Abstract
Acinetobacter baumannii is a significant contributor to intensive care unit (ICU) mortality causing numerous types of infection in this susceptible ICU population, most notably ventilator-associated pneumonia. The substantial disease burden attributed to A. baumannii and the rapid acquisition of antibiotic resistance make this bacterium a serious health care threat. A. baumannii is equipped to tolerate the hostile host environment through modification of its metabolism and nutritional needs. Among these adaptations is the evolution of mechanisms to acquire nutrient metals that are sequestered by the host as a defense against infection. Although all bacteria require nutrient metals, there is diversity in the particular metal needs among species and within varying tissue types and bacterial lifecycles. A. baumannii is well-equipped with the metal homeostatic systems required for the colonization of a diverse array of tissues. Specifically, iron and zinc homeostasis is important for A. baumannii interactions with biotic surfaces and for growth within vertebrates. This review discusses what is currently known regarding the interaction of A. baumannii with vertebrate cells with a particular emphasis on the contributions of metal homeostasis systems. Overall, published research supports the utility of exploiting these systems as targets for the development of much-needed antimicrobials against this emerging infectious threat.
Keywords: Acinetobacter, pathogenesis, iron, zinc, adherence, persistence
Introduction
The genus Acinetobacter is comprised of a number of Gram-negative species that are ubiquitous in the environment. Notably, some of the species within this genus have emerged as opportunistic human pathogens. A. baumannii is the most notorious among Acinetobacter species for causing a wide range of hospital-acquired infections. This organism is particularly problematic in intensive care units (ICUs) and can cause urinary tract infections, wound and burn infections, sepsis, ventilator-associated pneumonia, meningitis, and osteomyelitis (Davis et al., 2005; Peleg et al., 2008; Doyle et al., 2011). There have been numerous reports of A. baumannii wound infections in military personnel returning from Iraq and Afghanistan (Whitman, 2007; Calhoun et al., 2008; Sebeny et al., 2008). Moreover, A. baumannii causes community-acquired infections, predominately in Asia and tropic Australia (Falagas et al., 2007). A. baumannii infection is facilitated through biofilm formation on indwelling medical devices and other hospital surfaces and the ability to withstand desiccation and disinfection (Vidal et al., 1996; Wendt et al., 1997; Neely, 2000; Tomaras et al., 2003; Kawamura-Sato et al., 2010; Pour et al., 2011). In addition to the wide repertoire of diseases caused by A. baumannii, this organism has acquired antibiotic resistance at alarming rates. In fact, multi-drug resistance has become commonplace, and pan-drug resistant strains are increasingly identified. This is reflected in part by the fact that A. baumannii displays significant genomic plasticity and intraspecies heterogeneity (Fournier et al., 2006; Vallenet et al., 2008; Adams et al., 2010; Sahl et al., 2011, 2013). These facts highlight the necessity to develop new antimicrobials against A. baumannii.
A. baumannii can adapt to the hostile host environment through modification of its metabolism and nutritional needs. Like all organisms, A. baumannii requires nutrient metals to survive. Generally these essential metals include iron, zinc, manganese, copper, magnesium, and nickel, which serve as co-factors for a variety of fundamental cellular processes. Furthermore, A. baumannii displays diverse tissue tropism and has evolved mechanisms to acquire nutrient metals in various niches within the host. Although the lifecycle of A. baumannii in these tissues is not well-defined, several bacterial factors have been identified that are required for host cell interactions during infection. This review discusses A. baumannii interactions with host cells with a particular emphasis on the importance of nutrient metal homeostasis to the pathogenesis of A. baumannii infection.
Nutritional immunity
In order to adapt to different host niches, A. baumannii must possess metabolic flexibility and employ specialized systems for nutrient acquisition and homeostasis. Humans maintain extracellular free metals at low levels through several mechanisms including intracellular localization and the expression of metal-binding proteins. This provides protection against invading pathogens that must acquire metals from their hosts. Therefore, metal limitation is considered a host defense mechanism and has been termed nutritional immunity (Weinberg, 1975). Research into nutritional immunity has primarily focused on iron. Most iron in vertebrates is stored within heme, the oxygen-carrying cofactor of the protein hemoglobin, which is stored within erythrocytes. In addition to hemoglobin, the host has other iron transport and storage proteins that reduce iron availability in the vertebrate host, including myoglobin, transferrin, lactoferrin, hemopexin, and ferritin.
Vertebrates also limit non-iron metals to defend against infection. Calprotectin (CP) is a heterodimer of the two S100 proteins, S100A8 and S100A9, and exhibits high affinity binding for zinc and manganese. Importantly, CP displays antimicrobial activity against several pathogens, including A. baumannii, via its metal-sequestering properties (Corbin et al., 2008; Urban et al., 2009; McCormick et al., 2010; Bianchi et al., 2011; Damo et al., 2013). CP is frequently identified at sites of inflammation, including in the lungs during A. baumannii pneumonia where CP expression tracks with the progression and resolution of infection (Hood et al., 2012; Moore et al., 2013). In addition to CP, other S100 proteins have antimicrobial activity and the ability to sequester metals including S100A7 (psoriacin), S100A12, and S100A15 (Glaser et al., 2005; Buchau et al., 2007; Lee and Eckert, 2007; Michalek et al., 2009; Pietzsch and Hoppmann, 2009). Other than CP, these extracellular metal-binding proteins have yet to be investigated for their role in protection against A. baumannii infections.
Finally, in order to limit metal availability to invading pathogens, vertebrate cells efflux iron and manganese out of the phagosomal compartment into the cytoplasm via NRAMP1 (Jabado et al., 2000; Peracino et al., 2006; Cellier et al., 2007). Phagosomal zinc levels are decreased via ZIP8-mediated transport into the cytoplasm (Begum et al., 2002; Aydemir et al., 2009). Cytoplasmic zinc levels are then reduced via cellular extrusion by ZnT transporters (Kitamura et al., 2006; Murakami and Hirano, 2008). Vertebrate systems such as these are important to protect against intracellular bacterial growth but have yet to be studied in the context of A. baumannii infection.
Iron and A. baumannii
In response to low iron availability within the host, both Gram-negative and Gram-positive bacteria employ the iron-dependent repressor ferric uptake regulator (Fur), which regulates gene expression through binding of a conserved Fur box DNA sequence upstream of target genes. A. baumannii Fur has been identified and is expressed within the A. baumannii strain BM2580 and has 63% identity to the Escherichia coli Fur (Daniel et al., 1999). Furthermore, Fur boxes have been identified within the genomes of A. baumannii strains ATCC 17978 and ATCC 19606T (Mihara et al., 2004; Eijkelkamp et al., 2011a). A. baumannii responds to iron starvation by modifying gene expression for many predicted iron-related genes, as well as for genes involved in various processes such as respiration, biofilm formation, and motility, highlighting the importance of iron levels to A. baumannii virulence (Eijkelkamp et al., 2011a; Nwugo et al., 2011). This observation is consistent with the function of Fur in other bacterial species where Fur has been reported to regulate similar processes including genes required for virulence (Troxell and Hassan, 2013). Although not yet demonstrated for A. baumannii, in several organisms Zur is required for pathogenesis in in vivo models of infection, including Staphylococcus aureus, Vibrio cholera, Salmonella enterica serovar Typhimurium, and Helicobacter pylori (Bury-Mone et al., 2004; Mey et al., 2005; Gancz et al., 2006; Velayudhan et al., 2007; Curtiss et al., 2009; Torres et al., 2010; Troxell et al., 2011). Numerous studies have investigated the response of various A. baumannii isolates to iron-limiting conditions and demonstrated changes in outer membrane protein composition and/or secretion of iron-chelating siderophores, which are discussed further below (Echenique et al., 1992; Actis et al., 1993; Yamamoto et al., 1994; Goel et al., 1998; Daniel et al., 1999; Dorsey et al., 2003a). Interestingly, among A. baumannii strains, there is significant diversity in the numbers and types of these iron uptake and utilization systems that are expressed (Yamamoto et al., 1994; Dorsey et al., 2003a; Zimbler et al., 2009; Antunes et al., 2011).
