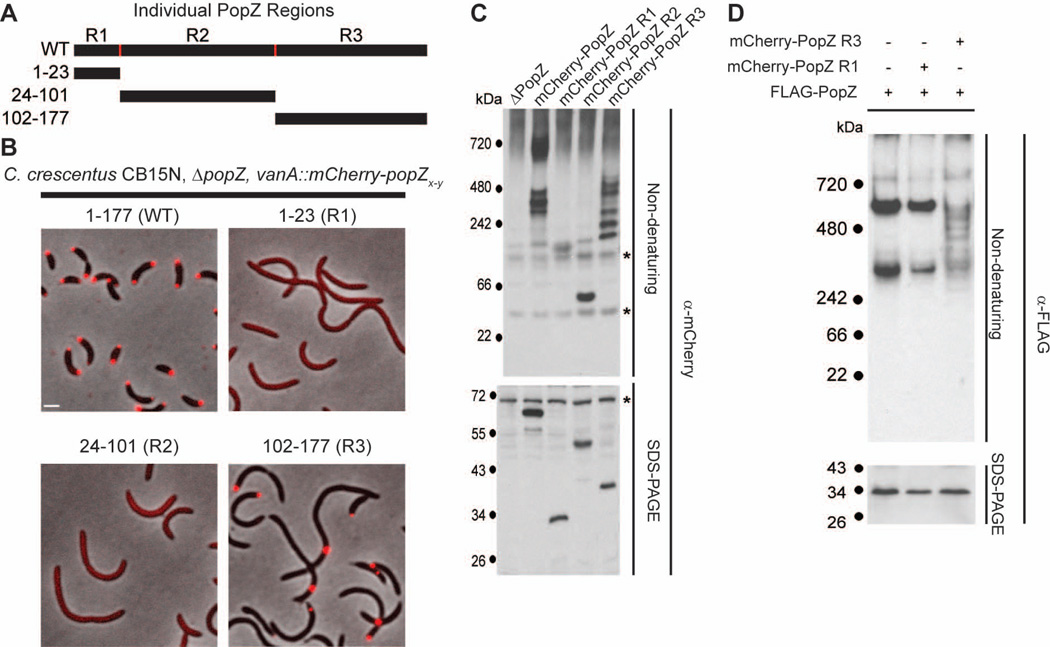
An important aspect of PopZ’s polar organizing function is its ability to dynamically localize to the cell poles. To determine if any of the three distinct regions of PopZ were sufficient for polar localization, we expressed each individual region as a fusion protein with the fluorescent localization marker mCherry at the N-terminus (Figure 2A,B). Strains were constructed in a ΔpopZ knockout background in order to eliminate the possibility that the labeled proteins could be influenced by interacting with endogenous PopZ. Whereas mCherry-R1 and mCherry-R2 were distributed diffusely in the cytoplasm, mCherry-R3 was found in tightly localized foci, suggesting that a localization determinant for PopZ lies within the C-terminal region. In contrast to the mixture of monopolar and bipolar distribution patterns observed in the full length mCherry-PopZ control strain, 97% of cells expressing mCherry-R3 had a single focus. Moreover, the single focus was found away from the pole in 47% of cells, whereas non-polar foci were never observed in cells expressing full length mCherry-PopZ. Cells expressing mCherry-R3 were filamentous, indicating that this fusion protein is unable to complement the cell division defect associated with the loss of full length PopZ. Our results suggest that R3 is sufficient for sub-cellular accumulation, and the R1 and R2 regions include additional activities that are required for bipolar localization and for supporting normal cell division.
Figure 2.
The C-terminal region of PopZ is sufficient for sub-cellular localization and oligomer formation.
A. Schematic of individual PopZ regions.
B. Images of strains expressing full length mCherry-PopZ (GB1078), mCherry-PopZ R1 (GB1079), mCherry-PopZ R2 (GB1080), and mCherry-PopZ R3 (GB750) in a ΔpopZ background. mCherry fluorescence (red) overlays the phase contrast image (grayscale). Bar = 1 µm.
C. Electrophoretic migration of individual PopZ regions. Whole cell lysates of strains expressing no PopZ (GB255), mCherry-PopZ (GB1078), mCherry-PopZ R1(GB1079), mCherry-PopZ R2 (GB1080), and mCherry-PopZ R3 (GB750) were resolved on native and denaturing gels, then probed with anti-mCherry sera by immunoblotting. Cross-reactive bands are indicated by an asterisk.
D. Electrophoretic migration of co-expressed PopZ variants. A PopZ-FLAG fusion was expressed by itself (GB135) and in parallel with either mCherry-PopZ R1 (GB757) or mCherry-PopZ R3 (GB758). Whole cell lysates were resolved on native and denaturing gels, then probed with anti-FLAG sera by immunoblotting.
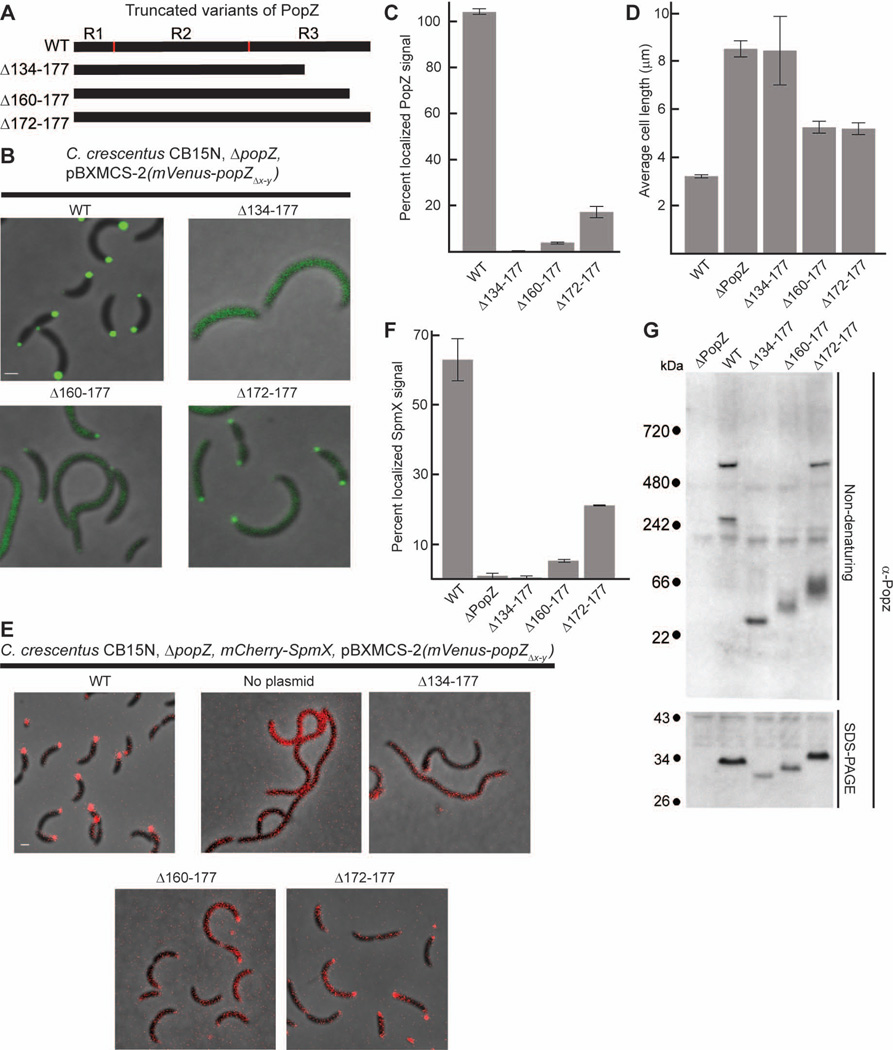
To determine if R3 is necessary for polar localization, we created a series of mutations in which portions of the C-terminus were removed (Figure3A). We combined each mutation with an N-terminal Venus tag to produce a fluorescently labeled fusion protein in a ΔpopZ strain background (Figure 3B). We found that the shortest truncation variant, Venus-PopZΔ134–177, which lacks the last two predicted alpha helical regions within R3, was diffuse in the cytoplasm. Venus-PopZΔ160–177, which lacks the last 18 amino acids of R3, was also delocalized, indicating that the final predicted alpha helical region is critical for efficient sub-cellular accumulation. Venus-PopZΔ172–177, which lacks the last six amino acids of R3, exhibited distinct polar foci within a large pool of diffusely distributed protein. Using computational methods to quantify the amount of localized protein, we found that 0% of Venus-PopZΔ134–177, 4% of Venus-PopZΔ160–177, and 17% of Venus-PopZΔ172–177 accumulated as polar foci, compared with 100% polar localization in cells that express full length Venus PopZ (Figure 3C).
Figure 3.
The C-terminal region of PopZ is necessary for sub-cellular localization and oligomer formation.
A. Schematic of PopZ truncations.
B. Images of strains expressing full length mVenus-PopZ (AP323) (WT), mVenus-PopZΔ134–177 (AP303), mVenus-PopZΔ160–177 (AP322), and mVenus-PopZΔ172–177 (AP305) in a ΔpopZ background. Venus fluorescence (green) overlays the phase contrast image (grayscale).
C. Percent of polar localized PopZ signal in B.
D. Average cell length of strains expressing WT PopZ (GB699), no PopZ (GB255), PopZΔ134–177 (GB885), PopZΔ160–177 (GB886), and PopZΔ172–177 (GB888) in a ΔpopZ background.
E. Localization of SpmX-mCherry in PopZ variant backgrounds. Strains in B are modified to express SpmX-mCherry from the chromosomal spmX promoter. Images from AP253, AP236, AP280, AP300, and AP282 are presented. SpmX-mCherry fluorescence (red) overlays the phase contrast image (grayscale).
F. Percent of polar localized SpmX-mCherry signal in PopZ variant strains, shown in E.
G. Electrophoretic migration of PopZ truncations. Whole cell lysates of strains in panel D were resolved on native and denaturing gels, then probed with anti-PopZ sera by immunoblotting.
In images, Bar = 1µm. In graphs, error bars represent SEM from 2 separate experiments of 30–60 cells each.
