Abstract
Traditionally, transfection complexes are typically formed by bulk mixing, producing particles with high polydispersity and limited control over vector size. Herein, we demonstrate the use of a commercial micro-reactor to assemble pDNA:cationic cyclodextrin:pendant polymer nanoparticles using a layer-by-layer approach. Our studies reveal that the particles formulated via microfluidic assembly have much smaller sizes, lower polydispersity, lower ζ-potentials, and comparable cell viability and transfection profiles in HeLa cells than bulk mixed particles. The complexes also show a flow rate-dependent stability, with particles formed at slower flow rates giving rise to more stable complexes as determined by heparin challenge. Our findings suggest that microfluidic reactors offer an attractive method for assembling reproducible, size-controlled complexes from multi-component transfection complex assemblies.
Gene therapy is a promising alternative to conventional chemotherapy due to its high specificity and modest off-target effects.1 While various viral vectors exist for the delivery of nucleic acids, they suffer from issues such as safety, scalability and immunogenicity. Non-viral carriers that can condense the nucleic acids into ≤ 200 nm positively charged nanocomplexes are gaining prominence because of their modest immunogenicity and ease of production.2,3 Many different carrier designs based on cationic polypeptides,4 cationic lipids,5 polymers6–8 and cyclodextrins9 nanoparticles have been developed. While most of these materials are able to effectively condense and deliver pDNA, they still exert relatively little control over the vector assembly process and final dimensions. This introduces large polydispersities and little control over initial physical characteristics such as diameter or ζ-potential of the transfection complexes.10 Vortex mixing and/or bulk mixing is the most common process used by researchers to assemble such nanoparticle complexes, but these complexation methods have high inherent variability due to gradients in concentration and temperature that are formed during the mixing process. Such batch-to-batch inconsistency can lead to poor reproducibility and false negative results in biological experiments.11
Microfluidics-based techniques such as flash nanoprecipitation (FNP)12–14 and hydrodynamic flow focusing (HFF)15–17 have been demonstrated as robust techniques for the formulation of drug-loaded polymeric nanoparticles, but similar control is yet to be shown for the formulation of nucleic acid cargo. Microfluidic formulation of nucleic acid-loaded nanoparticles has been previously performed for the rapid generation of libraries of siRNA-containing lipid nanoparticles18 and pDNA-containing cyclodextrin-based supramolecular nanoparticles.19 While these techniques are excellent tools for high throughput screening of nucleic acid nanoparticles, control over the physical properties of the nanoparticle complexes, in most cases, has not been demonstrated. Recently, Cullis, et al., have demonstrated the formulation of limit–sized lipid nanoparticles and lipid–siRNA nanoparticles using rapid microfluidic mixing of lipids dissolved in an ethanol stream.20,21 The authors demonstrated that microfluidic mixing was a powerful technique for formulation of both polar and non-polar core structures. They also showed that they could achieve particle sizes as small as 20 – 50 nm with encapsulated siRNA using this technique. Leong, et al., have developed the technique of microfluidics-assisted confinement of complexes in picoliter oil droplets. They demonstrated a reduction of size, PDI, ζ-potential and cytotoxicity, as well as higher transfection efficiency than the bulk mixed counterparts.22 The ability to tune the physical characteristics based on flow rate alone, in a manner similar to FNP or HFF, has still not been demonstrated. Herein, we demonstrate the flow rate-dependent control over particle characteristics such as diameter, polydispersity index (PDI), ζ-potential, and decomplexation rate. To control these properties, we formulated the transfection complexes in a commercially available glass Chemtrix microreactor (Figure 1). Our results suggest that the microreactor provides improved control over the physical characteristics of the nanoparticles generated under a wide range of feed conditions and flow rates. We also show that the flow mixing technique is a reproducible, operator-independent, and potentially scalable technique that can improve the uniformity and performance of nucleic acid nanoparticle complexes for transfection.