There are several generally conserved Gram-negative mechanisms for capturing iron or iron-protein complexes through specific receptors that are utilized by A. baumannii. Many bacterial pathogens lyse erythrocytes in order to initiate the coordinated process of freeing heme to extract the bound iron as a source of this essential metal. A. baumannii encodes a phospholipase C and other hemolysin-related genes that can lyse horse erythrocytes (Vallenet et al., 2008; Camarena et al., 2010; Antunes et al., 2011). Following erythrocyte lysis, bacteria can then capture iron-bound heme via specialized heme uptake systems (Runyen-Janecky, 2013). Among A. baumannii sequenced strains, there are at least two identified gene clusters encoding putative heme uptake systems, one of which is highly conserved among the sequenced strains (Zimbler et al., 2009; Antunes et al., 2011). The process of iron extraction from heme has not yet been described for A. baumannii.
Bacteria can also acquire iron from other host iron-binding proteins through the use of secreted siderophores. A. baumannii encodes several iron-scavenging siderophores, and at least five gene clusters for siderophore synthesis and transport have been discovered among the sequenced strains. The process of encoding more than one siderophore is not unique to A. baumannii, as several other organisms encode more than one siderophore including E. coli, Pseudomonas aeruginosa, Yersinia pestis, Mycobacterium tuberculosis, and S. aureus (Hammer and Skaar, 2011; Chaturvedi et al., 2012; Rakin et al., 2012; Li et al., 2013; Saha et al., 2013). The first identified A. baumannii siderophore cluster, comprised of 10 ORFs, was discovered in strain 8399 and several other clinical isolates from Oregon (Echenique et al., 1992; Dorsey et al., 2003b). This 8399 cluster is responsible for the production of a catechol siderophore that chelates oxidized ferric iron from transferrin and restores A. baumannii growth when iron is limiting. Other bacteria that produce catechol siderophores include Bacillus subtilis, Streptomyces, S. aureus, E. coli, V. cholera, Y. pestis, and P. aeruginosa (May et al., 2001; Chu et al., 2010; Saha et al., 2013). Specifically, the genes in the cluster encode proteins with similarity to B. subtilis DhbB, DhbE, DhbC, DhbA, DhbF, that are related to the Ent proteins required for enterobactin synthesis in E. coli (Rowland et al., 1996; Dorsey et al., 2003b). This cluster also encodes proteins with similarity to the Y. pestis putative enterobactin biosynthesis protein EntD, a Y. pestis iron-regulated efflux protein P114, E. coli siderophore efflux protein EntS, and the E. coli enterobactin degradation protein Fes (Armstrong et al., 1989; Coderre and Earhart, 1989; Brickman and McIntosh, 1992; Parkhill et al., 2001; Furrer et al., 2002). Finally, an ORF encoding a 73 kDa outer membrane protein OM73 was identified in the cluster and has homology to the E. coli CirA colicin receptor protein (Nau and Konisky, 1989). OM73 is surface-exposed, Fur- and iron-regulated, and contains a TonB box, which is consistent with described siderophore receptors (Dorsey et al., 2003b).
The most-studied A. baumannii siderophore gene cluster encodes the siderophore acinetobactin, which is found in numerous clinical isolates and in all sequenced genomes except A. baumannii SDF (Yamamoto et al., 1994; Dorsey et al., 2004; Mihara et al., 2004). The structure of acinetobactin is composed of equimolar amounts of 2,3-dihydrobenzoic acid (DHBA), threonine, and N-hydroxyhistamine (Yamamoto et al., 1994). Hydroxamate or mixed-type siderophores are also found in a wide array of bacterial species (Chu et al., 2010; Saha et al., 2013). In fact, acinetobactin is structurally related to the Vibrio anguillarum siderophore anguibactin, and acinetobactin only differs by the presence of an oxazoline ring instead of thiazoline ring (Yamamoto et al., 1994). Acinetobactin can functionally replace anguibactin for iron acquisition in a V. anguillarum mutant that cannot produce anguibactin (Dorsey et al., 2004). In addition to being regulated by iron levels, the acinetobactin gene cluster is regulated by Fur, and Fur boxes have been identified within the cluster (Mihara et al., 2004). Homology analyses identified three putative systems encoded within the acinetobactin cluster: basABCDEFGHIJ for A. baumannii acinetobactin synthesis, bauABCDEF for A. baumannii acinetobactin utilization, and barAB for A. baumannii acinetobactin release (Mihara et al., 2004). Of note, basF and basJ genes in the Fur-regulated cluster are homologs of the E. coli entC and entB, which are required for the production of DHBA; however, uniquely, A. baumannii encodes the entA homolog elsewhere in the genome (Mihara et al., 2004; Penwell et al., 2012). Although entA is found outside of the acinetobactin cluster in all strains investigated, the genomic context surrounding the entA gene differs by strain, highlighting the variability, and plasticity of the A. baumannii genome. EntA in A. baumannii ATCC 19606T is required for production of DHBA and for iron acquisition (Penwell et al., 2012). Analysis of a basD mutant demonstrated that no acinetobactin is produced and these mutants have a growth defect in iron-limiting conditions, consistent with the predicted function in acinetobactin synthesis (Dorsey et al., 2004; Mihara et al., 2004). The putative function of the bauABCDEF system in acinetobactin uptake was demonstrated through the use of bauA and bauD mutants, which can produce acinetobactin but cannot grow in conditions where ferric iron or transferrin-bound iron are the sole iron source (Dorsey et al., 2004; Mihara et al., 2004). Moreover, BauA has homology to FatA, which is the ferric-anguibactin outer membrane receptor belonging to the family of TonB-dependent receptors (described below), and BauA can be recognized by anti-FatA antibodies (Dorsey et al., 2004).
The third described siderophore biosynthesis cluster encodes a system that produces six siderophores termed fimsbactins A-F found in A. baumannii ATCC 17978 and ADP1 (Proschak et al., 2013). A fourth siderophore gene cluster (ACICU1672-1683) is conserved among the sequenced strains, except SDF; however, this cluster has yet to be empirically studied (Antunes et al., 2011; Eijkelkamp et al., 2011a). The final identified siderophore gene cluster (ABAYE1888-1889) contains two genes encoding an isochorismatase and a 2,3-dihydro-2,3-hydroxybenzoate dehydrogenase (Eijkelkamp et al., 2011a). These enzymes produce 2,3-dihydroxybenzoate, which is an iron-binding molecule but also a precursor for more complex siderophores. This fifth cluster is not conserved among all strains and remains to be experimentally assessed (Eijkelkamp et al., 2011a). Elucidation of the functions of the other siderophore gene clusters will provide insight into why A. baumannii exhibits such significant diversity in its iron acquisition systems.
The ability of a siderophore-iron complex to be transported into Gram-negative bacterial cells is reliant on an outer membrane TonB-dependent receptor, named such for its dependence on a TonB/ExbB/ExbD energy-transducing system. Many bacteria encode multiple TonB proteins required under different conditions and/or for varying purposes extending beyond siderophore transport to include the transport of heme, maltose, vitamin B12, and nickel (Lewis et al., 1997; Chimento et al., 2003; Neugebauer et al., 2005; Schauer et al., 2007; Krewulak and Vogel, 2011). In fact, A. baumannii strains contain from 8 to 22 predicted TonB-dependent receptors in the genome. The TonB-dependent receptor BauA is involved in acinetobactin transport, but the functions of the remaining TonB-dependent receptors are not known. Three TonB systems are conserved in all sequenced A. baumannii genomes, and recent work demonstrates that the genes encoding each of these systems were likely horizontally acquired from distinct sources (Zimbler et al., 2013). These three systems in A. baumannii ATCC 19606T are tonB1/exbB1/exbD1.1/exbD1.2, tonB2, and tonB3/exbB3/exbD3. The expression and function of each A. baumannii TonB system is variable. Of these systems, only tonB3 is up-regulated under iron-limiting conditions, indicating a link to iron homeostasis. Additionally, only TonB2 and TonB3 can functionally complement the iron-limited growth defect of an E. coli tonB mutant. Interestingly, both ExbB1/ExbD1.1/ExbD1.2 and ExbB3/ExbD3 are required for complementation of E. coli exbBD tolQR mutant growth. Within A. baumannii, tonB1, tonB2, and tonB1 tonB2 mutants are deficient for growth in iron-starved conditions as compared to wild-type and are less efficient at acinetobactin and iron transport. A tonB3 mutant has not been created, which may indicate that this gene is required for growth (Zimbler et al., 2013).