In parallel, we assessed protein function by asking if an untagged copy of the mutated PopZ could complement the cell division phenotype of the popZ knockout. We quantified this activity by measuring average cell length. We found that cells expressing any of the three truncated proteins were elongated relative to the wildtype control, indicating that they do not fully complement the loss of PopZ (Figure 3D). Since PopZ contributes to the timing and placement of the cytokinetic Z-ring via centromere anchoring at the new pole (Thanbichler, 2009), we asked if the increased cell length is associated with a centromere anchoring defect. We measured the distance between chromosome centromeres and cell poles in our PopZ mutant strains and our full length PopZ control strain, and found that the average distance corresponded closely with the differences in their average cell lengths (Supplementary Figure S3).
We also quantified the localization of SpmX, a member of the cell cycle regulatory network that accumulates at the stalked pole in wildtype cells (Radhakrishnan et al., 2008), and is de-localized in the absence of PopZ (Bowman et al., 2010). We found that strains expressing the Venus-tagged PopZ truncation mutants were defective in recruiting SpmX-mCherry to the cell poles (Figure 3E), and the percent of localized SpmX-mCherry was roughly proportional to the percent of polar localized Venus-PopZ (Compare Figure 3F to 3C). To test the possibility that de-localized PopZ fragments have dominant negative effects, we co-expressed Venus-PopZΔ134–177 with untagged wildtype PopZ or mCherry-PopZ protein. Cell length and mCherry-PopZ localization were normal in these strains even though the localization of Venus-PopZΔ134–177 remained diffuse, indicating that the first 133 amino acids of PopZ are not dominant negative when free in the cytoplasm (Supplementary Figure S4). Together, our analysis of isolated PopZ regions and C-terminal truncation mutants show that the C-terminal region of PopZ (R3) is necessary and sufficient for subcellular accumulation, but not sufficient for cell cycle - controlled bi-polar distribution. In strains where mutant variants of PopZ were mostly diffuse in the cytoplasm, the polar organizing activity of PopZ was impaired, leading to defects in cell division.
The R2 middle region of PopZ is a required spacer between the R1 N and R3 C terminal regions
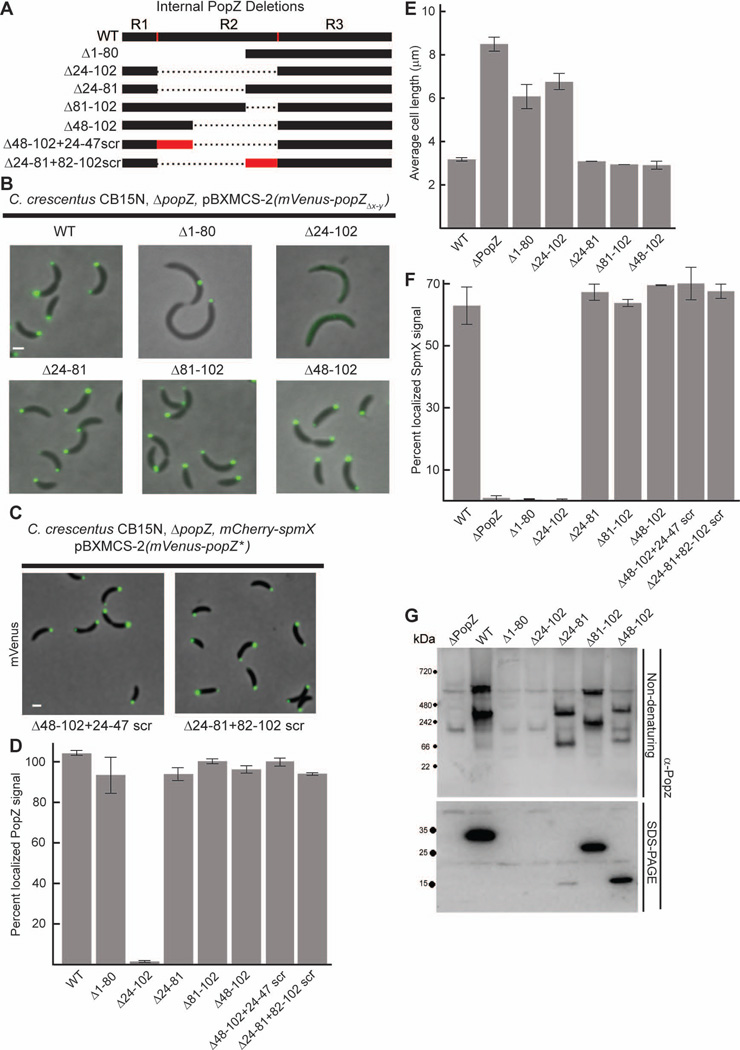
We added regions of R1/R2 back to R3 in order to determine which parts are needed for restoring functionality (Figure 4A). The constructs were expressed as N-terminal Venus-tagged fusion proteins in a ΔpopZ strain background (Figure 4B). We lengthened the C terminal region by a twenty one amino acid portion of R2 and found that Venus-PopZΔ1–80 accumulated as a single sub-cellular focus, similar to mCherry-R3, and its expression did not compliment the cell division defect of the ΔpopZ strain background. When we fused the R1 region directly to R3, thereby creating a construct that contained the most conserved regions of the protein without any intervening R2 sequence, we found that Venus-PopZΔ24–102 was diffuse throughout the cytoplasm. However, when R1 was added together with a twenty amino acid section from R2, this variant, Venus-PopZΔ24–81, regained bi-polar localization. Thus, a large part of the R2 region is dispensable for the localization activity of PopZ.
Figure 4.
The middle region of PopZ (R2) is a required spacer between N and C termini.
A. Schematic of internal PopZ deletions.
B. Images of strains expressing mVenus-PopZ (AP323) (WT), mVenus-PopZΔ1–80 (AP321), mVenus-PopZΔ24–102 (AP324), mVenus-PopZΔ24–81 (AP327), mVenus-PopZΔ81–102 (AP320), and mVenus-PopZΔ48–102 (AP301) in a ΔpopZ background. Venus fluorescence (green) overlays the phase contrast image (grayscale).
C. Images of strains expressing scrambled R2 regions in a ΔpopZ spmX-mCherry background. mVenus-PopZΔ48–102+24–47scr (AP343) has the linker sequence EEDPPPADAAAPPAPEAVPPEPPE and mVenus-PopZΔ24–81+82–102scr (AP344) has the linker sequence PDDEPEFETPPPERAYAPSVA. Venus fluorescence (green) overlays the phase contrast image (grayscale).
D. Percent of polar localized PopZ signal in B and C.
E. Average cell length of strains expressing WT PopZ (GB699), no PopZ (GB255), PopZΔ1–80 (GB1115), PopZΔ24–102 (GB1117), PopZΔ24–81 (GB1116), PopZΔ81–102 (GB1119), and PopZΔ48–102 (GB1118) in a ΔpopZ background.
F. Percent of polar localized SpmX-mCherry signal in PopZ variant strains. Strains in B are modified to express SpmX-mCherry from the chromosomal spmX promoter. Data from AP253, AP236, AP299, AP254, AP257, AP298, AP342, AP343, and AP344 are presented. Corresponding images are in Supplementary Figure S5.
G. Electrophoretic migration of PopZ variants. Whole cell lysates of strains in panel E were resolved on native and denaturing gels, then probed with anti-PopZ sera by immunoblotting.
In images, Bar = 1µm. In graphs, error bars represent SEM from 2 separate experiments of 30–60 cells each.
To determine if specific sequences within amino acids 81–102 of R2 are required for PopZ localization, we created a Venus tagged construct that encoded all of PopZ except for this region. We found that Venus-PopZΔ81–102 accumulated at both cell poles, indicating that this section of R2 is not a requirement for polar localization. We then asked if we could substitute a different region of R2 for making a localization-competent PopZ variant. To do this, we linked R1 and R3 with amino acids 24–47 from R2. This construct, Venus-PopZΔ48–102, appeared similar to Venus-PopZΔ24–81 in localization. Our observations that amino acids 24–47 and 81–102 are independently sufficient as a link between R1 and R3, together with the fact that the R2 region is not well conserved among Alphaproteobacterial homologs, suggested that this region tolerates a large amount of variability in both length and sequence. To determine if the sequence can be varied, we randomly scrambled amino acids 24–47 and 82–102 and asked if these altered sequences could still function as linkers between R1 and R3. When tagged with N-terminal Venus, these two PopZ variants, Venus-PopZ24–47scr+Δ48–102 and Venus-PopZΔ24–81+82–102scr were localized to both cell poles (Figure 4C,D).
We then asked if the localization patterns of our R2/R3 replacement mutants correlated with PopZ function. The R2 deletion mutants that exhibited a normal bi-polar localization pattern were the same length as the wildtype control strain (Figure 4E), and showed no difference in the amount of SpmX-mCherry localization (Figure 4F). In every case where PopZ localization was defective, the cells were elongated and the polar accumulation of SpmX-mCherry was reduced compared to the wildtype Venus-PopZ expressing control strain. Interestingly, the Venus-PopZΔ1–80 variant, which fails to accumulate at a second cell pole but is not defective in polar accumulation per se, exhibited defects that were comparable to the fully de-localized Venus-PopZΔ24–102 variant. This indicates that the N-terminal 80 amino acids are required for the polar organization activities of PopZ, and is consistent with the idea that dynamic, bi-polar localization is critical for normal function. Overall, our internal deletion experiments suggest that R2 acts a spacer for separating the two evolutionarily conserved modules encoded within R1 and R3, and that it can be functionally replaced by a short linker containing a random mixture of prolines, alanines, and negatively charged amino acids.