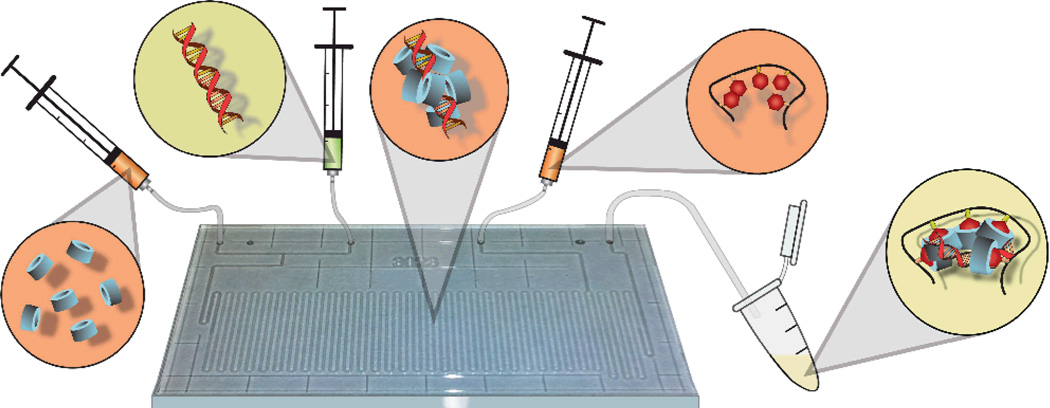
Figure 1.
Conceptual diagram of HA-Ad:CD-PEI:pDNA transfection complex preparation by microfluidic assembly. CD-PEI (far left) is first mixed with pDNA (mid-left); intermediate polyplexes (center) are mixed with HA-Ad (mid-right), resulting in the final nanoparticles of controlled diameter, composition, and low polydispersity.
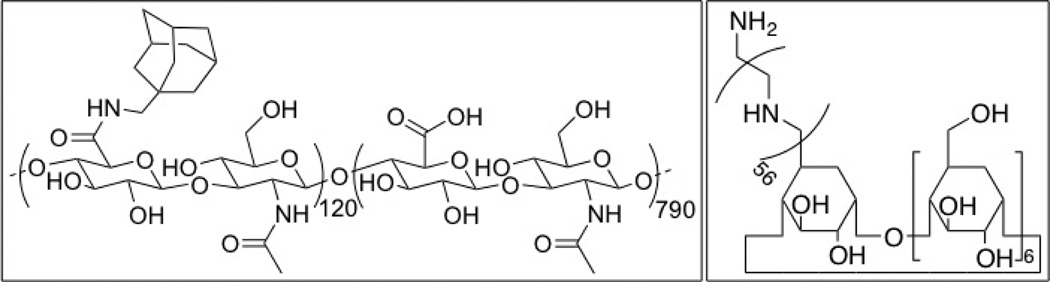
As an extension of our previously reported poly(vinyl alcohol)-based pendant polymer designs,23,24 we developed a new hyaluronic acid (HA)-based pendant polymer system capable of forming complexes with cationic cyclodextrins and pDNA. HA (350 kD) was chosen for its high water solubility, CD44-targeting capabilities, and biocompatibility.25,26 Adamantane-conjugated (HA-Ad) was synthesized via EDC mediated coupling between HA and adamantane methylamine (Supporting Information). Poly(ethyleneimine) (PEI) of molecular weight 2.5 kDa was used to introduce a single modification on the β-cyclodextrin 1° hydroxyl rim. Briefly, β-cyclodextrin was converted to the monotosylated form27 and was further converted to CD-PEI after isolation of the pure 6-monotosyl-β-cyclodextrin28 by treatment with an excess of PEI2.5k. The CD-PEI product was purified by precipitation in ether and exhaustive dialysis against DMSO and water. The HA-Ad and CD-PEI components (Figure 2) were then used for assembly of the pDNA nanoparticle complexes via bulk or flow mixing techniques. For bulk mixed complexes, the CD-PEI and pDNA were mixed first and incubated for 1 h before the HA-Ad was added with vortex mixing and further incubation for 1 h. At the same time, the pDNA, CD-PEI and HA-Ad component solutions were assembled under different flow mixing conditions using the Chemtrix microreactor as described below (Figure 1).
