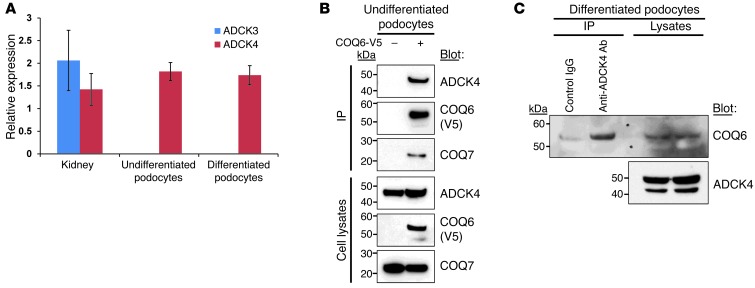
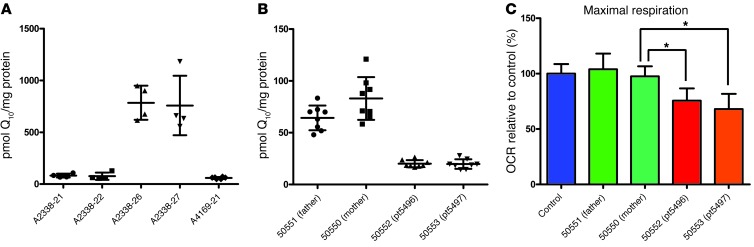
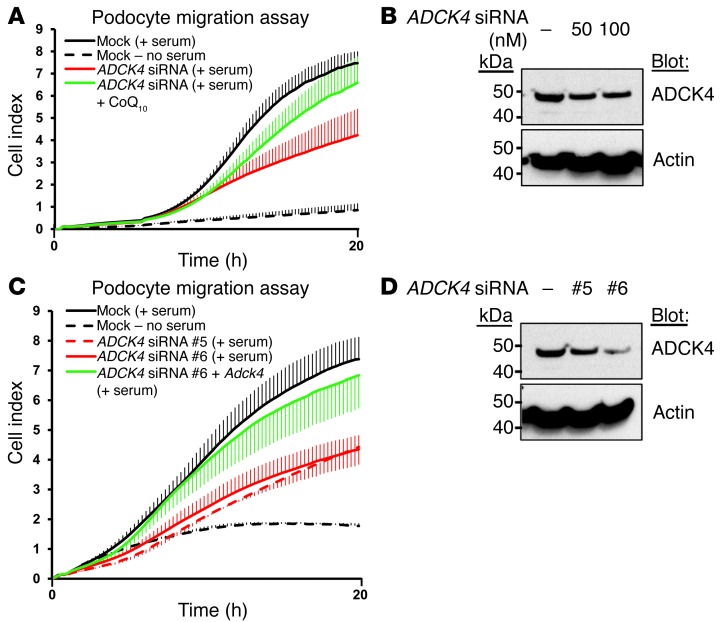
Shazia Ashraf
Shazia Ashraf
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,
Heon Yung Gee
Heon Yung Gee
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,
Stephanie Woerner
Stephanie Woerner
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
2,3,
Letian X Xie
Letian X Xie
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
4,
Virginia Vega-Warner
Virginia Vega-Warner
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
5,
Svjetlana Lovric
Svjetlana Lovric
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,
Humphrey Fang
Humphrey Fang
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,
Xuewen Song
Xuewen Song
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
6,
Daniel C Cattran
Daniel C Cattran
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
6,
Carmen Avila-Casado
Carmen Avila-Casado
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
7,
Andrew D Paterson
Andrew D Paterson
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
8,
Patrick Nitschké
Patrick Nitschké
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
9,
Christine Bole-Feysot
Christine Bole-Feysot
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
10,
Pierre Cochat
Pierre Cochat
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
11,
Julian Esteve-Rudd
Julian Esteve-Rudd
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
12,
Birgit Haberberger
Birgit Haberberger
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
13,14,
Susan J Allen
Susan J Allen
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
5,
Weibin Zhou
Weibin Zhou
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
5,
Rannar Airik
Rannar Airik
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,
Edgar A Otto
Edgar A Otto
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
5,
Moumita Barua
Moumita Barua
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
15,
Mohamed H Al-Hamed
Mohamed H Al-Hamed
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
16,
Jameela A Kari
Jameela A Kari
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
17,
Jonathan Evans
Jonathan Evans
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
18,
Agnieszka Bierzynska
Agnieszka Bierzynska
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
19,
Moin A Saleem
Moin A Saleem
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
19,
Detlef Böckenhauer
Detlef Böckenhauer
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
20,
Robert Kleta
Robert Kleta
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
20,
Sherif El Desoky
Sherif El Desoky
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
17,
Duygu O Hacihamdioglu
Duygu O Hacihamdioglu
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
21,
Faysal Gok
Faysal Gok
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
21,
Joseph Washburn
Joseph Washburn
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
22,
Roger C Wiggins
Roger C Wiggins
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
23,
Murim Choi
Murim Choi
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
24,25,
Richard P Lifton
Richard P Lifton
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
24,25,
Shawn Levy
Shawn Levy
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
26,
Zhe Han
Zhe Han
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
27,
Leonardo Salviati
Leonardo Salviati
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
28,
Holger Prokisch
Holger Prokisch
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
13,14,
David S Williams
David S Williams
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
12,
Martin Pollak
Martin Pollak
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
11,
Catherine F Clarke
Catherine F Clarke
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
4,
York Pei
York Pei
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
6,
Corinne Antignac
Corinne Antignac
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
2,3,29,
Friedhelm Hildebrandt
Friedhelm Hildebrandt
1Division of Nephrology, Department of Medicine, Boston
Children’s Hospital, Harvard Medical School, Boston, Massachusetts, USA.
