Abstract
We used next-generation sequencing to characterize the genomes of nine species of Orobanchaceae of known phylogenetic relationships, different life forms, and including a polyploid species. The study species are the autotrophic, nonparasitic Lindenbergia philippensis, the hemiparasitic Schwalbea americana, and seven nonphotosynthetic parasitic species of Orobanche (Orobanche crenata, Orobanche cumana, Orobanche gracilis (tetraploid), and Orobanche pancicii) and Phelipanche (Phelipanche lavandulacea, Phelipanche purpurea, and Phelipanche ramosa). Ty3/Gypsy elements comprise 1.93%–28.34% of the nine genomes and Ty1/Copia elements comprise 8.09%–22.83%. When compared with L. philippensis and S. americana, the nonphotosynthetic species contain higher proportions of repetitive DNA sequences, perhaps reflecting relaxed selection on genome size in parasitic organisms. Among the parasitic species, those in the genus Orobanche have smaller genomes but higher proportions of repetitive DNA than those in Phelipanche, mostly due to a diversification of repeats and an accumulation of Ty3/Gypsy elements. Genome downsizing in the tetraploid O. gracilis probably led to sequence loss across most repeat types.
Keywords: next-generation sequencing, polyploidy, genome size, genome downsizing, transposable elements, LTR retrotransposons, Ty3/Gypsy, Orobanche, Phelipanche, Orobanchaceae
Introduction
Plant genome evolution is significantly shaped by repetitive sequences, which can make up to 97% of the nuclear genome (Flavell et al. 1974; Murray et al. 1981; Leitch and Leitch 2008). Only a few of these repetitive sequences, such as tRNA genes and telomeric sequences, have well-defined functions, yet repetitive sequences are largely responsible for the 2,000-fold variation in nuclear haploid genome size (1C value) in angiosperms alone (Greilhuber et al. 2006; Leitch and Leitch 2008). An increase in genome size caused by tandemly repeated DNA families and transposon accumulation is well documented, and some broad patterns have been established (Bennetzen and Kellogg 1997; Bennetzen et al. 2005; Vitte and Panaud 2005; Hawkins et al. 2008; Leitch et al. 2008). For example, large genome size is correlated with slow meristem growth rates in the roots, with parasitic eudicots, such as Viscum, that do not have roots, apparently escaping this limitation (Gruner et al. 2010).
Despite the prominent role of repetitive DNA in plant genomes, the evolution and significance of different proportions and kinds of repetitive DNA are still little understood. This is due to the sheer amount of repetitive sequence within most genomes and the limited number of well-characterized element groups (Kalendar et al. 2004; Neumann et al. 2006; Manetti et al. 2007; Raskina et al. 2008). Recent advances in sequencing technologies, such as 454 pyrosequencing, now allow a comprehensive characterization of repetitive DNAs with information both on the types of repetitive sequences and on their relative proportions (Macas et al. 2007; Swaminathan et al. 2007; Wicker et al. 2009; Hribova et al. 2010; Kelly and Leitch 2011; Renny-Byfield et al. 2011). To better understand the role of repetitive sequences in the evolution of plant genomes requires “comparative” characterization in a phylogenetic framework. Toward this goal, we carried out an analysis of the repetitive DNA in a clade of broomrapes or Orobanchaceae.
Orobanchaceae contain c. 2000 species in 89 genera and are the largest family of parasitic flowering plants, with possibly several transitions toward parasitism, accompanied by partial or complete loss of photosynthetic ability (Bennett and Mathews 2006). This study system is set apart by a considerable variation in genome size combined with limited chromosome number variation; nevertheless, we included at least one tetraploid species. Nine species were selected for 454 pyrosequencing based on the previous work that sets up expectations about the dynamics of their genomes (Schneeweiss, Palomeque et al. 2004; Weiss-Schneeweiss et al. 2006; Park et al. 2007). We sampled seven holoparasites from the genera Phelipanche and Orobanche, the hemiparasitic species Schwalbea americana, and the autotrophic Lindenbergia philippensis. The latter is among the earliest diverging Orobanchaceae, while the monotypic genus Schwalbea is a member of the earliest diverging lineage of hemiparasites in the family (Bennett and Mathews 2006).