Figure 2.
Chemical structures of (left) HA-Ad and (right) CD-PEI.
The flow reactor (Figure S1) chosen provided sufficient length and volume to enable efficient mixing of the feed solutions. This reactor is equipped with 2 input channels, 1 quench channel, and 1 output channel with a total reactor volume of 10 µL and reactor length of 60 µm. CD-PEI and pDNA (pAcGFP1, Clontech) were introduced in separate input channels while HA-Ad was added via the quench channel. This setup replicates the stepwise assembly process used in bulk mixing by allowing the pDNA and CD-PEI to first yield pDNA:CD-PEI polyplexes before adding HA to consolidate the initially formed condensed pDNA core to generate the final transfection complexes. All the individual components were diluted in sterile, filtered, nanopure water instead of buffer to ensure that buffer counterions did not interfere with complex formation. Transfection complexes were assembled at N/P ratios of 10, 20 and 30, since these were the ratios shown to have optimal activity for the bulk mixed complexes in initial screening experiments. Flow mixing was carried out at pDNA flow rates of 5, 10, and 20 µL/min. CD-PEI and HA-Ad flow rates were adjusted accordingly to afford the required N/P ratios.
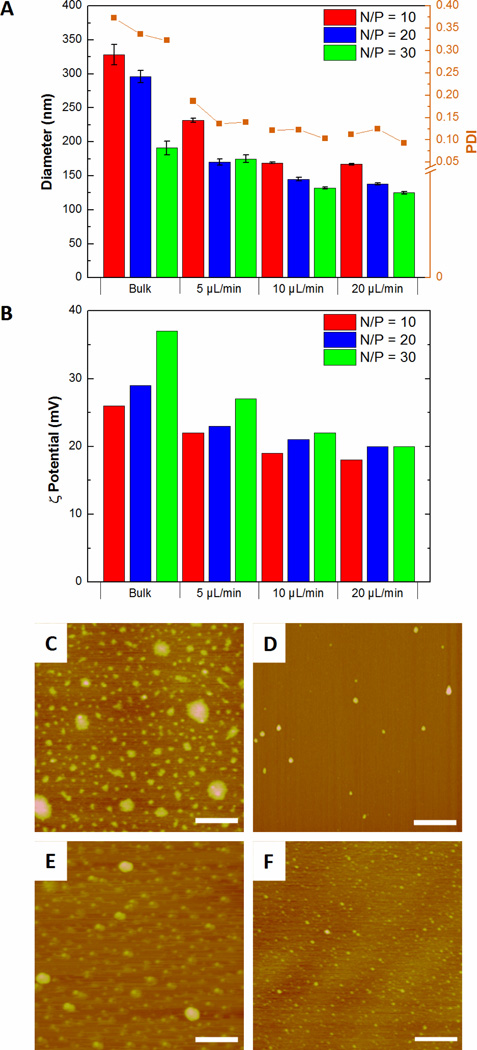
Our studies revealed that higher flow rates resulted in smaller complexes and lower PDIs than bulk mixed particles at the same N/P ratios (Figures 3A). Flow mixing also produced particles with lower ζ-potentials than the bulk mixed particles of the same N/P ratio (Figure 3B). AFM analysis of the complexes showed that flow mixed complexes were smaller and more monodisperse than the corresponding bulk mixed particles (Figure 3C – 3F), with average diameters of 50 ± 10 nm for flow mixed transfection complexes and 120 ± 20 nm for bulk mixed complexes of the same composition (Figure S3).
Figure 3.
(A) Diameter and polydispersity indices (PDI); (B) ζ-potentials; and AFM images of transfection complexes produced at N/P = 20 by (C) bulk mixing; and flow mixing at pDNA flow rates of (D) 5, (E) 10 and (F) 20µL/min. Scale bar = 500 nm.