In certain ecological niches, including the intracellular compartment, bacteria may be exposed to reduced ferrous iron as an iron source. Therefore, many bacteria encode ferrous iron uptake systems, most notably FeoAB transporters, which are required for iron acquisition and pathogenesis of numerous bacteria including E. coli, Shigella flexneri, H. pylori, and Legionella pneumophila (Stojiljkovic et al., 1993; Velayudhan et al., 2000; Robey and Cianciotto, 2002; Runyen-Janecky et al., 2003; Cartron et al., 2006). A. baumannii encodes putative ferrous iron import systems, FeoAB with its regulator FeoC, and at least one FeoB has been identified in all sequenced strains along with a FeoA and FeoC (Antunes et al., 2011). Finally, once in the bacterial cell, iron must be distributed to the cytosolic iron pool or incorporated into proteins requiring iron cofactors. One common class of iron metallocenter found in a variety of proteins are iron-sulfur clusters, whose formation is dependent on a metallocenter assembly scaffold of three main types: NIF, ISC, and SUF (Bandyopadhyay et al., 2008a). The Nfu proteins are a class of scaffold proteins outside of the three main iron-sulfur cluster types and have been described in eukaryotes, E. coli, and Azotobacter vinelandii (Angelini et al., 2008; Bandyopadhyay et al., 2008a,b; Py et al., 2012). A. baumannii NfuA is a cytoplasmic protein that binds iron and is predicted to function in iron-sulfur (Fe-S) cluster formation (Zimbler et al., 2012). Consistent with this function, NfuA is required for growth in low iron and in conditions of oxidative stress (Zimbler et al., 2012).
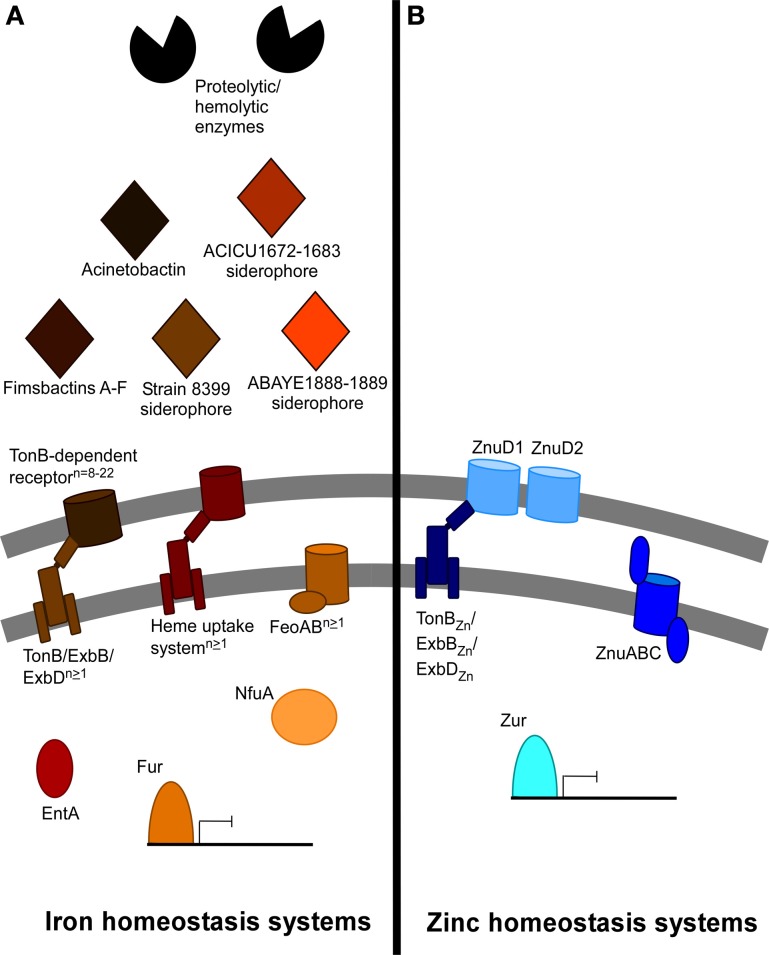
At least several of the above iron acquisition and metabolism systems are required for A. baumannii infection. Acinetobactin synthesis and uptake proteins BasD and BauA, respectively, are required for A. baumannii virulence in a Galleria mellonella larvae infection model and in a mouse model of systemic infection (Gaddy et al., 2012). Consistent with this, the DHBA production protein EntA and the Fe-S cluster protein NfuA are also required for virulence in the G. mellonella infection model (Penwell et al., 2012; Zimbler et al., 2012). Finally, TonB1 and TonB2 together but not individually are required for full virulence of A. baumannii ATCC 19606T in the G. mellonella infection model (Zimbler et al., 2013). It has been reported that clinical A. baumannii strains display an enhanced ability to resist iron starvation when compared to a non-human isolate (Antunes et al., 2011). Together these results support the importance of iron homeostasis to the success of A. baumannii in the host. A summary of identified and putative iron homeostasis systems in A. baumannii is depicted in Figure 1A.
Figure 1.
Summary of predicted and described iron and non-iron metal acquisition systems in A. baumannii. The iron and non-iron systems encoded within the A. baumannii genome vary dramatically between different strains. (A) This panel depicts the iron acquisition and utilization systems identified or described among the different A. baumannii strains. Secretion of enzymes such as phospholipase C and others may contribute to hemolysis and hemoglobin release from red blood cells. At least five clusters of genes for siderophore synthesis and secretion are dispersed among different strains. These include acinetobactin, fimsbactins A-F, the strain 8399 (om73-entD) siderophore, as well as siderophores produced by the ACICU1672-1683 and the ABAYE1888-1899 clusters. EntA is required for iron acquisition due to its involvement in the biosynthesis of the acinetobactin precursor 2,3-dihydroxybenzoic acid. Siderophores are recognized by TonB-dependent receptors, of which 8–22 have been discovered among the sequenced strains. At least one of multiple encoded TonB/ExbB/ExbD systems generates and provides energy to the TonB-dependent receptors, and at least one FeoAB ferrous iron transport system has been identified in all sequenced strains. Finally, 1–2 heme uptake systems have been identified among A. baumannii strains. NfuA is a Fe-S cluster protein required for iron utilization in the cytoplasm. Regulation of these numerous systems likely depends on the conserved Fur repressor. (B) This panel depicts the zinc acquisition and metabolism systems described in A. baumannii ATCC 17978. Outer membrane transport of zinc may occur via two zinc-regulated TonB-dependent receptors, energized by a zinc-regulated TonB/ExbB/ExbD system. Inner membrane transport is mediated by the ZnuABC transporter. Zur is a conserved repressor that controls the expression of znuABC and likely the other identified zinc systems.
Non-iron metals and A. baumannii
Non-iron metal homeostasis in A. baumannii is less well-understood; however, systems for zinc uptake and utilization have been investigated. A. baumannii encodes a conserved inner membrane ABC zinc transporter called ZnuABC. ZnuABC systems have been identified in numerous bacterial species such as Campylobacter jejuni, Salmonella, Neisseria meningitidis, E. coli, and Y. pestis and in some cases demonstrated to be required for virulence (Patzer and Hantke, 1998; Campoy et al., 2002; Ammendola et al., 2007; Davis et al., 2009; Stork et al., 2010). The A. baumannii ZnuABC transporter is up-regulated when A. baumannii is starved for zinc and in murine lungs during A. baumannii pneumonia (Hood et al., 2012). ZnuB, the inner membrane permease, is required for growth in zinc-limiting conditions and for bacterial growth in the mouse pneumonia model (Hood et al., 2012). A. baumannii also encodes the Fur family zinc uptake regulator Zur, a zinc-sensing repressor that recognizes a conserved Zur box DNA sequence upstream of target genes when zinc-bound. A putative list of Zur target genes has been identified and include candidate outer membrane zinc transporters ZnuD1 and ZnuD2, a TonB/ExbB/ExbD system, and genes involved in intracellular zinc homeostasis (Hood et al., 2012). These findings are consistent with Zur regulons in other bacteria such as M. tuberculosis, N. meningitidis, Corynebacterium glutamicum, and Y. pestis (Maciag et al., 2007; Li et al., 2009; Schroder et al., 2010; Pawlik et al., 2012). Furthermore, the expression of znuA, znuB, znuC, znuD1, znuD2, and tonB increases when zinc is limiting. The putative outer membrane transporters ZnuD1 and ZnuD2 have homology to the ZnuD described in N. meningitidis, which was demonstrated to be involved in both zinc and heme acquisition (Stork et al., 2010; Kumar et al., 2012). Although TonB-dependent receptors have been shown to be involved in zinc acquisition, no TonB system has yet been demonstrated to be directly involved in zinc acquisition through the transfer of energy to TonB-dependent receptors. Additional work is required to clarify functions for these proteins and their role in the varying in vivo niches of A. baumannii. Finally, further studies are needed to identify and characterize bacterial systems for import and utilization of other non-iron metals. For example, studies investigating the role of the host protein CP in the sequestration of zinc and manganese from bacterial pathogens demonstrate a requirement of manganese for A. baumannii growth (Damo et al., 2013). In S. aureus, CP-mediated manganese sequestration inhibits manganese-dependent superoxide defenses, and it is possible that similar inhibition is occurring in A. baumannii (Kehl-Fie et al., 2011). A summary of identified and putative zinc homeostasis systems within A. baumannii is depicted in Figure 1B.