The N-terminal region of PopZ facilitates cell cycle-controlled bipolar distribution
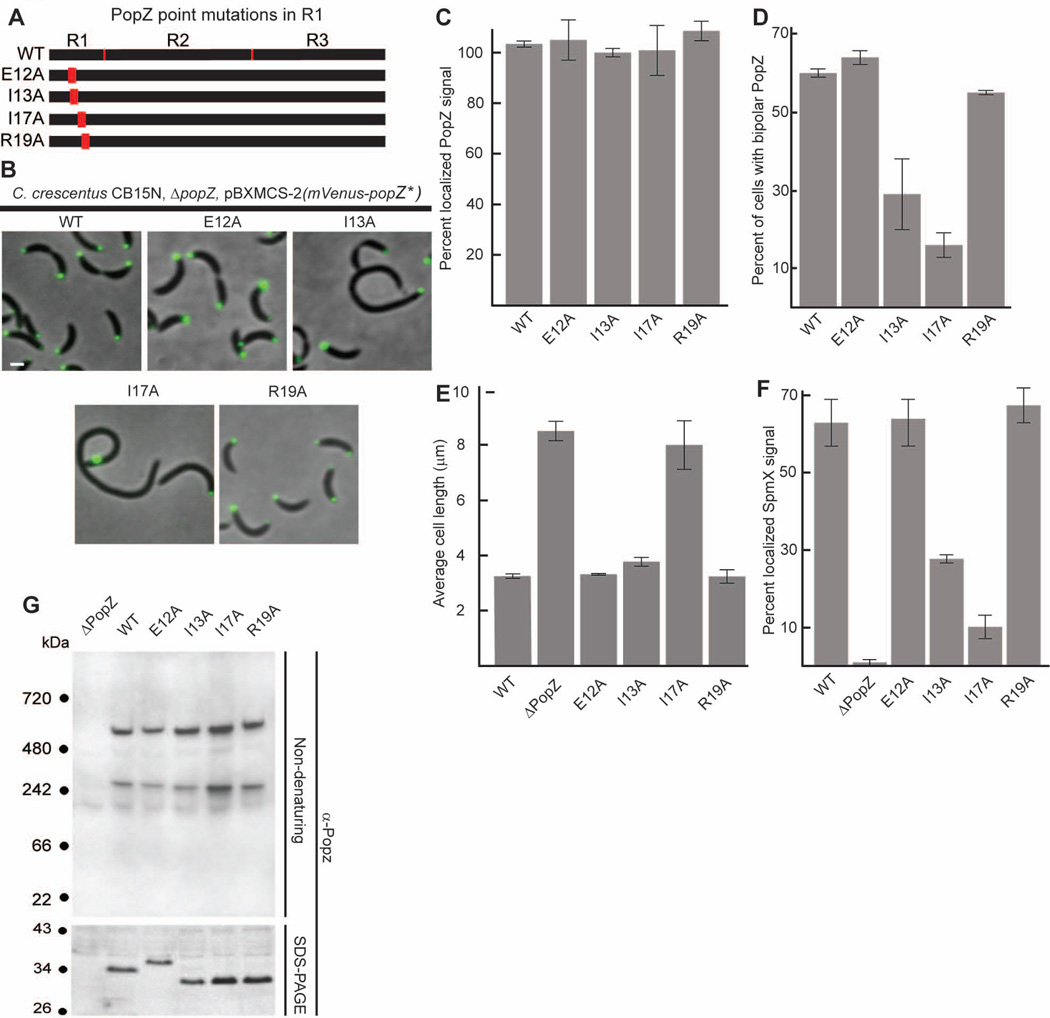
To investigate the role of the conserved 23 amino acid region at the N terminus of PopZ, we substituted four of the most highly conserved amino acids in this region with alanine (Figure 5A). We observed the sub-cellular localization of these mutated proteins by expressing them as fusions tagged with N-terminal Venus in a ΔpopZ background (Figure 5B,C). All of the strains produced bright Venus-PopZ polar foci and cells with bi-polar PopZ localization. However, among cells expressing the Venus-PopZ I13A and Venus-PopZ I17A variants, the percentage with bi-polar Venus-PopZ distribution was greatly reduced compared to the wildtype control (Figure 5D). Moreover, single foci were occasionally observed away from a cell pole (14% of all cells for PopZ I17A, 5% for PopZ I13A), as we observed in cells expressing PopZ variants with no N-terminal region (47% of cells expressing mCherry-R3, 39% of cells expressing Venus-PopZΔ1–80).
Figure 5.
The N-terminal R1 region of PopZ is necessary for bi-polar distribution.
A. Schematic of N-terminal PopZ amino acid substitutions.
B. Images of strains expressing mVenus-PopZ (AP323) (WT), mVenus-PopZ E12A (AP316), mVenus-PopZ I13A (AP313), mVenus-PopZ I17A (AP314), and mVenus-PopZ R19A (AP315) in a ΔpopZ background. Venus fluorescence (green) overlays the phase contrast image (grayscale).
C. Percent of polar localized PopZ signal in B.D. Average cell length in strains expressing WT PopZ (GB699), no PopZ (GB255), PopZ E12A (GB899), PopZ I13A (GB897), PopZ I17A (GB898), and PopZ R19A (GB900).
E. Percent of polar localized SpmX-mCherry signal in PopZ variant strains. Strains in B are modified to express SpmX-mCherry from the chromosomal spmX promoter. Data from AP253, AP236, AP292, AP293, AP290, and AP291 are presented. Corresponding images are in Supplementary Figure S5.F. Percent of cells exhibiting bi-polar localization of Venus-PopZ, in strains shown in panel B.
G. Electrophoretic migration of PopZ N-terminal mutant proteins. Whole cell lysates of strains in panel D were resolved on native and denaturing gels, then probed with anti-PopZ sera by immunoblotting.
In images, Bar = 1µm. In graphs, error bars represent SEM from 2 separate experiments of 30–60 cells each.
We assessed polar organization activity of the N-terminal mutant proteins by asking if they could complement the cell division phenotype of the popZ knockout (Figure 5E) and support SpmX polar localization (Figure 5F). Strains expressing Venus-PopZ E12A and Venus-PopZ R19A, which were similar to wildtype Venus-PopZ in polar distribution, had normal SpmX-mCherry localization, and strains expressing the corresponding untagged versions of these mutant proteins had cell lengths that were not significantly different than wildtype. Strains expressing Venus-PopZ I13A and Venus-PopZ I17A, which had relatively severe defects in bi-polar localization, also exhibited defects in polar recruitment of SpmX-mCherry. Since the relatively faint SpmX-mCherry foci were found at the same poles as the mutant forms of Venus-PopZ, co-localization of these PopZ variants with SpmX-mCherry was not sufficient for robust recruitment. Cells expressing PopZ I13A were on average slightly longer than wildtype, and cells expressing the I17A variant, which produced the strongest defects in Venus-PopZ and SpmX-mCherry polar localization, were more than twice as long as the wildtype control strain. Together, our results indicate that the evolutionarily conserved region within R1 is critical for controlling PopZ distribution between cell poles, and for PopZ’s function as a polar organizing protein in C. crescentus.
The C-terminus of PopZ is necessary for oligomeric assembly
We asked if the localization patterns of our mutated PopZ proteins correlated with their ability to form oligomeric complexes. Native gel electrophoresis on whole cell lysates is useful in detecting protein oligomerization (Gallagher 2001), and wildtype PopZ, despite a nominal molecular mass of 19 kDa and highly negative charge, runs as two slowly migrating bands in this assay, consistent with its being present in larger complexes (Bowman et al., 2008). In these gels, a ladder of standard proteins is included as a reference, but because the mobility of proteins and protein complexes are strongly influenced by charge and hydrodynamic radius in addition to molecular weight, the molecular mass of the PopZ complexes cannot be accurately determined with this method.
In comparing the mobility of wildtype PopZ with C-terminal truncations (Figure 3G), we found that the most diffusely localized forms, PopZΔ134–177 and PopZΔ160–177, did not run like either of the slowly migrating bands in wildtype lysates, but exhibited prominent bands migrating below the 66 kDa standard. Such a large mobility shift cannot be explained by the slight change in isoelectric point (pl from 4.0 to 3.8) or molecular mass (a loss of 2 kDa between PopZΔ160–177 and wildtype, as demonstrated by SDS-PAGE, lower panel of Figure 3G), suggesting that the last 16 amino acids of PopZ include a region that is necessary for the assembly of oligomeric forms. With the loss of only six amino acids, PopZΔ172–177 also exhibited anomalous migration on native gels. The majority of the mutant protein migrated with the 66 kDa standard, and the remainder ran in a single band, which corresponded to the slowest migrating band produced in wildtype cells. The faster migrating of the two PopZ bands observed in wildtype lysates was not detected in PopZΔ172–177 samples. In order to match the localization pattern directly with oligomerization state, we performed native gel electrophoresis on lysates from strains expressing Venus-tagged PopZ mutant proteins. We observed that the fusion proteins behaved similarly to un-tagged protein, with proportional shifts for the increase in molecular weight (Supplementary Figure S6). Thus, the N-terminal fluorescent protein tag used in our localization experiments had little effect on the influence of C-terminal mutations in oligomeric assembly. Overall, our native gel analysis of these PopZ variants indicated that the loss of oligomerization was correlated with diffuse sub-cellular localization.
The C-terminal region of PopZ is sufficient for oligomeric assembly
To determine if the localization patterns of regions R1, R2, and R3 correlate with their capacity for oligomeric complex formation, we measured the oligomerization state of PopZ fragments in cells expressing mCherry-R1, mCherry-R2, mCherry-R3, compared with a control strain expressing full length mCherry-PopZ (Figure 2C). We found that mCherry-R3 and full length mCherry-PopZ ran as multiple bands on native gels with relatively slow mobility, indicating oligomer formation. The bands in the mCherry-R3 lysates were more numerous and more widely distributed than those in the full length mCherry-PopZ lysates, suggesting that the assembly of oligomeric complexes by the truncated protein is less rigidly defined. In contrast, mCherry-R1 and mCherry-R2 ran as single bands with faster mobility, suggesting an absence of oligomer formation. mCherry-R1 (pl 5.3, 30.0 kDa) ran slower than mCherry-R2 (pl 4.6, 35.6 kDa), a difference that may be attributable to the influence of charge. When we resolved the same lysates under denaturing conditions, we observed that all four mCherry fusion proteins ran as a single predominant band. This supports our conclusion that full length mCherry-popZ and mCherry-R3 are present in multiple oligomeric forms, as the complex banding pattern cannot be attributed to truncated monomers arising from protein degradation. The relatively slow migration of mCherry-R2 during SDS-PAGE is a probably consequence of the distribution of charged residues in the primary sequence. Long stretches of acidic amino acids can block SDS binding, thereby reducing mobility under denaturing conditions (Shi et al., 2012).