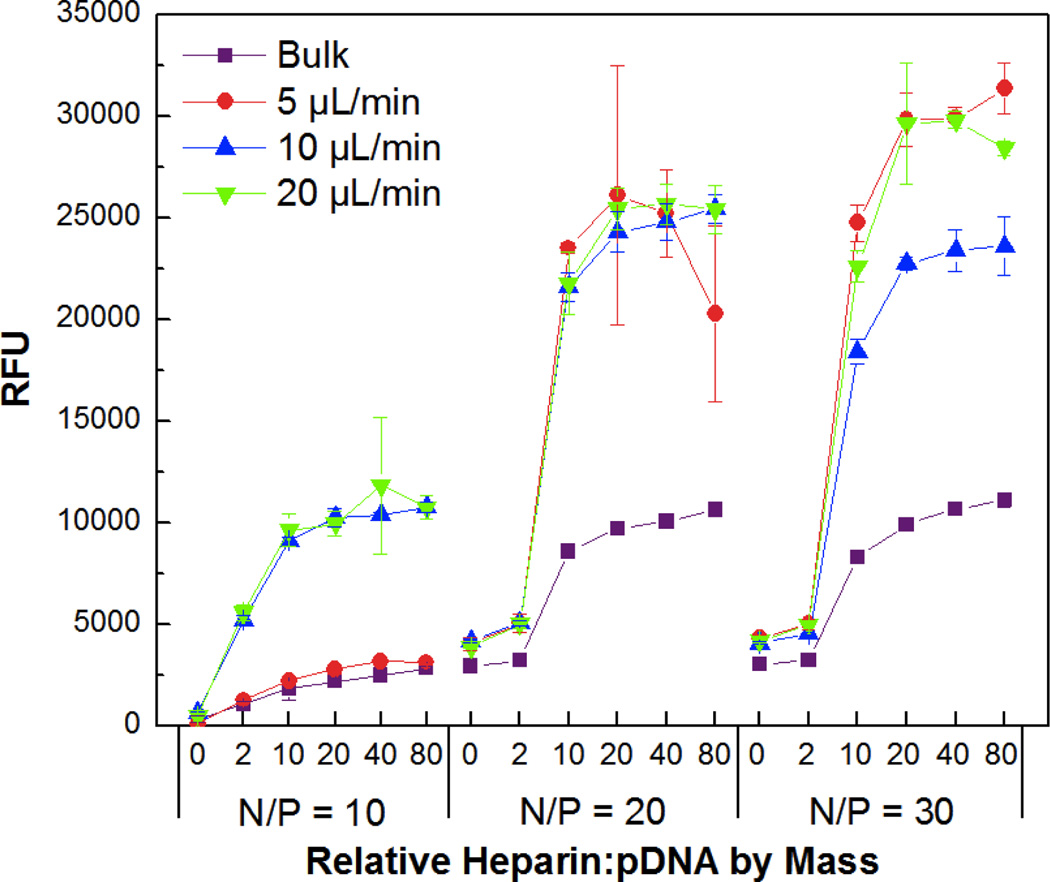
The relative complexation ability and colloidal stability of the transfection complexes were studied by gel retardation (Figure S4) and PicoGreen competitive binding assays (Figure 4), respectively. PicoGreen is a highly sensitive dye that fluoresces upon intercalation into dsDNA, but remains non-fluorescent when the DNA is tightly condensed and inaccessible for dye binding. The bulk mixed and flow mixed complexes were incubated for 30 min with increasing amounts of the negatively charged polysaccharide heparin to promote polyion exchange of CD-PEI:HA-Ad from the pDNA core. This was followed by addition of the dye and fluorescence analysis. Interestingly, while the gel shift assay indicated very similar complexation profiles for the bulk mixed and flow mixed complexes, the PicoGreen assay showed that the decomplexation characteristics of the various complexes were very different.
Figure 4.
PicoGreen competitive binding assay showing colloidal stability of bulk and flow mixed pDNA:CD-PEI:HA-Ad complexes assembled at different N/P ratios and flow rates in the presence of increasing amounts of heparin (0, 2, 10, 20, 40, and 80 times the mass of pDNA used for complex formation).