Adherence of A. baumannii to host cells
During infection, A. baumannii colonizes several niches and therefore interacts with and likely adheres to numerous cell types. A. baumannii has been shown to adhere to various cell types including human lung, laryngeal, nasopharyngeal, and cervical epithelial cells, Candida albicans filaments, and Acanthamoeba castellanii amoebal cells (Lee et al., 2006; Choi et al., 2008; Gaddy et al., 2009; Eijkelkamp et al., 2011b; Tamang et al., 2011). Furthermore, different strains of A. baumannii display varying capacities for cell adherence, and not all strains encode the same attachment machinery (Lee et al., 2006, 2008; de Breij et al., 2010; Eijkelkamp et al., 2011b). A. baumannii associates with epithelial cells by means of fimbria on the bacterial cell surface; however, additional bacterial proteins have been implicated in host epithelial cell attachment (Lee et al., 2006, 2008). A. baumannii outer membrane protein A (OmpA) is required for adherence to C. albicans filaments and epithelial cells (Choi et al., 2008; Gaddy et al., 2009). Notably, OmpA is enriched in bacterial membranes when iron is replete (Nwugo et al., 2011). Thus, it is possible that fluctuations in iron availability within host niches regulate the expression of OmpA, and possibly other OMPs, which promotes binding to host cells within a diverse array of tissue types. Furthermore, the ability of OmpA to mediate host cell binding is due at least in part to interactions with fibronectin (Smani et al., 2012b). Fibronectin also interacts with A. baumannii EF-Tu, Omp33, an unnamed TonB-dependent copper receptor, and a 34 kDa outer membrane protein (Gaddy et al., 2009; Dallo et al., 2012; Smani et al., 2012b, 2013). Interestingly, TonB2 a protein important for A. baumannii iron homeostasis, is also required for A. baumannii binding to fibronectin and epithelial cells (Zimbler et al., 2013). Beyond fibronectin, the autotransporter protein Ata mediates adhesion to Type IV collagen, and outer membrane protein ChoP interacts with the host protein platelet activating factor receptor (PFAR) (Bentancor et al., 2012; Smani et al., 2012a). It is possible that iron availability regulates outer membrane protein levels, such as OmpA, to levels that would promote binding to host cells. Overall, the number of factors interacting with host cells suggests that A. baumannii employs a multi-faceted strategy to host cell adherence that likely facilitates its capacity to thrive in a diverse array of tissue types.
A. baumannii invasion of host cells
Following attachment, A. baumannii can invade host cells. Susceptible cell types include human lung, laryngeal, and cervical epithelial cells, and A. castellanii cells, yet sensitivity to invasion varies by cell type (Choi et al., 2008; Gaddy et al., 2009; Tamang et al., 2011). Likewise, different strains of A. baumannii have varying invasive potential (Choi et al., 2008). A. baumannii enters epithelial cells by way of a microfilament- and microtubule-dependent, zipper-like mechanism and upon internalization, localizes to membrane-bound vacuoles (Choi et al., 2008). Clathrin and β-arrestins are also engaged during the uptake of A. baumannii into lung epithelial cells (Smani et al., 2012a). A. baumannii can then persist within host cells; however, no intracellular replication has been reported. Bacterial proteins that are required for invasion and intracellular persistence include OmpA, Omp33, blue-light-sensing A (BlsA), and phospholipase D (Choi et al., 2008; Gaddy et al., 2009; Jacobs et al., 2010; Mussi et al., 2010; Smani et al., 2013). Several iron-related genes are also required for intracellular survival. Acinetobactin synthesis and uptake proteins BasD and BauA are expressed by intracellular A. baumannii and are required for survival within epithelial cells (Gaddy et al., 2012). Likewise, EntA and NfuA are both necessary for A. baumannii intracellular persistence (Penwell et al., 2012; Zimbler et al., 2012). The requirement for NfuA may be due to its role in the alleviation of oxidative damage or indirectly through the formation of Fe-S clusters on NfuA targets that are required for intracellular persistence (Zimbler et al., 2012). Together these results demonstrate the importance of iron homeostasis for A. baumannii intracellular survival and support the idea that the intracellular compartment is an iron-starved environment.
Physiological relevance of A. baumannii intracellular persistence
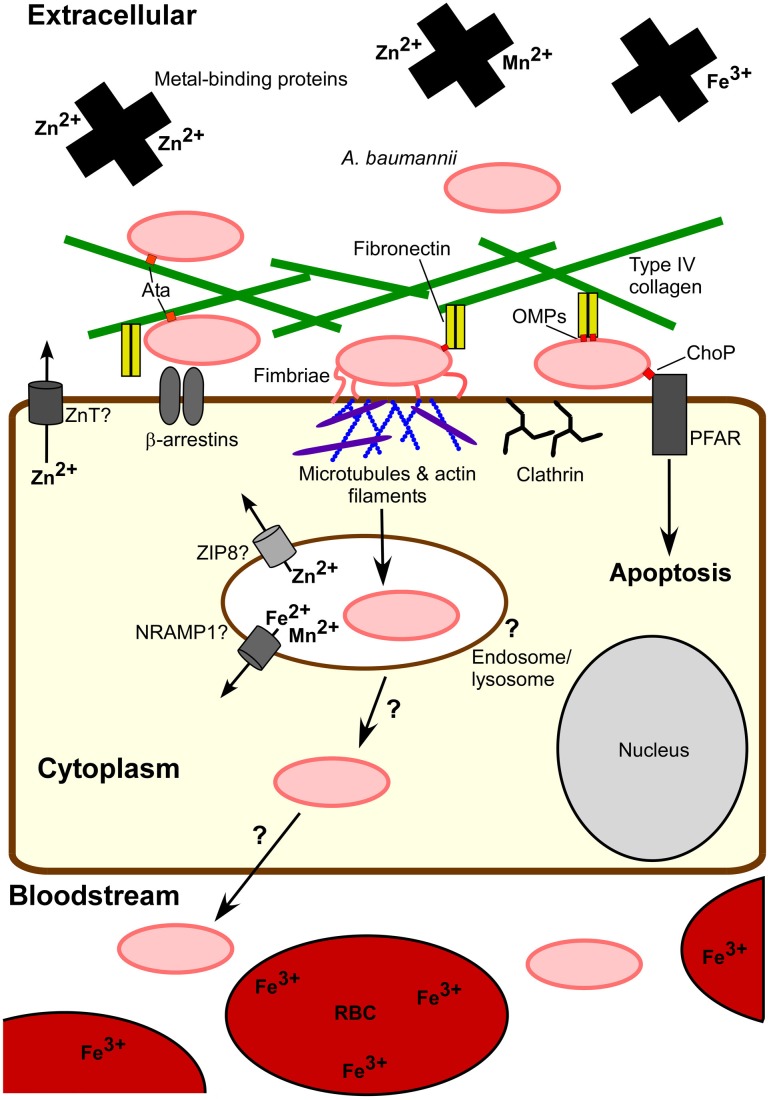
A better understanding of both bacterial and host factors involved in invasion and intracellular persistence will help to elucidate the role of these processes during infection. The fact that A. baumannii can be killed by macrophages and other phagocytic cells, and the lack of evidence for intracellular replication or long term intracellular survival direct us away from the designation of A. baumannii as a predominately intracellular pathogen. Nonetheless, the ability to invade and persist within eukaryotic cells advocates that A. baumannii's intracellular phase is physiologically relevant. First, A. baumannii entry into host cells may promote invasive disease. Although A. baumannii can survive within host cells, contact with cells triggers an apoptotic cell death, which is mediated at least in part by PARP and the iron-regulated protein OmpA (Choi et al., 2005; Gaddy et al., 2009; Smani et al., 2012a). Apoptosis may facilitate A. baumannii passage through the cell layer by eliminating the cellular barrier. One possibility is that intracellular passage may provide access to the basal side of cells through which A. baumannii can transit across to the underlying tissue. Additionally, A. baumannii stimulates a robust innate immune response; thus, relocating within host cells may afford A. baumannii a means to evade host immune attack. Figure 2 illustrates known aspects of A. baumannii-host cell interactions and highlights current questions regarding the intracellular lifecycle of A. baumannii with particular focus on the interplay between host and bacterial metal homeostatic mechanisms.