To confirm that the slowly migrating bands formed by mCherry-R3 are formed through self-association of PopZ protein sequences, we performed a mixed oligomerization experiment in which full length PopZ is offered as a binding partner (Figure 2D). To do this, we expressed FLAG-PopZ, which could be uniquely identified with a FLAG specific antibody, together with mCherry-R3 or mCherry-R1. If FLAG-PopZ co-assembled with mCherry-R3 or mCherry-R1, we would expect a shift in the mobility of the FLAG-PopZ oligomers on native gels, due to the incorporation of mCherry-tagged sub-units with a different size and shape. When expressed by itself, FLAG-PopZ behaved like untagged PopZ, forming slowly migrating bands on native gels. This pattern was unchanged when FLAG-PopZ was co-expressed with mCherry-R1, indicating that these proteins do not co-assemble. However, FLAG-PopZ migrated in a complex ladder pattern when co-expressed with mCherry-R3, suggesting that multiple hetero-oligomeric species were formed. The additional bands were not caused by protein degradation, as the size of FLAG-PopZ monomers on denaturing gels was not altered by the expression of mCherry-R3. Cumulatively, our gel electrophoresis experiments indicate that mCherry-R3 can self-assemble into oligomeric complexes and co-assemble with full length PopZ. Together with fluorescence localization data, the results indicate a correlation between oligomeric assembly and sub-cellular localization.
Mutations in the N-terminal and middle regions of PopZ do not affect oligomeric assembly
We also examined the oligomerization capacity of untagged PopZ variants that had point mutations in the N-terminal region or internal deletions in the R2 region. None of the point mutations in the N-terminal domain affected the mobility of PopZ in native gels (Figure 5G), indicating that these perturbations do not affect oligomeric assembly. Notably, the normal oligomerization of PopZ I17A protein shows that oligomer formation per se is not sufficient for bi-polar distribution in cells. Among the mutant variants with internal deletions in the R2 region, we were unable to detect any signal on western blots for strains expressing PopZ Δ1–80 and PopZ Δ24–102, and the signal from PopZ Δ24–81 on denaturing gels was weak compared to PopZ Δ81–102 and PopZ Δ48–102 (Figure 4G). Our inability to detect some of these proteins by western suggests that the anti-PopZ antibody specifically recognizes epitopes within R2 region, and this is supported by our inability to detect Venus-tagged versions of the same variants (Supplementary Figure S6), which are clearly visible by fluorescence microscopy. On native gels, PopZ Δ24–81, PopZ Δ81–102, and PopZ Δ48–102 all ran as two distinct bands, in a pattern that is similar to wildtype PopZ except that the bands migrate more quickly through the gel (Figure 4G). The PopZ Δ24–81 and PopZ Δ48–102 variants migrated faster than PopZ Δ81–102, a difference that correlates roughly with the varying lengths of R2. These patterns suggest normal oligomer formation, and this is consistent with our observation that these variants are functional in vivo (Figure 4B–F). Overall, our native gel analysis of PopZ variants in regions 1 and 2 found no evidence that these regions are important for oligomeric assembly.
Point mutations in the C-terminal domain affect oligomerization and polar localization
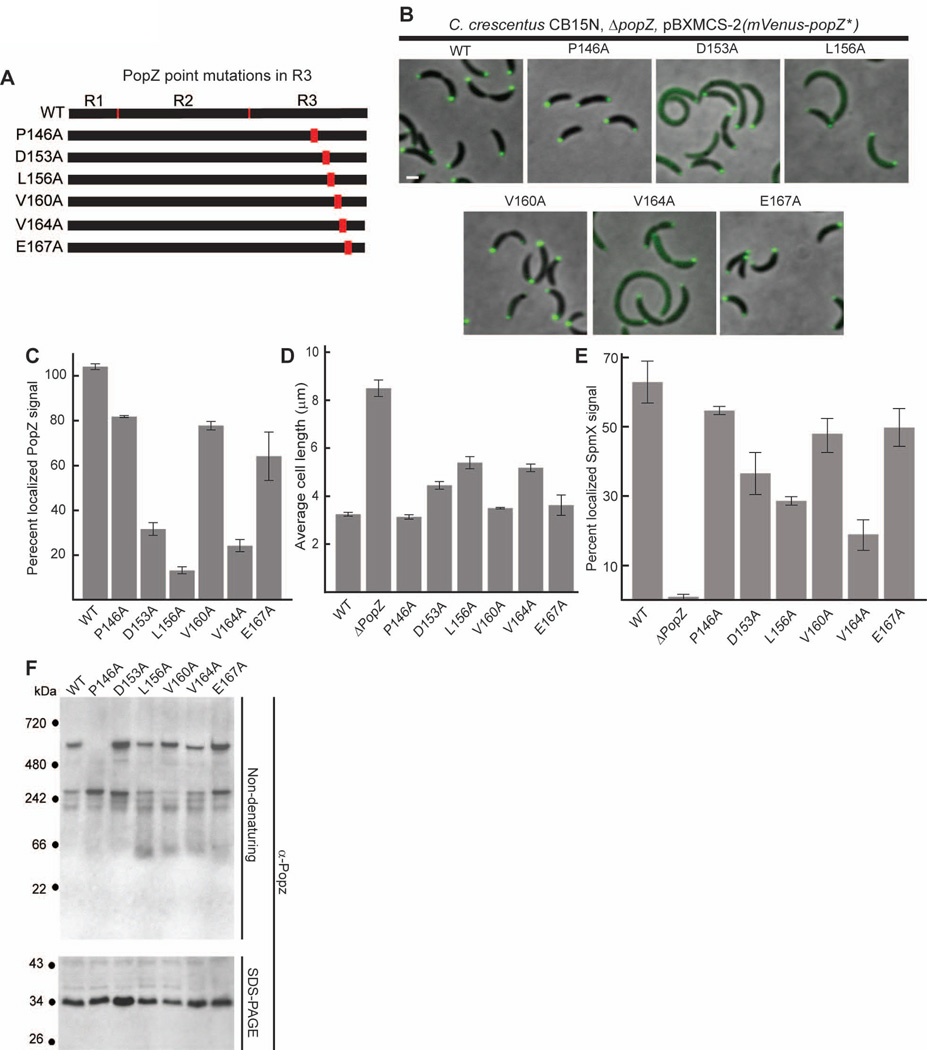
To further investigate the relationships between sub-cellular localization, oligomeric assembly, and cell function, we created a series of strains bearing point mutations in the C-terminal region of PopZ (Figure 6A). We chose six highly conserved residues and converted each into an alanine residue. The sub-cellular localization of the mutant proteins was observed by expressing them as fusion proteins with N-terminal Venus (Figure 6 B,C). We quantified the polar recruitment of SpmX-mCherry in strains expressing these Venus-tagged variants, and the average cell length and PopZ oligomerization state in strains expressing untagged forms of mutant protein. Our cell length analysis revealed that three out of the six point mutations in R3 (D153A, L156A, V164A) caused cell division defects, indicating impaired PopZ function (Figure 6D). These mutations also reduced PopZ localization at cell poles (Figure 6 C), and polar recruitment of SpmX-mCherry (Figure 6E). For the other three point mutants (P146A, V160A, E167A), the majority of Venus tagged protein was localized to both cell poles, cell length was not significantly perturbed, and SpmX-mCherry recruitment was closer to the normal level. Thus, in our set of R3 point mutants, we observed a consistent correlation between polar localization and functional activity.
Figure 6.
Amino acid substitutions in the C-terminal R3 region of PopZ affect both localization and oligomer formation.
A. Schematic of C-terminal PopZ amino acid substitutions.
B. Images of strains expressing mVenus-PopZ (AP323) (WT), mVenus-PopZ P146A (AP306), mVenus-PopZ D153A (AP308), mVenus-PopZ L156A (AP309), mVenus-PopZ V160A (AP310), mVenus-PopZ V164A (AP311), and mVenus-PopZ E167A (AP312) in a ΔpopZ background.
C. Percent of polar localized PopZ signal in B. D. Average cell length of strains expressing WT PopZ (GB699), no PopZ (GB255), PopZ P146A (GB890), PopZ D153A (GB892), PopZ L156A (GB893), PopZ V160A (GB894), PopZ V164A (GB895), and PopZ E167A (GB896) in a ΔpopZ background.
E. Percent of polar localized SpmX-mCherry signal in PopZ variant strains. Strains in B are modified to express SpmX-mCherry from the chromosomal spmX promoter. Data from AP253, AP236, AP283, AP285, AP286, AP287, AP288, and AP289 are presented. Corresponding images are in Supplementary Figure S5.
F. Electrophoretic migration of PopZ N-terminal mutant proteins. Whole cell lysates of strains in panel D were resolved on native and denaturing gels, then probed with anti-PopZ sera by immunoblotting.
In images, Bar = 1µm. In graphs, error bars represent SEM from 2 separate experiments of 30–60 cells each.
We assayed the oligomerization capacity of the R3 point mutant proteins using native gels (Figure 6F). Among the three that are mostly diffuse in the cytoplasm, two (L156A and V164A) exhibited defects in oligomerization. Both formed the two slowly migrating bands observed in wildtype cell lysates, but some of the protein also accumulated in a form that ran faster than the 66 kDa standard. The other mislocalized mutant protein, D153A, exhibited robust oligomer formation that appeared very similar to wildtype, thus the normal pattern of PopZ oligomerization is not sufficient for polar accumulation.
Among the three point mutations in R3 that complement the cell division defect of ΔpopZ and produce protein that localizes efficiently to the cell poles, V160A and P146A, exhibited alternative oligomerization profiles (Figure 6F). PopZ V160A accumulated predominantly in the slowest migrating form, and some of the protein ran below the 66 kDa marker. This oligomerization profile is quite similar to that of PopZΔ172–177. The P146A mutant protein exhibited the inverse phenotype, accumulating exclusively in the faster migrating form. Thus, either of the oligomeric forms can be compatible with robust PopZ activity. Together, our oligomerization, localization, and cell length analyses indicate that the formation of at least one of the two oligomeric species is necessary, but not sufficient for bi-polar localization and function in vivo.