The PicoGreen studies indicated that complexes formulated by flow mixing, in general, were more susceptible to disassembly in the presence of polyanions than those prepared by bulk mixing. Competitive binding assays also showed that the transfection complexes prepared by flow mixing at higher flow rates were prone to disassemble more readily than those produced at lower flow rates. These data indicate that the complexation ability of all the pDNA:CD-PEI:HA-Ad ratios and flow rates are nearly the same, however, disassembly is more facile in the presence of a negatively charged polymer challenge when the transfection complexes have been prepared by flow mixing. This can be attributed to the shorter interaction time between the individual components when they are flow mixed than occurs for bulk mixed complexes. The longer interaction times in the bulk mixed case enables multiple encounters with other particles, leading to larger and more highly entangled ionomer complexes with higher polydispersity. These findings suggest that the flow mixing strategy demonstrated here gives us control over properties such as diameter, PDI and ζ-potential, as well as a less described characteristic, the disassembly propensity of the complexes.
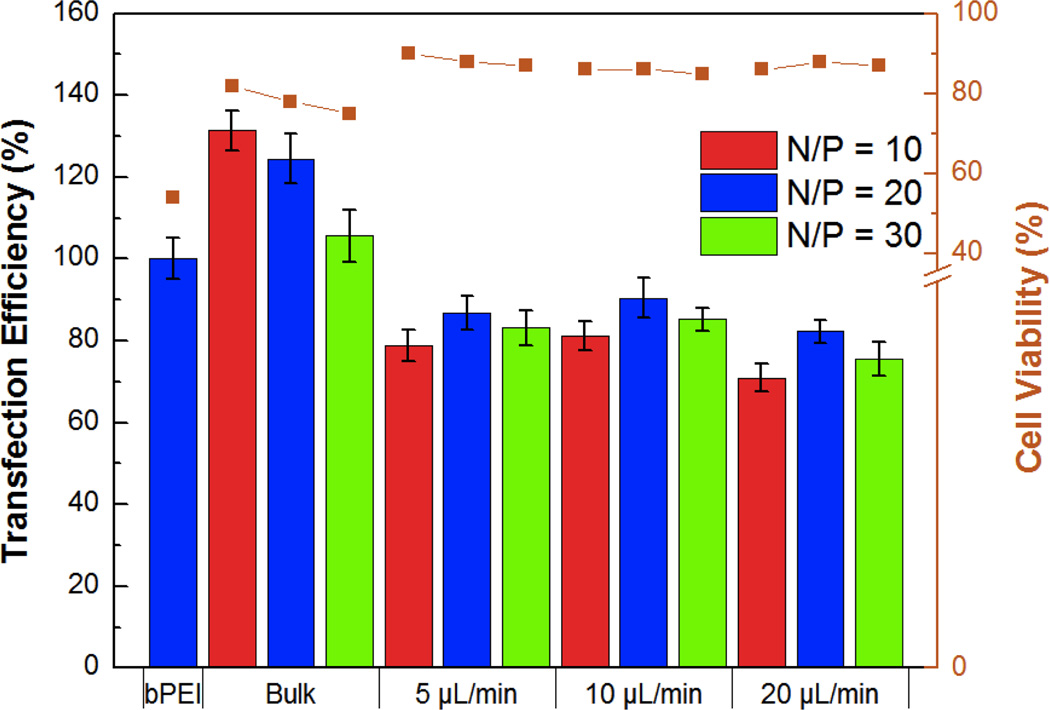
The MTS cell viability assay was performed to compare the HA-Ad and CD-PEI materials using bPEI as control. These studies indicated that CD-PEI and the CD-PEI:HA-Ad complexes had negligible effects on cell viability, even at concentrations as high as 1mM, whereas bPEI had an LD50 of ~ 0.15mM (Figure S5). The relative transfection efficiencies and cell viabilities of the bulk and flow mixed complexes was then studied. Upon incubation with HeLa cells, flow mixed complexes demonstrated improved cell viability (10 – 20%), but lower transfection efficiency (20 – 30%), than the bulk mixed complexes (Figure 5). We infer from these data that the flow mixed complexes undergo reduced cellular uptake due to their lower ζ-potential and reduced stability in the presence of polyanions present in the culture media. Cell viability was also evaluated using the 7AAD viability stain during the transfection experiment. We found that cells treated with flow mixed complexes had higher cell viabilities (85 – 90%) than bulk mixed complexes (70 – 80%; Figure 5).