Figure 2.
A. baumannii-host cell interactions. Extracellular A. baumannii experiences a metal-limited environment at least in part due to metal-binding host proteins and intracellular localization of metals, e.g., red blood cells (RBCs). A. baumannii attaches to host cells through interactions between fibronectin and bacterial outer membrane proteins (OMPs), such as OmpA, as well as Type IV collagen via Ata. These OMPs may bind to other host cell proteins, for example the interaction of OMP ChoP with platelet activating factor receptor (PFAR). Fimbriae also contribute to eukaryotic cell adherence. Invasion into epithelial cells occurs via host cell actin filament and microtubule reorganization, recruitment of β-arrestin and clathrin, and a bacterial uptake via a zipper-like mechanism. Although A. baumannii can persist intracellularly and includes entry into endosomes, the intracellular trafficking of A. baumannii is not clear. Eukaryotic cell proteins NRAMP1, ZIP8, ZnTs, and other transporters pump essential metals out of endocytic vesicles and the cytoplasm, limiting the intracellular pool available to pathogens. The physiological relevance of the intracellular lifecycle of A. baumannii remains to be understood. One consequence of host cell exposure to A. baumannii is apoptosis. It is possible that cell death and/or host cell invasion serve to permit A. baumannii dissemination to deeper tissues leading to invasive disease.
Conclusions
Many vertebrates sequester nutrient metals as a defense mechanism against invading pathogens through the use of several transport and storage proteins. In order to obtain metals in the face of host metal-limiting strategies A. baumannii employs specialized systems that are required for growth in numerous models of A. baumannii infection. Future work is required to define the specific bacterial strategies for maintenance of metal homeostasis in the different host niches that A. baumannii occupies. During A. baumannii infection, the host rapidly deploys phagocytic cells and other effectors to clear infection. While A. baumannii has not been observed to persist within these phagocytic cells, A. baumannii does bind to fibronectin and epithelial cells, permitting bacterial invasion and intracellular persistence. A predominately intracellular lifecycle for A. baumannii is not supported by current studies, but future research will clarify the role of A. baumannii host cell invasion and intracellular persistence. Importantly, elucidation of A. baumannii mechanisms for metal transport and maintenance and their contributions to A. baumannii success within the various infection sites may reveal targets for the generation of new antimicrobials against this widely antibiotic-resistant organism.
Authors contributions
Brittany L. Mortensen reviewed the literature and contributed to writing and revising this manuscript. Eric P. Skaar contributed to writing and revising this manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the members of the Skaar lab for their editorial comments on the manuscript. Work in the Skaar lab is supported by grant numbers AI091771, AI069233, and AF073843 from the National Institutes of Health (NIH). Eric P. Skaar is a Burroughs Wellcome Fund Investigator in the Pathogenesis of Infectious Diseases. Brittany L. Mortensen is supported by grant number F32-AI108192 from the National Institute of Allergy and Infectious Disease (NIAID). The content of this article does not necessarily represent the views of the NIH or NIAID and is solely the responsibility of the authors.
References
- Actis L. A., Tolmasky M. E., Crosa L. M., Crosa J. H. (1993). Effect of iron-limiting conditions on growth of clinical isolates of Acinetobacter baumannii. J. Clin. Microbiol. 31, 2812–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. D., Chan E. R., Molyneaux N. D., Bonomo R. A. (2010). Genomewide analysis of divergence of antibiotic resistance determinants in closely related isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 54, 3569–3577 10.1128/AAC.00057-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammendola S., Pasquali P., Pistoia C., Petrucci P., Petrarca P., Rotillio G., et al. (2007). High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infect. Immun. 75, 5867–5876 10.1128/IAI.00559-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S., Gerez C., Ollagnier-de Choudens S., Sanakis Y., Fontecave M., Barras F., et al. (2008). NfuA, a new factor required for maturing Fe/S proteins in Escherichia coli under oxidative stress and iron starvation conditions. J. Biol. Chem. 283, 14084–14091 10.1074/jbc.M709405200 [DOI] [PubMed] [Google Scholar]
- Antunes L. C., Imperi F., Towner K. J., Visca P. (2011). Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 162, 279–284 10.1016/j.resmic.2010.10.010 [DOI] [PubMed] [Google Scholar]
- Armstrong S. K., Pettis G. S., Forrester L. J., McIntosh M. A. (1989). The Escherichia coli enterobactin biosynthesis gene, entD: nucleotide sequence and membrane localization of its protein product. Mol. Microbiol. 3, 757–766 10.1111/j.1365-2958.1989.tb00224.x [DOI] [PubMed] [Google Scholar]
- Aydemir T. B., Liuzzi J. P., McClellan S., Cousins R. J. (2009). Zinc transporter ZIP8 (SLC39A8) and zinc influence IFN-gamma expression in activated human T cells. J. Leukoc. Biol. 86, 337–348 10.1189/jlb.1208759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Chandramouli K., Johnson M. K. (2008a). Iron-sulfur cluster biosynthesis. Biochem. Soc. Trans. 36, 1112–1119 10.1042/BST0361112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S., Naik S. G., O'Carroll I. P., Huynh B. H., Dean D. R., Johnson M. K., et al. (2008b). A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 283, 14092–14099 10.1074/jbc.M709161200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum N. A., Kobayashi M., Moriwaki Y., Matsumoto M., Toyoshima K., Seya T. (2002). Mycobacterium bovis BCG cell wall and lipopolysaccharide induce a novel gene, BIGM103, encoding a 7-TM protein: identification of a new protein family having Zn-transporter and Zn-metalloprotease signatures. Genomics 80, 630–645 10.1006/geno.2002.7000 [DOI] [PubMed] [Google Scholar]
- Bentancor L. V., Camacho-Peiro A., Bozkurt-Guzel C., Pier G. B., Maira-Litran T. (2012). Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 194, 3950–3960 10.1128/JB.06769-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M., Niemiec M. J., Siler U., Urban C. F., Reichenbach J. (2011). Restoration of anti-Aspergillus defense by neutrophil extracellular traps in human chronic granulomatous disease after gene therapy is calprotectin-dependent. J. Allergy Clin. Immunol. 127, 1243–1252 10.1016/j.jaci.2011.01.021 [DOI] [PubMed] [Google Scholar]
- Brickman T. J., McIntosh M. A. (1992). Overexpression and purification of ferric enterobactin esterase from Escherichia coli. Demonstration of enzymatic hydrolysis of enterobactin and its iron complex. J. Biol. Chem. 267, 12350–12355 [PubMed] [Google Scholar]
- Buchau A. S., Hassan M., Kukova G., Lewerenz V., Kellermann S., Wurthner J. U., et al. (2007). S100A15, an antimicrobial protein of the skin: regulation by E. coli through Toll-like receptor 4. J. Invest. Dermatol. 127, 2596–2604 10.1038/sj.jid.5700946 [DOI] [PubMed] [Google Scholar]
- Bury-Mone S., Thiberge J. M., Contreras M., Maitournam A., Labigne A., de Reuse H. (2004). Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53, 623–638 10.1111/j.1365-2958.2004.04137.x [DOI] [PubMed] [Google Scholar]
- Calhoun J. H., Murray C. K., Manring M. M. (2008). Multidrug-resistant organisms in military wounds from Iraq and Afghanistan. Clin. Orthop. Relat. Res. 466, 1356–1362 10.1007/s11999-008-0212-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarena L., Bruno V., Euskirchen G., Poggio S., Snyder M. (2010). Molecular mechanisms of ethanol-induced pathogenesis revealed by RNA-sequencing. PLoS Pathog. 6:e1000834 10.1371/journal.ppat.1000834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoy S., Jara M., Busquets N., Perez de Rozas A. M., Badiola I., Barbe J. (2002). Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infect. Immun. 70, 4721–4725 10.1128/IAI.70.8.4721-4725.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartron M. L., Maddocks S., Gillingham P., Craven C. J., Andrews S. C. (2006). Feo–transport of ferrous iron into bacteria. Biometals 19, 143–157 10.1007/s10534-006-0003-2 [DOI] [PubMed] [Google Scholar]