Previous studies on Orobanchaceae that include most of our nine focal species provide information on phylogenetic relationships (Young et al. 1999; Schneeweiss, Colwell et al. 2004; Wolfe et al. 2005; Bennett and Mathews 2006; Park et al. 2008), life history (Schneeweiss 2007), chromosome numbers, genome sizes (Schneeweiss, Palomeque et al. 2004; Weiss-Schneeweiss et al. 2006), and retroelement evolution (Park et al. 2007). A survey of long terminal repeat (LTR) retroelement diversity in Orobanchaceae also found more phylogenetic structure in the elements isolated from Orobanche species than in those isolated from Phelipanche, indicative of more recent or more pronounced retroelement activity in the former genus (Park et al. 2007). In addition, Orobanche is more variable in genome size and ploidy level (Schneeweiss, Palomeque et al. 2004; Weiss-Schneeweiss et al. 2006). All this suggests that the Orobanche genome is more dynamic than that of Phelipanche, and therefore, the hypothesis we wanted to test was that this is largely due to labile repetitive DNA fractions in Orobanche.
Besides comparing the types and proportions of repetitive DNA in Orobanche and Phelipanche (with Schwalbea and Lindenbergia as the outgroups), we were interested in changes in sequence type and copy number following polyploidy. Polyploids can undergo genome downsizing (Doležel et al. 1998; Leitch and Bennett 2004; Beaulieu et al. 2009; Renny-Byfield et al. 2011), probably involving mechanisms such as retrotransposition and DNA deletion (Leitch and Leitch 2008). Of our nine study species, Orobanche gracilis has tetraploid and hexaploid forms, and both forms have low monoploid genome sizes (Weiss-Schneeweiss et al. 2006; this study), indicating genome downsizing. Thus, we analyzed whether there was preferential loss or retention of particular types of repetitive elements. The only comparable study so far, focusing on the young allopolyploid hybrid Nicotiana tabacum, found that the most common repeat types in the parental species were the ones most affected by loss, which set up an expectation to be tested in O. gracilis.
Materials and Methods
Plant Material
The chromosome numbers, 1C values, and other genomic features of the nine focal species are listed in table 1. Orobanche crenata Forssk. is distributed in the Mediterranean region and the Near East. It is an important pest species that mainly attacks legume hosts including many, usually annual, crop species (Teryokhin 1997). The sequenced sample came from the Bonn Botanical Garden, where it was parasitizing Vicia faba (voucher: S. Wicke OC41, Bonn university herbarium).
Table 1. Genomic and Sequencing Features of the Studied Species of Orobanche, Phelipanche, Schwalbea, and Lindenbergia.
| Species | Chromosome Number (2n) |
1C Value (pg) |
Genome Size (Gb) |
Sequenced Read Numbera |
Total Read Length (Mb)a |
454-Pyrosequencing Genome Coverage (%) |
Repetitive DNA Cluster Number |
|---|---|---|---|---|---|---|---|
| O. crenata | 38b | 2.84c | 2.78c | 1,397,401 | 555.14 | 20 | 223 |
| O. cumana | 38b | 1.45c | 1.42c | 1,030,193 | 341.48 | 23 | 230 |
| O. gracilis | 76b | 2.10d | 2.05d | 1,130,469 | 411.15 | 20 | 250 |
| O. pancicii | 38e | 3.24d | 3.17d | 1,397,998 | 492.19 | 16 | 251 |
| P. lavandulacea | 24b | 4.38c | 4.29c | 1,285,657 | 480.59 | 11 | 189 |
| P. purpurea | 24b | 4.42c | 4.32c | 965,667 | 239.97 | 6 | 195 |
| P. ramosa | 24b | 4.34c | 4.25c | 1,482,417 | 510.89 | 12 | 190 |
| S. americana | 36f | 0.57d | 0.56d | 479,780 | 122.60 | 22 | 211 |
| L. philippensis | 32d | 0.46d | 0.45d | 310,533 | 75.98 | 17 | 167 |
After removing mitochondrial and chloroplast contamination.
Schneeweiss et al. (2004a).
This study.