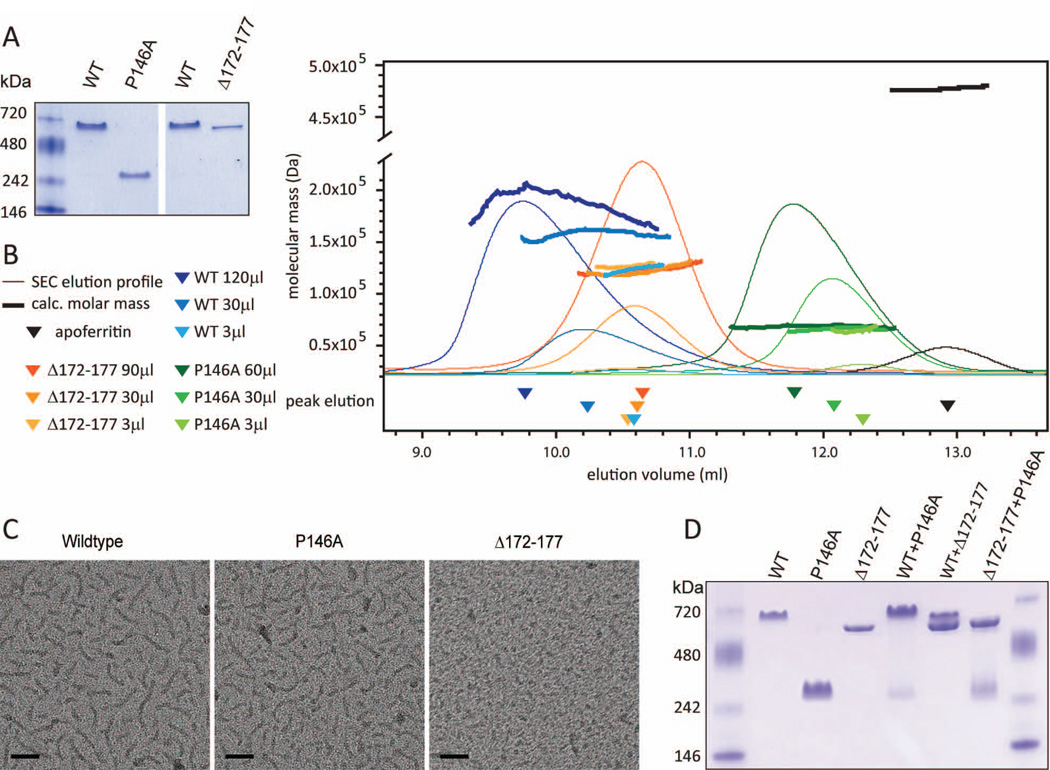
PopZ oligomers form highly elongated structures
We used a combination of biophysical methods to obtain an accurate assessment of the molecular mass, size, shape, and fold of purified PopZ protein. Our analyses included wildtype protein and two mutant proteins that, in vivo, had exhibited well-defined defects in their oligomerization profiles. We chose the PopZ P146A mutant protein because it assembles exclusively into the faster migrating of the two oligomeric forms (Figure 6F), and PopZ Δ172–177 because it preferentially assembles into the slower migrating form (Figure 3G). When we ran recombinantly expressed and purified proteins on native gels (Figure 7A), we found that the oligomerization states of PopZ Δ172–177 and PopZ P146A were consistent with what we observed in C. crescentus cell lysates. As we did not observe any protein running as quickly migrating monomers in either preparation, the oligomeric form of PopZ appears to be stabilized in these conditions. Purified wildtype PopZ ran as a single band, with a mobility that was similar to the slower of the two oligomeric species found in C. crescentus whole cell lysates. This is consistent with previous studies, which have shown that two oligomeric forms can be observed in E. coli at low expression levels, and that only one form is produced at preparatory expression levels (Bowman et al., 2008; Ebersbach et al., 2008). Surprisingly, purified wildtype PopZ ran slightly slower than purified PopZΔ172–177, even though these proteins appeared to have identical mobility in C. crescentus lysates (Figure 3G).
Figure 7.
Biophysical analysis of PopZ complexes.
A. Recombinant wildtype 6His-PopZ protein and the indicated 6-His tagged mutant forms were purified from E. coli, resolved by native gel electrophoresis, and subsequently stained with Coomassie blue.
B. SEC-LS analysis. The mobility of 6His-tagged proteins (X-axis) was determined by measuring protein concentration in fractions eluting from the column (narrow lines). The peak concentration is indicated by a triangle below the X-axis. Fractions in the vicinity of the peak were analyzed by MALLS to determine the molecular mass of the particles (Y-axis, thick lines). Three separate column runs were performed for each 6His-PopZ variant, with increasing amounts of protein loaded. Darker shading represents data from runs with more protein. In a separate run, apoferritin was used as a standard.
C. TEM microscopy on purified 6His tagged proteins, prepared at a concentration of 0.11 mg/ml. Scale bar 50nm.
D. Co-assembly of purified proteins. 6-His tagged proteins were denatured in 5M urea and then refolded by dialysis before running on a native gel and staining with Coomassie blue. Proteins were mixed at the following ratios: 2:1 WT:P146A (lane 5), 2:1 WT:Δ172–177 (lane 6), 2:1 Δ172–177:P164A (lane 7).
We used SEC/MALLS (size exclusion chromatography followed by multiangle laser light scattering) to obtain information on molecular mass, size, and shape of the PopZ oligomers (Figure 7B, Table 1). In SEC, the mobility of the protein is determined by hydrodynamic size, which is a function of mass and shape. The light scattering analysis provides a shape independent method for determining molecular mass within 5% accuracy (Folta-Stogniew and K R Williams 1999) and orthogonal methods for analyzing size and shape (Wyatt 1993). We found that all variants of PopZ protein eluted from the SEC column in order of their apparent sizes as determined by native gel electrophoresis. They also eluted ahead of Apoferritin, a globular protein of 475 kDa that is also included as a standard in our native gels. The molecular mass measurements indicated that PopZ oligomers are no more than 200 kDa (Table 1), thus their rapid elution compared to this spherical shaped standard suggests that they have elongated structures.
Table 1.
Size and shape of PopZ complexes determined by SEC-LS
| sample | Concentration at peak elution (mg/ml) |
Calculated Molecular weight (kDa)a |
calculated Rg (nm)b |
calculated Rh (average, nm)c |
|---|---|---|---|---|
| 6His-WT | 0.007 | 122 (+/− 5%) | 8.6 | |
| 6His-WT | 0.059 | 163 (+/− 5%) | 12.0 | |
| 6His-WT | 0.230 | 202 (+/− 5%) | 18.0 | 11.5 (+/− 0.3) |
| 6His-Δ172–177 | 0.008 | 128 (+/− 5%) | 9.3 | |
| 6His-Δ172–177 | 0.085 | 121 (+/− 5%) | 9.7 | |
| 6His-Δ172–177 | 0.253 | 127 (+/− 5%) | 9.3 | 9.1 (+/− 0.3) |
| 6His-P146A | 0.014 | 64 (+/− 5%) | 10.0 | |
| 6His-P146A | 0.131 | 66 (+/− 5%) | 8.0 | |
| 6His-P146A | 0.22 | 71 (+/− 5%) | 13.0 | 6.8 (+/− 0.3) |
| apoferritin | 475 (+/− 5%) | 6.4 | 6.9 (+/− 0.3) |
Average value determined from all incident light angles.
Determined from first order Zimm fitting, excluding low angle (<45° ) signal coming from large particles.
Due the limited sensitivity of dynamic light scattering, this value could only be determined in fractions with high protein concentrations.
Light scattering analysis confirmed that the unusual behavior of PopZ during SEC is due to the shape of PopZ complexes. We used multiple angle light scattering (MALLS) to obtain the root mean square radius (rms radius, or Rg), which represents the average distance of all points in the particle from the center of mass. We also used Dynamic Light Scattering (DLS), to obtain the hydrodynamic radius (Rh), which is equivalent to the radius of a hard sphere that would diffuse at the same rate. The Rg and Rh values for wildtype PopZ, PopZ P146A, and PopZΔ172–177 were all equivalent to or higher than that of apoferritin, which is a few times larger in mass (Table 1), indicating that PopZ and the mutated variants have an extended shape. The expression ρ, defined as the ratio of Rg to Rh, has been used to characterize elongated molecules in solution (Burchard et al., 1980). The computed value of ρ for wildtype PopZ as well as PopZΔ172–177and PopZ P146A is 1.0 or greater, indicating that PopZ structures behave like semi-flexible rods (Hearst and WH Stockmayer 1962). There was an indication of even more extended configurations (i.e. higher ρ values) in the wildtype and the P146A samples, suggesting that in vivo functionality is in some way correlated with further elongated structures.
Because fully denatured proteins also exhibit rod-like characteristics during sizing on SEC (Corbett and Roche 1984; Wilkins et al., 1999), we analyzed our protein samples by circular dichroism to ask if they had elements of secondary structure. We observed a strong alpha helix signature in each case (Supplementary Figure S7), indicating that significant portions the proteins are structured. The spectra also included characteristics of natively unfolded proteins, which have disordered regions that can take on extended configurations (Chemes et al., 2012; Fink 2005). The alpha helical signature in PopZΔ172–177 was somewhat reduced compared to the other samples, suggesting that this truncation affects the stability of a helical element.
Functional PopZ filaments are formed through higher-order interactions between trimers and hexamers
Additional information about PopZ assembly was derived from SEC/MALLS analyses performed at various protein concentrations (Figure 7B, Table 1). For stable oligomers, elution time from the SEC column and molecular mass are independent of the protein concentration in the eluting peak. We observed this behavior for PopZΔ172–177, and the experimentally determined molecular mass (125 kDa +/− 5%) indicates that it is a hexamer (The Mw of PopZΔ172–177 monomer is 20.7 kDa). In contrast, the calculated molecular mass and elution time of PopZ P146A exhibited concentration dependent effects, suggesting some variability in oligomeric structure. At the lowest concentration, we found that the molecular mass of PopZ P146A was 61 kDa (+/− 5%), consistent with it being a trimer (The Mw of PopZ P146A monomer is 21.2 kDa). With increasing protein concentration, PopZ P146A showed a 16% increase in molecular mass and the elution time in SEC was shifted forward correspondingly.