- Cellier M. F., Courville P., Campion C. (2007). Nramp1 phagocyte intracellular metal withdrawal defense. Microbes Infect. 9, 1662–1670 10.1016/j.micinf.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Chaturvedi K. S., Hung C. S., Crowley J. R., Stapleton A. E., Henderson J. P. (2012). The siderophore yersiniabactin binds copper to protect pathogens during infection. Nat. Chem. Biol. 8, 731–736 10.1038/nchembio.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimento D. P., Kadner R. J., Wiener M. C. (2003). The Escherichia coli outer membrane cobalamin transporter BtuB: structural analysis of calcium and substrate binding, and identification of orthologous transporters by sequence/structure conservation. J. Mol. Biol. 332, 999–1014 10.1016/j.jmb.2003.07.005 [DOI] [PubMed] [Google Scholar]
- Choi C. H., Lee E. Y., Lee Y. C., Park T. I., Kim H. J., Hyun S. H., et al. (2005). Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 7, 1127–1138 10.1111/j.1462-5822.2005.00538.x [DOI] [PubMed] [Google Scholar]
- Choi C. H., Lee J. S., Lee Y. C., Park T. I., Lee J. C. (2008). Acinetobacter baumannii invades epithelial cells and outer membrane protein A mediates interactions with epithelial cells. BMC Microbiol. 8:216 10.1186/1471-2180-8-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu B. C., Garcia-Herrero A., Johanson T. H., Krewulak K. D., Lau C. K., Peacock R. S., et al. (2010). Siderophore uptake in bacteria and the battle for iron with the host; a bird's eye view. Biometals 23, 601–611 10.1007/s10534-010-9361-x [DOI] [PubMed] [Google Scholar]
- Coderre P. E., Earhart C. F. (1989). The entD gene of the Escherichia coli K12 enterobactin gene cluster. J. Gen. Microbiol. 135, 3043–3055 10.1099/00221287-135-11-3043 [DOI] [PubMed] [Google Scholar]
- Corbin B. D., Seeley E. H., Raab A., Feldmann J., Miller M. R., Torres V. J., et al. (2008). Metal chelation and inhibition of bacterial growth in tissue abscesses. Science 319, 962–965 10.1126/science.1152449 [DOI] [PubMed] [Google Scholar]
- Curtiss R., 3rd., Wanda S. Y., Gunn B. M., Zhang X., Tinge S. A., Ananthnarayan V., et al. (2009). Salmonella enterica serovar typhimurium strains with regulated delayed attenuation in vivo. Infect. Immun. 77, 1071–1082 10.1128/IAI.00693-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallo S. F., Zhang B., Denno J., Hong S., Tsai A., Haskins W., et al. (2012). Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. ScientificWorldJournal 2012, 128705 10.1100/2012/128705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damo S. M., Kehl-Fie T. E., Sugitani N., Holt M. E., Rathi S., Murphy W. J., et al. (2013). Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens. Proc. Natl. Acad. Sci. U.S.A. 110, 3841–3846 10.1073/pnas.1220341110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel C., Haentjens S., Bissinger M. C., Courcol R. J. (1999). Characterization of the Acinetobacter baumannii Fur regulator: cloning and sequencing of the fur homolog gene. FEMS Microbiol. Lett. 170, 199–209 10.1016/S0378-1097(98)00533-3 [DOI] [PubMed] [Google Scholar]
- Davis K. A., Moran K. A., McAllister C. K., Gray P. J. (2005). Multidrug-resistant Acinetobacter extremity infections in soldiers. Emerg. Infect. Dis. 11, 1218–1224 10.3201/eid1108.050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L. M., Kakuda T., DiRita V. J. (2009). A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. J. Bacteriol. 191, 1631–1640 10.1128/JB.01394-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breij A., Dijkshoorn L., Lagendijk E., van der Meer J., Koster A., Bloemberg G., et al. (2010). Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS ONE 5:e10732 10.1371/journal.pone.0010732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C. W., Beglin M. S., Actis L. A. (2003a). Detection and analysis of iron uptake components expressed by Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. 41, 4188–4193 10.1128/JCM.41.9.4188-4193.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsey C. W., Tolmasky M. E., Crosa J. H., Actis L. A. (2003b). Genetic organization of an Acinetobacter baumannii chromosomal region harbouring genes related to siderophore biosynthesis and transport. Microbiology 149, 1227–1238 10.1099/mic.0.26204-0 [DOI] [PubMed] [Google Scholar]
- Dorsey C. W., Tomaras A. P., Connerly P. L., Tolmasky M. E., Crosa J. H., Actis L. A. (2004). The siderophore-mediated iron acquisition systems of Acinetobacter baumannii ATCC 19606 and Vibrio anguillarum 775 are structurally and functionally related. Microbiology 150, 3657–3667 10.1099/mic.0.27371-0 [DOI] [PubMed] [Google Scholar]
- Doyle J. S., Buising K. L., Thursky K. A., Worth L. J., Richards M. J. (2011). Epidemiology of infections acquired in intensive care units. Semin. Respir. Crit. Care Med. 32, 115–138 10.1055/s-0031-1275525 [DOI] [PubMed] [Google Scholar]
- Echenique J. R., Arienti H., Tolmasky M. E., Read R. R., Staneloni R. J., Crosa J. H., et al. (1992). Characterization of a high-affinity iron transport system in Acinetobacter baumannii. J. Bacteriol. 174, 7670–7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Hassan K. A., Paulsen I. T., Brown M. H. (2011a). Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics 12:126 10.1186/1471-2164-12-126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijkelkamp B. A., Stroeher U. H., Hassan K. A., Papadimitrious M. S., Paulsen I. T., Brown M. H., et al. (2011b). Adherence and motility characteristics of clinical Acinetobacter baumannii isolates. FEMS Microbiol. Lett. 323, 44–51 10.1111/j.1574-6968.2011.02362.x [DOI] [PubMed] [Google Scholar]
- Falagas M. E., Karveli E. A., Kelesidis I., Kelesidis T. (2007). Community-acquired Acinetobacter infections. Eur. J. Clin. Microbiol. Infect. Dis. 26, 857–68 10.1007/s10096-007-0365-6 [DOI] [PubMed] [Google Scholar]
- Fournier P. E., Vallenet D., Barbe V., Audic S., Ogata H., Poirel L., et al. (2006). Comparative genomics of multidrug resistance in Acinetobacter baumannii. PLoS Genet. 2:e7 10.1371/journal.pgen.0020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furrer J. L., Sanders D. N., Hook-Barnard I. G., McIntosh M. A. (2002). Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44, 1225–1234 10.1046/j.1365-2958.2002.02885.x [DOI] [PubMed] [Google Scholar]
- Gaddy J. A., Arivett B. A., McConnell M. J., Lopez-Rojas R., Pachon J., Actis L. A. (2012). Role of acinetobactin-mediated iron Acquisition functions in the interaction of Acinetobacter baumannii Strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect. Immun. 80, 1015–1024 10.1128/IAI.06279-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddy J. A., Tomaras A. P., Actis L. A. (2009). The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect. Immun. 77, 3150–3160 10.1128/IAI.00096-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancz H., Censini S., Merrell D. S. (2006). Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 602–614 10.1128/IAI.74.1.602-614.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser R., Harder J., Lange H., Bartels J., Christophers E., Schroder J. M. (2005). Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nat. Immunol. 6, 57–64 10.1038/ni1142 [DOI] [PubMed] [Google Scholar]
- Goel V. K., Kapil A., Das B., Rao D. N. (1998). Influence of iron on growth and extracellular products of Acinetobacter baumannii. Jpn. J. Med. Sci. Biol. 51, 25–33 [DOI] [PubMed] [Google Scholar]