H. Weiss-Schneeweiss, University of Vienna, personal communication, September 2011.
Orobanche cumana Wallr. is a close relative of Orobanche cernua (Teryokhin 1997; they are sometimes treated as conspecific) and is widely distributed in the Mediterranean region to Central Asia. It parasitizes mainly Asteraceae, including annual crop species, such as sunflower (Helianthus annuus; Teryokhin 1997). The sequenced sample came from the Bonn Botanical Garden where it was parasitizing H. annuus (voucher: S. Wicke OC16/17).
O. gracilis Beck is distributed in the Mediterranean northward to southern Central Europe. It grows exclusively on (semi-)shrubby Fabaceae. The sequenced sample was collected in Lower Austria where it was parasitizing a species of Chamaecytisus (voucher: G. Schneeweiss 7, Vienna university herbarium), and its 1C value was determined for this study (see below).
Orobanche pancicii Beck is distributed in the Balkan Peninsula northwards to the Eastern Alps. It parasitizes Knautia and possibly also Scabiosa species (Dipsacaceae). The sequenced sample was collected in Styria, Austria, where it was parasitizing Knautia drymeia (voucher: G. Schneeweiss 42). The 1C value of the sequenced sample was determined for this study (see below).
Phelipanche lavandulacea Pomel is a Mediterranean species restricted to a single perennial host, Bituminaria bituminosa (Fabaceae). The sequenced sample was collected in the Toscana in Italy (voucher: Schönswetter and Tribsch 12761, Vienna university herbarium).
Phelipanche purpurea (Jacq.) Soják is widely distributed in southern and central Europe to Central Asia. It often occurs in slightly disturbed habitats, and its exclusive host species are perennial Achillea and Artemisia species (both Asteraceae). The sequenced sample came from the Bonn Botanical Garden, where it was parasitizing a species of Achillea (voucher: S. Wicke Op38/39).
Phelipanche ramosa (L.) Pomel is native to the Mediterranean region and Near Asia but has been introduced worldwide. It grows on a broad range of usually annual hosts, including the crops such as tobacco (N. tabacum, Solanaceae), tomato (S. lycopersicum, Solanaceae), hemp (C. sativa, Cannabaceae), and cabbage (B. oleracea, Brassicaceae; Teryokhin 1997). The sequenced sample came from the Bonn Botanical Garden, where it was growing on tomato (voucher: S. Wicke Pr52/53).
Lindenbergia philippensis (Cham. and Schltd.) Benth. is an autotrophic, nonparasitic species reported from Bangladesh, India, Burma, Thailand, Cambodia, Laos, Vietnam, tropical China, and the Philippines. The sequenced sample came from a plant cultivated at PennState University (voucher: S. Wicke LP60/LP61). The 1C value and chromosome number of the species were determined for this study (below), using offspring grown from seeds of the PennState plants.
S. americana L. is a hemiparasite from the southeastern Coastal Plain of the United States of America. It is a fire-dependent non-host-specific species that today is endangered, but historically ranged from New York to Texas (Norden and Kirkman 2004). The sequenced sample was provided by C. DePamphilis, PennState; that for the C-value measurement by M. Wenzel from the ex situ conservation collection of the Atlanta Botanical Garden (voucher: M. Wenzel s.n., 1 August 2011, Munich University Herbarium).
Genome Size Estimation and Cytological Analysis
The 1C values of O. gracilis, O. pancicii, L. philippensis, and S. americana were measured using flow cytometry with propidium iodide (PI) as the DNA stain and Solanum pseudocapsicum (1C = 1.29 pg, Temsch et al. 2010) as the standard plant (the method is described in detail in the study by Temsch et al. 2010). Fresh leaf, root, or carpel material was co-chopped together with the standard organism in Otto’s buffer I (Otto et al. 1981) according to the chopping instructions of Galbraith et al. (1983). The resulting suspension was filtered (30 μm nylon mesh), treated with RNase, and incubated in PI containing Otto’s buffer II (Otto et al. 1981). A CyFlow ML flow cytometer (Partec, Muenster, Germany) equipped with a green laser (100 mW, 532 nm, Cobolt Samba, Cobolt, Stockholm, Sweden) was used for the fluorescence measurements, with 5,000 particles measured per run and three runs performed per plant preparation. The C-value was calculated according to the formula: 1C valueObject = (mean G1 nuclei fluorescence intensityObject/mean G1 nuclei fluorescence intensityStandard) × 1C valueStandard. The peak coefficient of variation percentages usually were <5%, but reached 7% in S. americana roots perhaps due to the presence of polyphenolic compounds (Greilhuber 1988).