A more pronounced concentration effect was observed for wildtype PopZ. At the lowest concentration, the calculated molecular mass and elution time for wildtype PopZ was nearly identical to PopZΔ172–177, suggesting that it may also form a hexamer. This is consistent with our observation that wildtype and PopZΔ172–177 oligomers had the same mobility on native gels of whole cell lysates (Figure 3G), in which the concentration of PopZ was low compared to our purified preparations. With increasing protein concentrations, wildtype PopZ exhibited a 66% increase in calculated molecular mass and correspondingly large forward shifts in elution time. Since our calculation of molecular mass measures the average of all particles in the sample, the fractional increases that we observed at higher concentrations could indicate that the oligomers are beginning to assemble into higher-order structures, which exist in equilibrium with the base oligomeric form of the protein, i.e. hexamers for wildtype PopZ and trimers for PopZ P146A. This is supported by our multi-angle light scattering analysis, in which the non-linear relationship between incident light angle and scatter indicated some heterogeneity in particle size (Supplementary Figure S8). This analysis indicated that some of the protein is present in particles that are heavier than the base oligomer. Transient assembly and disassembly of higher-order protein complexes could also be responsible for the slightly reduced mobility of purified wildtype PopZ relative to PopZΔ172–177 on native gels (Figure 7A).
A previous study showed that purified PopZ forms filament structures that are directly visible by TEM (Bowman et al., 2008), and we reasoned that these could be the large particles indicated by our light scattering analysis. Our preparation of wildtype protein (Figure 7C) was visible as short filaments of 40–50 nanometers in length, as previously observed, a length that exceeds the population averaged value of Rg (Table 1) by approximately five fold. Thus, the protein visible in our TEM preparatory technique appears to be enriched for higher-order PopZ structures that are larger than individual hexamers and trimers, and which are present in lower abundance in our native gel and SEC-LS analyses. Since we did not observe any concentration-dependent effects on the mobility of PopZΔ172–177 hexamers by SEC-LS, we predicted that this mutant would be defective in assembling the higher-order structures. As expected, PopZΔ172–177 formed only small and amorphous particles by TEM. In contrast, PopZ P146A, which is functional in vivo and exhibits behavior that is consistent with higher-order assembly during SEC-LS, formed filamentous structures that resemble wildtype. Together, the data suggests a correlation between filament formation and functional activity in vivo. Given that PopZ behaves like an interconnected macromolecular complex in cells (Bowman et al., 2010; Ebersbach et al., 2008), it is reasonable to predict that this is the active form of PopZ in the cell.
Together, the data suggests that PopZΔ172–177, but not PopZ P146A, is defective in assembling oligomeric sub-units into higher-order structures. To test this hypothesis, we asked if PopZΔ172–177 and PopZ P146A exhibit differential ability to co-assemble with wildtype PopZ. We created mixed oligomers by combining proteins in their denatured state and allowing them to refold together during dialysis, and measured assembly by running the products on a native gel (Figure 7D). We found that combinations of wildtype and PopZ P146A protein ran as a single band that migrated to the same position as pure wildtype protein, indicating that the proteins are forming combined structures. The position of this band was super-shifted relative to PopZ Δ172–177. Given that these proteins differ by only 3.6 kDa in mass and 0.09 in pI, it is unlikely that the difference in migration is due to these physiochemical properties, and the slower migrating band is more likely retarded by the formation of higher-order structures, as we observed by SEC-LS and TEM. When we combined wildtype and PopZΔ172–177, the product ran as two bands. This is consistent with the formation of mixed hexamers, in which only those with a sufficient fraction of wildtype subunits can form higher-order, super-shifted structures. In the mixture of PopZ P146A and PopZΔ172–177 a significant fraction of the protein re-assembled as trimers, and the hexamer band exhibited a small amount of super-shifting. Overall, our data indicates that PopZ P146A can combine with wildtype PopZ to form supershifted oligomeric complexes, and that PopZΔ172–177is reduced in this capcity.
DISCUSSION
The PopZ protein localizes to the cell pole, where it functions as a polar organizer in recruiting different protein complexes at distinct times in the cell cycle. PopZ first assembles at the pole opposite the stalk following the initiation of DNA replication, where it is essential for the polar capture of the ParB/parS centromere during chromosome segregation (Bowman et al., 2008; Ebersbach et al., 2008) and consequently for the mid-cell deposition of the FtsZ cell division ring (Thanbichler 2009). It remains at this pole through the remainder of the cell cycle (Fig 1A), later releasing the centromere and recruiting a different set of proteins that are present at the stalked pole (Bowman et al., 2010). Electron microscopy has shown that PopZ is present in a macromolecular structure that fills the cytoplasm at cell poles, and it may build this structure by assembling into an interconnected filament network. Consistent with the self-assembly model, purified 19 kDa PopZ monomers migrate on native gels as higher-order structures (Bowman et al., 2008). Here, we have used a combination of genetic and biophysical approaches to investigate the remarkable biochemical properties of this small protein and find how they are related to the construction of polar macromolecular complexes in vivo.
Among the 23 different popZ mutants that we analyzed, 13 displayed significant defects in PopZ localization. Strains bearing these localization defective mutants also had a cell division phenotype, indicating that PopZ must be in the right place in order to function, consistent with PopZ’s role as a polar organizing factor. PopZ localization defects fell into two distinct categories: aberrant positional accumulation as a single sub-cellular focus, or an increase in diffuse distribution throughout the cytoplasm. This suggests that two different processes are at play in the PopZ localization process. One is the accumulation of PopZ into sub-cellular foci and the other controls the sites of accumulation so that PopZ localizes at the proper cell poles at the correct time in the cell cycle.
We identified three PopZ mutant proteins that formed improperly localized sub-cellular foci, usually at only one pole and less frequently at an interior location, which led to a filamentous cell division phenotype. Two of these, Venus-PopZΔ1–80 and mCherry-PopZΔ1–101, lacked the highly conserved 23 amino acid region in the N terminus and the internal linker. The other, I17A, contained a point mutation within the N-terminal region. The localization pattern of these mutant proteins in C. crescentus is similar to the localization of full length PopZ in E. coli (Bowman et al., 2008).Taken together, these observations suggest that the conserved N-terminus of PopZ facilitates the cell cycle-specific activation of bipolar localization through a mechanism present in C. crescentus but not in E. coli. A recent study has shown that the chromosome partitioning protein ParA, which is present in C. crescentus but not in E. coli, accumulates at the new pole during the time of PopZ localization and could act as a landmark for recruiting PopZ to this location (Laloux and Jacobs-Wagner 2013). Moreover, ParA was found to interact with the N-terminus of PopZ when the two proteins were expressed heterologously in E. coli. The mono-polar localization pattern observed for our Venus-PopZΔ1–80, mCherry-PopZΔ1–101, and Venus-PopZ I17A mutants is consistent with the ParA recruitment mechanism that was proposed. It is also possible that the packing of the N-terminal domain with respect to other regions of PopZ can influence the dynamics of structural assembly, which may affect the seeding of new foci. Some support for this is evident in the localization phenotypes of our PopZ linker region deletion mutants. PopZ tolerates large changes in the R2 sequence between the R1 N terminal region and the R3 C terminal region, but sub-cellular accumulation is lost if R2 becomes too short, perhaps because there is not enough conformational flexibility to form interconnected structures.
We found that the C terminal region of PopZ is critical for sub-cellular accumulation, as all mutations in this region resulted in protein with an elevated level of diffuse localization in the cytoplasm. Since the proteins that were partially localized retained the capacity for bi-polar distribution, the defect in these cases appears to be related to the interconnections necessary for holding PopZ in a cohesive complex, not in the use of poles as sites of accumulation. Many of the localization defective proteins also exhibited defects in oligomerization. Those that failed to produce any oligomers were diffuse, while those that exhibited aberrant oligomerization had reduced quantities of polar localization compared to wildtype PopZ. Together, these observations suggest that sub-cellular accumulation of PopZ is linked to oligomer formation, a conclusion that is supported by our observation that the R3 C-terminal region is sufficient for both of these functions. Laloux and Jacobs-Wagner (2013) performed a similar examination on the sub-cellular localization and oligomerization of PopZ fragments. Their results were in agreement with ours except that in their study, an R3 fragment tagged with C-terminal YFP formed oligomers that did not accumulate in intracellular foci. Although their construct included three fewer amino acids on the N-terminal side of R3, another possible explanation for this discrepancy is the effect of the C-terminal fluorescent protein tag, which could inhibit localization. In support of this, full length PopZ-YFP exhibits only 60% polar localization (Laloux and Jacobs-Wagner, 2013), and full length PopZ tagged with C-terminal mCherry fails to localize in cells (our unpublished data).
Our biophysical analysis of mutated PopZ proteins provided information that enables us to make confident predictions about oligomer size and shape. The truncated PopZΔ172–177 protein failed to form filaments in vitro and exhibited no concentration-dependent increase in molecular mass, indicating that this protein is ‘locked’ as an oligomer that does not form higher-order structures such as filaments and filament assemblies. Having a stable oligomeric form allowed us to make reliable measurements with laser scattering, and we determined that this oligomer is a rod-shaped structure that is comprised of six sub-units. In order to measure the base oligomer size of purified wildtype PopZ, we used low protein concentration to reduce interference from the assembly of higher-order forms. Again, we found that the oligomer is comprised of six sub-units.