- Hammer N. D., Skaar E. P. (2011). Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu. Rev. Microbiol. 65, 129–147 10.1146/annurev-micro-090110-102851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood M. I., Mortensen B. L., Moore J. L., Zhang Y., Kehl-Fie T. E., Sugitani N., et al. (2012). Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin-mediated zinc sequestration. PLoS Pathog. 8:e1003068 10.1371/journal.ppat.1003068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabado N., Jankowski A., Dougaparsad S., Picard V., Grinstein S., Gros P. (2000). Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192, 1237–1248 10.1084/jem.192.9.1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs A. C., Hood I., Boyd K. L., Olson P. D., Morrison J. M., Carson S., et al. (2010). Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect. Immun. 78, 1952–1962 10.1128/IAI.00889-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura-Sato K., Wachino J., Kondo T., Ito H., Arakawa Y. (2010). Correlation between reduced susceptibility to disinfectants and multidrug resistance among clinical isolates of Acinetobacter species. J. Antimicrob. Chemother. 65, 1975–1983 10.1093/jac/dkq227 [DOI] [PubMed] [Google Scholar]
- Kehl-Fie T. E., Chitayat S., Hood M. I., Damo S., Restrepo N., Garcia C., et al. (2011). Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe 10, 158–164 10.1016/j.chom.2011.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura H., Morikawa H., Kamon H., Iguchi M., Hojyo S., Fukada T., et al. (2006). Toll-like receptor-mediated regulation of zinc homeostasis influences dendritic cell function. Nat. Immunol. 7, 971–977 10.1038/ni1373 [DOI] [PubMed] [Google Scholar]
- Krewulak K. D., Vogel H. J. (2011). TonB or not TonB: is that the question? Biochem. Cell Biol. 89, 87–97 10.1139/o10-141 [DOI] [PubMed] [Google Scholar]
- Kumar P., Sannigrahi S., Tzeng Y. L. (2012). The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infect. Immun. 80, 657–667 10.1128/IAI.05208-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. W., Koh Y. M., Kim J., Lee J. C., Lee Y. C., Seol S. Y., et al. (2008). Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 14, 49–54 10.1111/j.1469-0691.2007.01842.x [DOI] [PubMed] [Google Scholar]
- Lee J. C., Koerten H., van den Broek P., Beekhuizen H., Wolterbeek R., van den Barselaar M., et al. (2006). Adherence of Acinetobacter baumannii strains to human bronchial epithelial cells. Res. Microbiol. 157, 360–366 10.1016/j.resmic.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Lee K. C., Eckert R. L. (2007). S100A7 (Psoriasin)–mechanism of antibacterial action in wounds. J. Invest. Dermatol. 127, 945–957 10.1038/sj.jid.5700663 [DOI] [PubMed] [Google Scholar]
- Lewis L. A., Gray E., Wang Y. P., Roe B. A., Dyer D. W. (1997). Molecular characterization of hpuAB, the haemoglobin-haptoglobin-utilization operon of Neisseria meningitidis. Mol. Microbiol. 23, 737–749 10.1046/j.1365-2958.1997.2501619.x [DOI] [PubMed] [Google Scholar]
- Li W., He J., Xie L., Chen T., Xie J. (2013). Comparative genomic insights into the biosynthesis and regulation of mycobacterial siderophores. Cell. Physiol. Biochem. 31, 1–13 10.1159/000343343 [DOI] [PubMed] [Google Scholar]
- Li Y., Qiu Y., Gao H., Guo Z., Han Y., Song Y., et al. (2009). Characterization of Zur-dependent genes and direct Zur targets in Yersinia pestis. BMC Microbiol. 9:128 10.1186/1471-2180-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag A., Dainese E., Rodriguez G. M., Milano A., Provvedi R., Pasca M. R., et al. (2007). Global analysis of the Mycobacterium tuberculosis Zur (FurB) regulon. J. Bacteriol. 189, 730–740 10.1128/JB.01190-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May J. J., Wendrich T. M., Marahiel M. A. (2001). The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J. Biol. Chem. 276, 7209–7217 10.1074/jbc.M009140200 [DOI] [PubMed] [Google Scholar]
- McCormick A., Heesemann L., Wagener J., Marcos V., Hartl D., Loeffler J., et al. (2010). NETs formed by human neutrophils inhibit growth of the pathogenic mold Aspergillus fumigatus. Microbes Infect. 12, 928–936 10.1016/j.micinf.2010.06.009 [DOI] [PubMed] [Google Scholar]
- Mey A. R., Wyckoff E. E., Kanukurthy V., Fisher C. R., Payne S. M. (2005). Iron and fur regulation in Vibrio cholerae and the role of fur in virulence. Infect. Immun. 73, 8167–8178 10.1128/IAI.73.12.8167-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalek M., Gelhaus C., Hecht O., Podschun R., Schroder J. M., Leippe M., et al. (2009). The human antimicrobial protein psoriasin acts by permeabilization of bacterial membranes. Dev. Comp. Immunol. 33, 740–746 10.1016/j.dci.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Mihara K., Tanabe T., Yamakawa Y., Funahashi T., Nakao H., Narimatsu S., et al. (2004). Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 150, 2587–2597 10.1099/mic.0.27141-0 [DOI] [PubMed] [Google Scholar]
- Moore J. L., Becker K. W., Nicklay J. J., Boyd K. L., Skaar E. P., Caprioli R. M. (2013). Imaging mass spectrometry for assessing temporal proteomics: analysis of calprotectin in Acinetobacter baumannii pulmonary infection. Proteomics. [Epub ahead of print]. 10.1002/pmic.201300046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M., Hirano T. (2008). Intracellular zinc homeostasis and zinc signaling. Cancer Sci. 99, 1515–1522 10.1111/j.1349-7006.2008.00854.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussi M. A., Gaddy J. A., Cabruja M., Arivett B. A., Viale A. M., Rasia R., et al. (2010). The opportunistic human pathogen Acinetobacter baumannii senses and responds to light. J. Bacteriol. 192, 6336–6345 10.1128/JB.00917-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nau C. D., Konisky J. (1989). Evolutionary relationship between the TonB-dependent outer membrane transport proteins: nucleotide and amino acid sequences of the Escherichia coli colicin I receptor gene. J. Bacteriol. 171, 1041–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely A. N. (2000). A survey of gram-negative bacteria survival on hospital fabrics and plastics. J. Burn Care Rehabil. 21, 523–527 10.1097/00004630-200021060-00009 [DOI] [PubMed] [Google Scholar]
- Neugebauer H., Herrmann C., Kammer W., Schwarz G., Nordheim A., Braun V. (2005). ExbBD-dependent transport of maltodextrins through the novel MalA protein across the outer membrane of Caulobacter crescentus. J. Bacteriol. 187, 8300–8311 10.1128/JB.187.24.8300-8311.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwugo C. C., Gaddy J. A., Zimbler D. L., Actis L. A. (2011). Deciphering the iron response in Acinetobacter baumannii: a proteomics approach. J. Proteomics 74, 44–58 10.1016/j.jprot.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J., Wren B. W., Thomson N. R., Titball R. W., Holden M. T., Prentice M. B., et al. (2001). Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413, 523–527 10.1038/35097083 [DOI] [PubMed] [Google Scholar]
- Patzer S. I., Hantke K. (1998). The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol. Microbiol. 28, 1199–1210 10.1046/j.1365-2958.1998.00883.x [DOI] [PubMed] [Google Scholar]
- Pawlik M. C., Hubert K., Joseph B., Claus H., Schoen C., Vogel U. (2012). The zinc-responsive regulon of Neisseria meningitidis comprises 17 genes under control of a Zur element. J. Bacteriol. 194, 6594–6603 10.1128/JB.01091-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg A. Y., Seifert H., Paterson D. L. (2008). Acinetobacter baumannii: emergence of a successful pathogen. Clin. Microbiol. Rev. 21, 538–582 10.1128/CMR.00058-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penwell W. F., Arivett B. A., Actis L. A. (2012). The Acinetobacter baumannii entA gene located outside the acinetobactin cluster is critical for siderophore production, iron acquisition and virulence. PLoS ONE 7:e36493 10.1371/journal.pone.0036493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peracino B., Wagner C., Balest A., Balbo A., Pergolizzi B., Noegel A. A., et al. (2006). Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic 7, 22–38 10.1111/j.1600-0854.2005.00356.x [DOI] [PubMed] [Google Scholar]
- Pietzsch J., Hoppmann S. (2009). Human S100A12: a novel key player in inflammation? Amino Acids 36, 381–389 10.1007/s00726-008-0097-7 [DOI] [PubMed] [Google Scholar]
- Pour N. K., Dusane D. H., Dhakephalkar P. K., Zamin F. R., Zinjarde S. S., Chopade B. A. (2011). Biofilm formation by Acinetobacter baumannii strains isolated from urinary tract infection and urinary catheters. FEMS Immunol. Med. Microbiol. 62, 328–338 10.1111/j.1574-695X.2011.00818.x [DOI] [PubMed] [Google Scholar]