DNA Extraction and 454 Sequencing
DNA isolation followed a standard cetyl trimethyl ammonium bromide (CTAB) extraction protocol (Doyle and Doyle 1987) with a low-salt CTAB buffer with low ethylenediaminetetraacetic acid concentration using 5 g of fresh flower material. After complete removal of RNA with DNAse-free RNAse A, DNA was precipitated overnight after addition of 0.5 volumes of cold (4°C) 7.5 M NaAc, and 2 volumes of ice-cold (−20°C) ultrapure ethanol and washed twice. Pellets of sufficient clarity were resuspended in 1.5 mM Tris–buffer (pH = 8.0) to a final concentration of 200–300 ng/μl. For each species, approximately 5 μg of genomic DNA was submitted for sequencing at the Core Facility Molecular Biology of the Centre of Medical Research, Medical University of Graz. DNA was randomly fragmented by nebulization, converted into a single strand 454 GS FLX compatible DNA library, and sequenced on one (or in two cases a half) picotiter plate on a 454 Genome Sequencer (Roche Diagnostics) using the recommended standard protocols and chemistry.
Data Analysis
Sequencing data were preprocessed to remove identical reads, which are artifacts of the 454 technology. Repeat sequence assembly was performed using a graph-based clustering approach as described in the study by Novak et al. (2010) on each of the nine species data sets. In this approach, reads from one species are subjected to a pairwise sequence comparison, and their mutual similarities are then represented as a graph in which the vertices corresponded to sequence reads; overlapping reads are connected with edges, and their similarity scores are expressed as edge weights. The graph structure was analyzed using custom-made programs to detect clusters of frequently connected nodes representing groups of similar sequences. These clusters, corresponding to families of genomic repeats, were separated and analyzed with respect to the number of reads they contained (which is proportional to their genomic abundance). Graphs of selected clusters were also visually examined using the SeqGrapheR program (Novak et al. 2010) to assess structure and variability of the repeats. In this case, distances between a given node (a single sequence) and other related nodes are determined, in part, by the bit-score (edge weight) of a basic local alignment search tool (Blast) analysis between sequences, with a Fruchterman–Reingold algorithm used to position the nodes. This results in more similar sequences being placed closer together, whereas more distantly related reads are placed further apart.
The reads within individual clusters were assembled into contigs using TIGR Gene Indices clustering tools (Pertea et al. 2003) with the -O′-p 80-o 40′ parameters, specifying overlap percent identity and minimal length cutoff for cap3 assembler. Repeat type identification was done by sequence-similarity searches of assembled contigs against GenBank using BlastN and BlastX (Altschul et al. 1990), by sequence-similarity searches of assembled contigs against Munich Information Center for Protein Sequences (MIPS) Repeat Element Database (accessed 2 January 2011; Spannagl et al. 2007) using TBlastX, by sequence-similarity searches of reads against MIPS Repeat Element Database using RepeatMasker (Smit et al. 1996), and by detection of conserved protein domains using RPS-Blast (Reversed Position Specific-Blast; Altschul et al. 1997). Satellites within contig sequences were identified using Tandem Repeats Finder (Benson 1999). Sequences that corresponded to putative mitochondrial and plastid contaminations were then eliminated. The genome proportion of each cluster was calculated as the percentage of reads.
To determine the distribution of the different repeats in Orobanche and Phelipanche, we built a combined data set comprising 2,450,000 reads (350,000 reads per species of length 300 bp) each labeled with a species code. To assess possible effects of different genome coverage, we built a second combined data set in which we used the same coverage (2.43%) for each species. Both data sets were analyzed as described above. We performed individual genome and combined data set screenings because each has specific advantages: A combined data set facilitates finding shared repeat families of unequal abundance among species. The individual genome screening, by contrast, facilitates detection of species-specific repeat families.