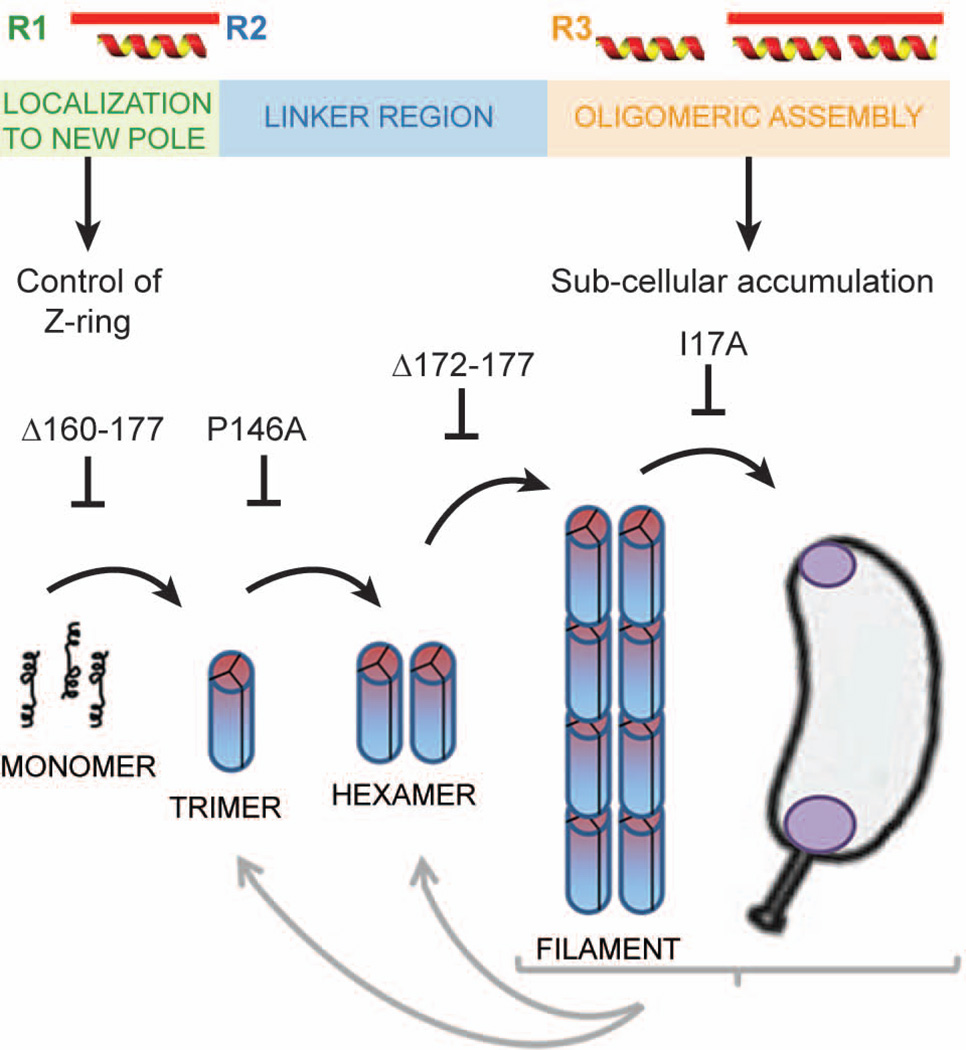
In wildtype cell lysates, PopZ runs as two distinct bands, but only the larger form is present in purified preparations. To characterize the faster migrating band, we analyzed PopZ P146A, which is stable in this form. At the lowest concentration measured, P146A was a trimer. Surprisingly, this half sized oligomer assembles into filaments and is fully functional in vivo. To explain this, we propose that the larger, hexameric form of wildtype PopZ is comprised of a dimer of trimers, and that the P146A mutation destabilizes the trimer-trimer interface. A model of the PopZ assembly process is presented in Figure 8B. Since the radius of the hexamer (as indicated by Rg and Rh) is nearly the same as that of the trimer, we conclude that trimers interact to form hexamers by side-to-side association. The width of the filaments observed by TEM (4–5 nm) is less than the estimated length of the oligomers, thus filaments are most likely formed by end-to-end interactions between hexamers. With regard to the assembly of PopZP146A into filaments, contacts made in these end-to-end interactions could stabilize the interface weakened by the P146A mutation, thereby resulting in normally shaped higher-order structures.
Fig. 8.
Functionally distinct regions of PopZ are responsible for control of cell division and the assembly of PopZ superstructures.
A. Schematic diagram of the PopZ protein sequence. Distinct functional regions, based on areas of conserved sequence, predicted positions of alpha helices, and our functional data, are color coded.
B. A model showing the relationship between PopZ assembly and sub-cellular localization. The first step is the self-association of monomers into rod-shaped trimers, which subsequently dimerize through lateral contact to form hexamers. End-to-end contacts between hexamers produce filaments, and in vivo, these filaments accumulate at cell poles. Each step in this process can be blocked by a mutation in PopZ, as indicated. Each of the forms is metastable and its frequency is influenced by protein concentration and conditions in the buffer or cell extract.
The behavior of PopZΔ172–177 suggests that this mutation does not affect associations between trimers, but destabilizes end-to-end contact between hexamers, thereby preventing filament assembly. We propose that the assembly of oligomers into relatively short filaments is an intermediate step in the formation of a much larger three-dimensional interconnected network that forms at the cell poles in vivo. We found evidence that individual oligomers can interact to form larger structures in our TEM images and in our biophysical analysis of purified protein. However, the 66% increase in average molecular mass that we observed at increased protein concentrations of wildtype protein in SEC-LS is not consistent with extensive polymerization, suggesting that purified PopZ exists in a dynamic equilibrium between individual oligomers and higher-order structures, and that almost all of the protein is present as uncomplexed oligomers at the lowest concentrations. A likely reason for the fact that PopZ runs as well-defined bands on native gels and single peaks by SEC is that individual oligomers and higher-ordered structures are in rapid equilibrium, and our separation methods are too slow to differentiate these dynamic states. Inter-oligomer interactions may be stabilized by conditions in vivo, at high protein concentration, and on the grid surface in our TEM preparations, shifting the equilibrium towards higher-ordered forms.
While PopZ has functional similarity to the DivIVA polar organizing protein in gram positive bacteria, this study has revealed some significant differences. Whereas DivIVA forms a tetrameric rod that is comprised mainly of a coiled coil (Oliva et al., 2010), PopZ forms trimers and hexamers that are not predicted to include any coiled coils. Thus, it is likely that the associations between monomers and the folds that are involved in producing elongated structures are not similar. Such a difference in structure could reflect different localization mechanisms. Whereas the curved structure of DivIVA oligomers may enable localization to curved membranes, PopZ may form superstructures in order to be excluded from DNA (Saberi and Emberly 2010; Ebersbach et al., 2008). Consistent with this, DivIVA-GFP has a cup shaped localization pattern around cell poles (Ramamurthi and Losick 2009; Lenarcic et al., 2009), but such a pattern is not clearly indicated in superresolution images of fluorescently tagged PopZ (Gahlmann et al., 2013) or in electron micrographs of wildtype C. crescentus (Bowman et al., 2010), where the PopZ domain appears to fill the entire area of the pole.
Our study shows that higher-order assembly within the C-terminal domain of PopZ plays an essential role in forming superstructures that are the polar localized, active form of PopZ in the cell. Notably, the assembled form of PopZ has a well-defined structure, as evident by the tight distribution of oligomer sizes. This is significantly different from examples of bacterial proteins that localize through aggregation, all of which are partially denatured due to temperature stress or mutation (Rokney et al., 2009; Winkler et al., 2010). PopZ may therefore be a “structured aggregate” like those formed by prion and prion-like proteins in eukaryotic cells (Eisenberg and Jucker 2012), one of a small number of cases in which the normal physiological activity of the protein is performed in the assembled conformational state (Fowler et al., 2007). The timing of superstructure assembly and its location to the cell pole are therefore under tight control in C. crescentus cells, and future studies are needed to understand how temporal and spatial cues are coordinated with higher-order assembly and heterologous interactions with other proteins.
Experimental Procedures
C. crescentus strains and cell growth
All C. crescentus strains used in this study are derived from the synchronizeable wild-type strain CB15N, and were grown at 28°C in PYE medium(0.2% bactopeptone, 0.1% yeast extract, 1 mM MgSO4, 0.5 mM CaCl2). When appropriate, popZ expression was stimulated by the addition of Na-vanillate at 500 µM for 120 min before imaging. When popZ was introduced on the pBXMCS-2 plasmid (Thanbichler et al., 2007), the leakiness of the un-induced xylX promoter was sufficient for expression. Strains were analyzed at mid-exponential phase of growth for imaging and at stationary phase for protein electrophoresis. Descriptions of strains and plasmids used in this study are in Supplementary Tables 1 and 2.
Gel electrophoresis and Western blotting
Whole cell lysates for protein analysis were prepared by pelleting 10 mL of stationary phase cultures. Pellets were resuspended in 10 mL of Co-IP Buffer (20 mM Hepes, 100 mM NaCl, 20% glycerol, pH 7.5). Cells were repelleted and resuspended in 1.5 mL of Co-IP buffer plus added protease inhibitors. Cells were lysed with a sonicator microtip (Branson) using three sets of ten 1 second pulses, and debris was pelleted by centrifugation at 13,000 x g for 10 minutes. 10 µg of total protein was loaded for electrophoresis. SDS-PAGE (buffered with 25mM Tris, 250 mM glycine, 0.5% SDS, pH 8.3) and native gel electrophoresis (buffered with 25mM Tris, 192 mM glycine, pH 8.8) used 12% or 4–20% Tris-glycine gels (Bio-Rad), respectively, followed by electrophoretic transfer to a PVDF membrane (Millipore). Immunoblotting was performed with anti-PopZ sera (Bowman et al., 2008) or with anti-FLAG (Sigma), followed by detection with chemiluminescent substrate (Pierce).
Light Microscopy and quantitative image analysis
Images were collected as described by Bowman et al. (2010). For all image panels, the brightness and contrast of the images were balanced identically. For computational image analyses, MicrobeTracker (Sliusarenko et al., 2011) was used to determine cell outlines from phase images, and length was determined from a line drawn along the long axis of the cell. SpotFinderZ was used to identify and outline fluorescent foci within the cells. To measure the amount of polar localization, the total intensity of these puncta was divided by the total fluorescence intensity within the cell. For particularly bright puncta the outline of the focus extended beyond the cell border, sometimes resulting in localization ratios that were slightly higher than 100%.