Figure 5.
Quantification of relative transfection efficiency and cytotoxicity of bulk and flow mixed pDNA:CD-PEI:HA-Ad complexes in HeLa cells with bPEI as a control. Incubation for 4 h in 10% serum-supplemented medium was followed by flow cytometry analysis at 24 h.
The presence of uncomplexed polycations in bulk mixed complexes has been shown to be the reason for their higher in vitro cellular uptake and transfection efficiency.29 These free polycations are also responsible for increased in vitro cytotoxicity.30 Since no free CD-PEI was detected in our transfection complexes, the lower transfection efficiency and improved cell viability of the flow mixed complexes may arise from the absence of similarly uncomplexed polycations.
While a significant amount of research has been carried out in the development of therapeutic genes and non-viral carriers, the absence of a robust operator-independent technique for the production of nucleic acid complexes is still needed. Unpackaging and escape of nucleic acids from transfection complexes that have been internalized is regarded as a crucial barrier to realizing more efficient gene delivery. Traditionally, transfection complex disassembly has relied on chemical approaches, such as pH-31 or enzyme-32responsive linkages, or physical approaches such as temperature cycling.33 Herein, we have demonstrated control over decomplexation properties based solely on the formulation parameter of mixing rate without making any changes to the carrier materials themselves.
Conclusions
While control over diameter and PDI has been demonstrated by use of microfluidics-assisted confinement, to our knowledge there exists no technique that has shown flow rate-dependent control over formulation of nucleic acid transfection complexes. The high reproducibility of this technique allows for more rigorous analysis of the dependence of transfection efficiency on particle diameter, PDI, ζ-potential and disassembly rate.
Supplementary Material
Acknowledgements
We would like to express our special thanks for the support of this work by NIH grant GM087016 and the Purdue Department of Chemistry. 1H NMR data were acquired in the Purdue Interdepartmental NMR Facility supported by NCI CCSG CA23168 to the Purdue University Center for Cancer Research. We would also like to thank Rob Reason for help with the illustrations.
Footnotes
Electronic Supplementary Information (ESI) available: synthesis procedures, 1H NMR characterization of compounds, gel shift assay, and flow cytometry raw data. See DOI: 10.1039/b000000x/
Notes and references
- 1.Mancuso K, Hauswirth WW, Li Q, Connor TB, Kuchenbecker JA, Mauck MC, Neitz J, Neitz M. Nature. 2009;461:784. doi: 10.1038/nature08401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waehler R, Russell SJ, Curiel DT. Nature Rev. Genet. 2007;8:573. doi: 10.1038/nrg2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehead KA, Langer R, Anderson DG. Nature Rev. Drug. Disc. 2009;8:129. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano T, Okuda T, Aoyagi H, Niidome T. J. Control. Rel. 2004;99:329. doi: 10.1016/j.jconrel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DWY, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ. Nature Biotech. 2010;20:172. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 6.Yu HJ, Wagner E. Curr. Op. Mol. Ther. 2009;11:165. [PubMed] [Google Scholar]
- 7.Vachutinsky Y, Kataoka K. Isr. J. Chem. 2010;50:175–184. [Google Scholar]
- 8.Zhang J, Lynn DM. Macromolecules. 2006;39:8928. doi: 10.1021/ma061815g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis ME, Zuckerman JE, Choi CHJ, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Nature. 2010;464:1067. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma VK, Thomas M, Klibanov AM. Biotechnol. Bioeng. 2005;90:614. doi: 10.1002/bit.20444. [DOI] [PubMed] [Google Scholar]