- Proschak A., Lubuta P., Grun P., Lohr F., Wilharm G., De Berardinis V., et al. (2013). Structure and biosynthesis of fimsbactins A-F, siderophores from Acinetobacter baumannii and Acinetobacter baylyi. Chembiochem 14, 633–638 10.1002/cbic.201200764 [DOI] [PubMed] [Google Scholar]
- Py B., Gerez C., Angelini S., Planel R., Vinella D., Loiseau L., et al. (2012). Molecular organization, biochemical function, cellular role and evolution of NfuA, an atypical Fe-S carrier. Mol. Microbiol. 86, 155–171 10.1111/j.1365-2958.2012.08181.x [DOI] [PubMed] [Google Scholar]
- Rakin A., Schneider L., Podladchikova O. (2012). Hunger for iron: the alternative siderophore iron scavenging systems in highly virulent Yersinia. Front. Cell. Infect. Microbiol. 2:151 10.3389/fcimb.2012.00151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey M., Cianciotto N. P. (2002). Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70, 5659–6569 10.1128/IAI.70.10.5659-5669.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland B. M., Grossman T. H., Osburne M. S., Taber H. W. (1996). Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene 178, 119–123 10.1016/0378-1119(96)00349-6 [DOI] [PubMed] [Google Scholar]
- Runyen-Janecky L. J. (2013). Role and regulation of heme iron acquisition in gram-negative pathogens. Front. Cell. Infect. Microbiol. 3:55 10.3389/fcimb.2013.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runyen-Janecky L. J., Reeves S. A., Gonzales E. G., Payne S. M. (2003). Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71, 1919–1928 10.1128/IAI.71.4.1919-1928.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R., Saha N., Donofrio R. S., Bestervelt L. L. (2013). Microbial siderophores: a mini review. J. Basic Microbiol. 53, 303–317 10.1002/jobm.201100552 [DOI] [PubMed] [Google Scholar]
- Sahl J. W., Gillece J. D., Schupp J. M., Waddell V. G., Driebe E. M., Engelthaler D. M., et al. (2013). Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PLoS ONE 8:e54287 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl J. W., Johnson J. K., Harris A. D., Phillippy A. M., Hsiao W. W., Thom K. A., et al. (2011). Genomic comparison of multi-drug resistant invasive and colonizing Acinetobacter baumannii isolated from diverse human body sites reveals genomic plasticity. BMC Genomics 12:291 10.1186/1471-2164-12-291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K., Gouget B., Carriere M., Labigne A., de Reuse H. (2007). Novel nickel transport mechanism across the bacterial outer membrane energized by the TonB/ExbB/ExbD machinery. Mol. Microbiol. 63, 1054–1068 10.1111/j.1365-2958.2006.05578.x [DOI] [PubMed] [Google Scholar]
- Schroder J., Jochmann N., Rodionov D. A., Tauch A. (2010). The Zur regulon of Corynebacterium glutamicum ATCC 13032. BMC Genomics 11:12 10.1186/1471-2164-11-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebeny P. J., Riddle M. S., Petersen K. (2008). Acinetobacter baumannii skin and soft-tissue infection associated with war trauma. Clin. Infect. Dis. 47, 444–449 10.1086/590568 [DOI] [PubMed] [Google Scholar]
- Smani Y., Docobo-Perez F., Lopez-Rojas R., Dominguez-Herrera J., Ibanez-Martinez J., Pachon J. (2012a). Platelet-activating factor receptor initiates contact of Acinetobacter baumannii expressing phosphorylcholine with host cells. J. Biol. Chem. 287, 26901–26910 10.1074/jbc.M112.344556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smani Y., McConnell M. J., Pachon J. (2012b). Role of fibronectin in the adhesion of Acinetobacter baumannii to host cells. PLoS ONE 7:e33073 10.1371/journal.pone.0033073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smani Y., Dominguez-Herrera J., Pachon J. (2013). Association of the outer membrane protein Omp33with fitness and virulence of Acinetobacter baumannii. J. Infect. Dis. 10, 1561–1570 10.1093/infdis/jit386 [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I., Cobeljic M., Hantke K. (1993). Escherichia coli K-12 ferrous iron uptake mutants are impaired in their ability to colonize the mouse intestine. FEMS Microbiol. Lett. 108, 111–115 10.1111/j.1574-6968.1993.tb06082.x [DOI] [PubMed] [Google Scholar]
- Stork M., Bos M. P., Jongerius I., de Kok N., Schilders I., Weynants V. E., et al. (2010). An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathog. 6:e1000969 10.1371/journal.ppat.1000969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamang M. D., Kim S., Kim S. M., Kong H. H., Kim J. (2011). Interaction of Acinetobacter baumannii 19606 and 1656-2 with Acanthamoeba castellanii. J. Microbiol. 49, 841–846 10.1007/s12275-011-1063-8 [DOI] [PubMed] [Google Scholar]
- Tomaras A. P., Dorsey C. W., Edelmann R. E., Actis L. A. (2003). Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149, 3473–3484 10.1099/mic.0.26541-0 [DOI] [PubMed] [Google Scholar]
- Torres V. J., Attia A. S., Mason W. J., Hood M. I., Corbin B. D., Beasley F. C., et al. (2010). Staphylococcus aureus fur regulates the expression of virulence factors that contribute to the pathogenesis of pneumonia. Infect. Immun. 78, 1618–1628 10.1128/IAI.01423-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Hassan H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Sikes M. L., Fink R. C., Vazquez-Torres A., Jones-Carson J., Hassan H. M. (2011). Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J. Bacteriol. 193, 497–505 10.1128/JB.00942-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban C. F., Ermert D., Schmid M., Abu-Abed U., Goosmann C., Nacken W., et al. (2009). Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathog. 5:e1000639 10.1371/journal.ppat.1000639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallenet D., Nordmann P., Barbe V., Poirel L., Mangenot S., Bataille E., et al. (2008). Comparative analysis of Acinetobacters: three genomes for three lifestyles. PLoS ONE 3:e1805 10.1371/journal.pone.0001805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayudhan J., Castor M., Richardson A., Main-Hester K. L., Fang F. C. (2007). The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63, 1495–1507 10.1111/j.1365-2958.2007.05600.x [DOI] [PubMed] [Google Scholar]
- Velayudhan J., Hughes N. J., McColm A. A., Bagshaw J., Clayton C. L., Andrews S. C., et al. (2000). Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37, 274–286 10.1046/j.1365-2958.2000.01987.x [DOI] [PubMed] [Google Scholar]
- Vidal R., Dominguez M., Urrutia H., Bello H., Gonzalez G., Garcia A., et al. (1996). Biofilm formation by Acinetobacter baumannii. Microbios 86, 49–58 [PubMed] [Google Scholar]
- Weinberg E. D. (1975). Nutritional immunity. Host's attempt to withold iron from microbial invaders. JAMA 231, 39–41 10.1001/jama.231.1.39 [DOI] [PubMed] [Google Scholar]
- Wendt C., Dietze B., Dietz E., Ruden H. (1997). Survival of Acinetobacter baumannii on dry surfaces. J. Clin. Microbiol. 35, 1394–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman T. J. (2007). Infection control challenges related to war wound infections in the ICU setting. J. Trauma 62, S53 10.1097/TA.0b013e318065aa71 [DOI] [PubMed] [Google Scholar]
- Yamamoto S., Okujo N., Sakakibara Y. (1994). Isolation and structure elucidation of acinetobactin, a novel siderophore from Acinetobacter baumannii. Arch. Microbiol. 162, 249–254 10.1007/s002030050133 [DOI] [PubMed] [Google Scholar]
- Zimbler D. L., Arivett B. A., Beckett A. C., Menke S. M., Actis L. A. (2013). Functional features of TonB energy transduction systems of Acinetobacter baumannii. Infect. Immun. 81, 3382–3394 10.1128/IAI.00540-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimbler D. L., Park T. M., Arivett B. A., Penwell W. F., Greer S. M., Woodruff T. M., et al. (2012). Stress response and virulence functions of the Acinetobacter baumannii NfuA Fe-S scaffold protein. J. Bacteriol. 194, 2884–28893 10.1128/JB.00213-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimbler D. L., Penwell W. F., Gaddy J. A., Menke S. M., Tomaras A. P., Connerly P. L., et al. (2009). Iron acquisition functions expressed by the human pathogen Acinetobacter baumannii. Biometals 22, 23–32 10.1007/s10534-008-9202-3 [DOI] [PubMed] [Google Scholar]