Protein expression and purification
For expression of 6His-PopZ and variants, E. coli Rosetta strains bearing pET28-6His-PopZ and mutants (Supplementary Table 1) were cultured at 37°C in LB supplemented with 30µg/ml kanamycin and 20µg/ml chloramphenicol. Mid-log phase cultures were shifted to 30°C and induced with 1mM IPTG for 2 hours before pelleting and storage at −80°C until use. Pellets were resuspended in Lysis/Wash buffer (100mM Na-phosphate pH8, 10mM Tris-Cl, 300mM NaCl, 8M urea, 20mM imidazole) supplemented with Roche Complete protease inhibitors. After solubilization, lysates were applied to Talon metal affinity resin (Clontech) and washed with Lysis/Wash buffer before elution in 100mM phosphate, 10mM Tris-Cl, 300mM NaCl, 8M urea, 250mM imidazole, pH8. Purified samples were refolded via dialysis against 20mM Tris-Cl pH8.5 at 4°C. Refolded protein samples displayed similar oligomeric assembly characteristics to natively-purified samples as judged by gel filtration, native gel electrophoresis, and electron microscopy (data not shown).
Mixed oligomerization assay
Refolding dialysis on mixtures of PopZ variants was performed to assess co-oligomerization of wild type 6His-PopZ and mutant proteins. Briefly, 6His-PopZ wild type, P146A, and S172 were purified as described above. Denatured samples were normalized to 0.3 mg/ml and equal volumes of the indicated samples (or elution buffer) were mixed, applied to mini Slide-a-Lyzer dialysis caps and dialyzed against 20mM Tris-Cl pH8.5 at 4°C for refolding. Approximately 0.2–0.4 µg of each mix was loaded onto a 4–15% Tris-Glycine native gel (BioRad) and run at 80V for 4 hours at 4°C before staining with Coomassie R250.
TEM
Proteins were diluted to a concentration of 0.110 mg/ml in buffer (20 mM Tris pH 8.5, 25mM NaCl), and 5µl was added to glow-discharged carbon-coated Formvar grids (Ted Pella 01753F). After 1.5 minutes incubation, the grids were floated on a large drop of buffer for 20 seconds before blotting with filter paper (Whatman #1). Before any drying could occur, grids were incubated in 5µl of uranyl acetate for 30 seconds. After another brief blotting, grids were imaged as described (Bowman et al. 2008).
SEC-LS
The light scattering data were collected using a Superose 610/300, HR Size Exclusion Chromatography (SEC) column (GE Healthcare, Piscataway, NJ), connected to High Performance Liquid Chromatography System (HPLC), Agilent 1200, (Agilent Technologies, Wilmington, DE) equipped with an autosampler. The elution from SEC was monitored by a photodiode array (PDA) UV/VIS detector (Agilent Technologies, Wilmington, DE), differential refractometer (OPTI-Lab rEx Wyatt Corp., Santa Barbara, CA), static and dynamic, multiangle laser light scattering (LS) detector (HELEOS II with QELS capability, Wyatt Corp., Santa Barbara, CA). The SEC-UV/LS/RI system was equilibrated in running buffer (150mM NaCl, 20mM Tris-Cl pH 8.5) at the flow rate of 0.3 ml/min. Two software packages were used for data collection and analysis: the Chemstation software (Agilent Technologies, Wilmington, DE) controlled the HPLC operation and data collection from the multi-wavelength UV/VIS detector, while the ASTRA software (Wyatt Corp., Santa Barbara, CA) collected data from the refractive index detector, the light scattering detectors, and recorded the UV trace at 280 nm sent from the PDA detector. The weight average molecular masses, Mw, were determined across the entire elution profile in the intervals of 1 sec from static LS measurement using ASTRA software as previously described (Folta-Stogniew and Williams, 1999). Hydrodynamic radii, Rh, were measured from an “on-line” dynamic LS measurement every 2 sec. The dynamic light scattering signal was analyzed by the method of cumulants (Koppel, 1972).
Circular dichrosim
Circular Dichroism (CD) measurements were performed using Chirascan (Applied Photophysics ;Leatherhead, United Kingdom) instrument with a 0.1 or 1 mm pathlength quartz cuvette The CD spectra of PopZ, WT (8.1 microM; monomer), PopZ P146A (5.7 microM) and PopZΔ172–177 (5.4 microM) in optically clear buffer (50 mM potassium borate,10 mM Sodium Fluoride, pH 7.5) were recorded from 180 to 260 nm. Each spectrum is an average of five scans with background of buffer alone subtracted. Secondary structure analysis of the spectra was performed using CDNN 2.1 software (developed by Gerald Böhm) supplied by Applied Photophysics.
Supplementary Material
Acknowledgements
We thank David Liberles for helpful advice. The work was supported by National Institute of Health Grants GM51426 and GB32506, and Department of Energy Grant DE-FG02-05ER64136 to LS. LRC was supported by Office of Basic Energy Sciences, Biological and Environmental Research, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
References Cited
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Gaietta GM, Fero M, Hong S-H, Jones Y, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Comolli LR, Zhu J, Eckart M, Koenig M, Downing KH, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GR, Lyuksyutova AI, Shapiro L. Bacterial polarity. Curr Opin Cell Biol. 2011;23:71–77. doi: 10.1016/j.ceb.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burchard W, Schmidt M, Stockmayer WH. Information on Polydispersity and Branching from Combined Quasi-Elastic and Intergrated Scattering. Macromolecules. 1980;13:1265–1272. [Google Scholar]
- Chemes LB, Alonso LG, Noval MG, Prat-Gay G, de Circular dichroism techniques for the analysis of intrinsically disordered proteins and domains. Methods Mol Biol. 2012;895:387–404. doi: 10.1007/978-1-61779-927-3_22. [DOI] [PubMed] [Google Scholar]
- Corbett RJ, Roche RS. Use of high-speed size-exclusion chromatography for the study of protein folding and stability. Biochemistry. 1984;23:1888–1894. doi: 10.1021/bi00303a047. [DOI] [PubMed] [Google Scholar]
- Donovan C, Sieger B, Krämer R, Bramkamp M. A synthetic Escherichia coli system identifies a conserved origin tethering factor in Actinobacteria . Mol Microbiol. 2012;84:105–116. doi: 10.1111/j.1365-2958.2012.08011.x. [DOI] [PubMed] [Google Scholar]
- Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter . Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid--from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Gahlmann A, Ptacin JL, Grover G, Quirin S, Diezmann ARS, von, Lee MK, et al. Quantitative multicolor subdiffraction imaging of bacterial protein ultrastructures in three dimensions. Nano Lett. 2013;13:987–993. doi: 10.1021/nl304071h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher SR. One-dimensional electrophoresis using nondenaturing conditions. Curr Protoc Mol Biol. 2001;Chapter 10 doi: 10.1002/0471142727.mb1002bs47. Unit 10.2B. [DOI] [PubMed] [Google Scholar]
- Hearst J, Stockmayer W. Sedimentation constants of broken chains and wormlike coils. The Journal of Chemical Physics. 1962;37:1425–1433. [Google Scholar]
- Kaval KG, Halbedel S. Architecturally the same, but playing a different game: the diverse species-specific roles of DivIVA proteins. Virulence. 2012;3:406–407. doi: 10.4161/viru.20747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiekebusch D, Michie KA, Essen L-O, Löwe J, Thanbichler M. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol Cell. 2012;46:245–259. doi: 10.1016/j.molcel.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick CL, Viollier PH. Decoding Caulobacter development. FEMS Microbiol Rev. 2012;36:193–205. doi: 10.1111/j.1574-6976.2011.00309.x. [DOI] [PubMed] [Google Scholar]
- Koppel DE. Analysis of macromolecular polydispersity in intensity correlation spectroscopy: the method of cumulants. Journal of Chemical Physics. 57:4814–4820. [Google Scholar]
- Kusek G, Campbell M, Doyle F, Tenenbaum SA, Kiebler M, Temple S. Asymmetric Segregation of the Double-Stranded RNA Binding Protein Staufen2 during Mammalian Neural Stem Cell Divisions Promotes Lineage Progression. Cell Stem Cell. 2012;11:505–516. doi: 10.1016/j.stem.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloux G, Jacobs-Wagner C. Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization. J Cell Biol. 2013;201:827–841. doi: 10.1083/jcb.201303036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, et al. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva MA, Halbedel S, Freund SM, Dutow P, Leonard TA, Veprintsev DB, et al. Features critical for membrane binding revealed by DivIVA crystal structure. EMBO J. 2010;29:1988–2001. doi: 10.1038/emboj.2010.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Thanbichler M, Viollier PH. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus . Genes Dev. 2008;22:212–225. doi: 10.1101/gad.1601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci USA. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokney A, Shagan M, Kessel M, Smith Y, Rosenshine I, Oppenheim AB. E. coli transports aggregated proteins to the poles by a specific and energy-dependent process. J Mol Biol. 2009;392:589–601. doi: 10.1016/j.jmb.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Saberi S, Emberly E. Chromosome driven spatial patterning of proteins in bacteria. PLoS Comput Biol. 2010;6:e1000986. doi: 10.1371/journal.pcbi.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mowery RA, Ashley J, Hentz M, Ramirez AJ, Bilgicer B, et al. Abnormal SDS-PAGE migration of cytosolic proteins can identify domains and mechanisms that control surfactant binding. Protein Sci. 2012;21:1197–1209. doi: 10.1002/pro.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80:612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg H, Kutejová E, Muchová K, Gregorini M, Lustig A, Müller SA, et al. Oligomeric structure of the Bacillus subtilis cell division protein DivIVA determined by transmission electron microscopy. Mol Microbiol. 2004;52:1281–1290. doi: 10.1111/j.1365-2958.2004.04074.x. [DOI] [PubMed] [Google Scholar]
- Thanbichler M. Spatial regulation in Caulobacter crescentus . Curr Opin Microbiol. 2009;12:715–721. doi: 10.1016/j.mib.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Thanbichler M, Iniesta AA, Shapiro L. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus . Nucleic Acids Res. 2007;35:e137. doi: 10.1093/nar/gkm818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter . Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- Winkler J, Seybert A, König L, Pruggnaller S, Haselmann U, Sourjik V, et al. Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 2010;29:910–923. doi: 10.1038/emboj.2009.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt P. Light Scattering and the absolute characterization of macromolecules. Chimica acta. 1993;272:1–40. [Google Scholar]
- Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–266. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.