2Inserm U983, Necker Hospital, Paris, France.
3Université Paris Descartes, Sorbonne Paris Cité, Imagine
Institute, Paris, France. 4Department of Chemistry and Biochemistry and
Molecular Biology Institute, UCLA, Los Angeles, California, USA.
5Department of Pediatrics, University of Michigan, Ann Arbor, Michigan,
USA. 6Division of Nephrology, University Health Network, and University
of Toronto, Toronto, Ontario, Canada. 7Department of Pathology,
University Health Network, Toronto, Ontario, Canada. 8Program in Genomic
Biology, Hospital for Sick Children, Toronto, Ontario, Canada.
9Bioinformatics Plateform, Université Paris Descartes, Paris,
France. 10Genomics Plateform, Imagine Institute, Université Paris
Descartes, Paris, France. 11Service de Néphrologie et
Rhumatologie Pédiatrique, Centre de référence des maladies
rénales rares, Hôpital Femme Mère Enfant, Hospices Civils de
Lyon, Bron, France. 12Jules Stein Eye Institute, UCLA School of Medicine,
Los Angeles, California, USA. 13Institute of Human Genetics, Helmholtz
Zentrum Munich, Neuherberg, Germany. 14Institute of Human Genetics,
Klinikum rechts der Isar, Technical University Munich, Munich, Germany.
15Division of Nephrology, Department of Medicine, Beth Israel
Deaconess Medical Center, Boston, Massachusetts, USA. 16Institute of
Genetic Medicine, Newcastle University, Central Parkway, Newcastle upon Tyne, United
Kingdom. 17Pediatrics Department, King Abdulaziz University, Jeddah,
Kingdom of Saudi Arabia. 18Nottingham University Hospitals, Nottingham,
United Kingdom. 19Children’s and Academic Renal Unit, University
of Bristol, Bristol, United Kingdom. 20UCL Institute of Child Health and
Paediatric Nephrology, Great Ormond Street Hospital, London, United Kingdom.
21Department of Pediatric Nephrology and Rheumatology, Gülhane
Military Academy of Medicine, School of Medicine, Etlik, Ankara, Turkey.
22Biomedical Research Core Facilities and 23Department
of Internal Medicine, Department of Pathology, and Department of Otolaryngology,
University of Michigan, Ann Arbor, Michigan, USA. 24Department of
Genetics, Howard Hughes Medical Institute, and 25Yale Center for
Mendelian Genomics, Yale University School of Medicine, New Haven, Connecticut, USA.
26HudsonAlpha Institute for Biotechnology, Huntsville, Alabama, USA.
27Department of Internal Medicine — Molecular Medicine and
Genetics, University of Michigan, Ann Arbor, Michigan, USA. 28Department
of Woman and Child Health, University of Padova and Istituto di Ricerca Pediatrica
Città della Speranza, Padova, Italy. 29Assistance Publique
– Hôpitaux de Paris, Department of Genetics, Necker Hospital, Paris,
France. 30Department of Human Genetics, University of Michigan, Ann
Arbor, Michigan, USA. 31Howard Hughes Medical Institute, Chevy Chase,
Maryland, USA.
1,30,31