- 11.Xu Y, Hui SW, Frederik P, Szoka FC. Biophys. J. 1995;77:341. doi: 10.1016/S0006-3495(99)76894-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar V, Adamson DH, Prud’homme RK. Small. 2010;6:2907. doi: 10.1002/smll.201001199. [DOI] [PubMed] [Google Scholar]
- 13.Gindy ME, Ji S, Hoye TR, Panagiotopoulos AZ, Prud’homme RK. Biomacromol. 2008;9:2705. doi: 10.1021/bm8002013. [DOI] [PubMed] [Google Scholar]
- 14.Gindy ME, Prud’homme RK. Expert Opin. Drug Del. 2009;6:865. doi: 10.1517/17425240902932908. [DOI] [PubMed] [Google Scholar]
- 15.Kolishetti N, Dhar S, Valencia PM, Lin LQ, Karnik R, Lippard SJ, Langer R, Farokhzad OC. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17939. doi: 10.1073/pnas.1011368107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee M, Valencia PM, Rodriguez MI, Langer R, Farokhzad OC, Karnik R. Adv. Mater. 2011;23:H79. doi: 10.1002/adma.201004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valencia PM, Basto PA, Zhang L, Rhee M, Langer R, Farokhzad OC, Karnik R. ACS Nano. 2010;4:1671. doi: 10.1021/nn901433u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG. J. Am. Chem. Soc. 2012;134:6948. doi: 10.1021/ja301621z. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Liu K, Chen KJ, Lu Y, Wang S, Lin WY, Guo F, Kamei K, Chen YC, Ohashi M, Wang M, Garcia MA, Zhao XZ, Shen CK, Tseng HR. ACS Nano. 2010;10:6235. doi: 10.1021/nn101908e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhigaltsev IV, Belliveau N, Hafez I, Leung AK, Huft J, Hansen C, Cullis PR. Langmuir. 2012;28:3633. doi: 10.1021/la204833h. [DOI] [PubMed] [Google Scholar]
- 21.Leung AK, Hafez IM, Baoukina S, Belliveau NM, Zhigaltsev IV, Afshinmanesh E, Tieleman DP, Hansen CL, Hope MJ, Cullis PR. J. Phys. Chem. C. 2012;116:18440. doi: 10.1021/jp303267y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho Y-P, Grigsby CL, Zhao F, Leong KW. Nano Lett. 2011;11:2178. doi: 10.1021/nl200862n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulkarni A, DeFrees K, Hyun SH, Thompson DH. J. Am. Chem. Soc. 2012;134:7596. doi: 10.1021/ja300690j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kulkarni A, Deng W, Hyun SH, Thompson DH. Bioconj. Chem. 2012;23:933. doi: 10.1021/bc2005158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim EK, Kim HO, Jang E, Park J, Lee K, Suh JS, Huh YM, Haam S. Biomaterials. 2011;32:7941. doi: 10.1016/j.biomaterials.2011.06.077. [DOI] [PubMed] [Google Scholar]
- 26.Platt VM, Szoka FC. Mol. Pharm. 2008;4:474. doi: 10.1021/mp800024g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang W, Ng S-C. Nat. Protoc. 2008;3:691. doi: 10.1038/nprot.2008.37. [DOI] [PubMed] [Google Scholar]
- 28.Kulkarni A, Badwaik V, DeFrees K, Schuldt RA, Gunasekera DS, Powers C, Vlahu A, VerHeul R, Thompson DH. doi: 10.1021/bm401096v. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Z, Gjetting T, Mattebjerg MA, Wu C, Andresen TL. Biomaterials. 2011;32:8626. doi: 10.1016/j.biomaterials.2011.07.044. [DOI] [PubMed] [Google Scholar]
- 30.Boeckle S, von Gersdorff K, van der Piepen S, Culmsee C, Wagner E, Ogris MJ. Gene Med. 2004;6:1102. doi: 10.1002/jgm.598. [DOI] [PubMed] [Google Scholar]
- 31.Boomer JA, Thompson DH, Sullivan S. Pharm. Res. 2002;19:1289. doi: 10.1023/a:1020342523694. [DOI] [PubMed] [Google Scholar]
- 32.Andresen TL, Thompson DH, Kaasgaard T. Mol. Membr. Biol. 2010;27:353. doi: 10.3109/09687688.2010.515950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Putnam D. Nat. Mater. 2006;5:439. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.