Phylogenetic Analysis
To place the study species in a phylogenetic context, they were added to the largest applicable matrix (Park et al. 2008), which relies on sequences from the plastid gene rps2 and the nuclear internal transcribed spacer region including the 5.8S gene. The only species not previously sequenced was O. pancicii, for which we obtained Internal Transcribed Spacer (ITS) and rps2 sequences from a silica-dried leaf of the voucher G. Schneeweiss 42 (Vienna University Herbarium), using standard methods (Park et al. 2008). ITS and rps2 sequences were also obtained from the O. gracilis 454-sequenced individual and appeared identical to those reported previously for another individual collected in Italy (DQ310030, AY209239). Tree searching and bootstrapping (with 100 replicates) relied on maximum likelihood under the GTR + G model of substitution, using RAxML version 7.2.8 (Stamatakis 2006).
Results
Genome Size Estimation
The genome sizes of the nine study species are shown in table 1. The 1C value of the only tetraploid species, O. gracilis, is 2.10 pg, congruent with previous estimates (1.66–2.45 pg; Weiss-Schneeweiss et al. 2006), and one of the diploid Orobanche species, O. pancicii, has the largest genome, with 3.24 pg. The 1C values of L. philippensis (0.46 pg) and S. americana (0.57 pg) are ~2.5–9.6× lower than those observed in the seven holoparasitic species.
Phylogenetic Analysis
A maximum likelihood tree for the Orobanchaceae that includes the nine study species is shown in figure 1. Species relationships are congruent with those obtained in previous studies (Park et al. 2008; some species-to-genus assignments have changed since that article, and here, we use the latest classification). The tree is rooted on the autotrophic L. philippensis based on the more comprehensive analysis of Park et al. (2008). The hemiparasite S. americana is part of a clade that branched off more recently than Lindenbergia, and Orobanche and Phelipanche are surprisingly distantly related to each other (fig. 1). Within Phelipanche, P. ramosa and P. lavandulacea are more closely related than to P. purpurea. Within Orobanche, relationships are poorly resolved by the relatively short sequences used here (1,210 aligned nucleotides of nuclear ITS and plastid rps2), but O. pancicii is close to O. crenata, and O. cumana is the most isolated of the four Orobanche species studied (in agreement with Manen et al. 2004; Schneeweiss, Colwell et al. 2004).
Fig. 1. Phylogenetic relationships in Orobanchaceae inferred from maximum likelihood analysis of a combined data set of plastid rps2 and nuclear ITS sequences.
The nine species analyzed in this study are indicated in bold. Statistical support (>60%) comes from parametric bootstrapping using 100 replicates.
454 High-Throughput DNA Sequencing
The 454 GS FLX Titanium sequencing returned from 347,565–1,508,792 reads per species with an average read length of 340 nt, resulting in 3.3 Gb of sequence data or 87–558 Mb of DNA sequence per species. Filtering for plastid contaminants resulted in 76–555 Mb of DNA sequence for each accession. This amounts to ~23% coverage of the O. cumana genome (table 1), ~20% coverage of the O. gracilis genome, ~20% coverage for O. crenata genome, ~16% coverage for O. pancicii, ~12% coverage for P. ramosa, ~11% coverage for P. lavandulacea, ~6% coverage for P. purpurea, ~22% coverage for S. americana, and ~17% coverage for L. philippensis.
Individual Genome Characterization
We subjected each of the nine 454 read data sets to cluster-based repeat identification, which partitioned the data into groups of overlapping reads representing individual repeat families as described in the study by Novak et al. (2010). Cluster number (for clusters comprising at least 0.01% of the examined reads per species) ranged from 167 clusters detected in L. philippensis to 251 in O. pancicii (table 1). Next, we assembled and annotated each cluster. Examples of the annotated outputs are shown in supplementary figure 1, where each node within a network corresponds to a single 454 read, and similar reads are placed more closely together than more distantly related sequences. The genome proportions of each type of repetitive DNA in the different species are shown in table 2. The genomic proportion of highly or moderately repetitive DNA appears highly variable among species, ranging from 24.75% in S. americana to 60.13% in O. gracilis. Except for the Penelope retrotransposons and the P transposon superfamilies, all repetitive DNA types and transposable element superfamilies described in plants (Wicker et al. 2007) were detected. Satellites, rDNA, LTR and LINE/SINE retrotransposons, Mutator and En-Spm transposons are widely distributed in the nine species, whereas the hAT, PIF/Harbinger, RC/Helitron, and Tc/Mariner transposons were detected in only a few.
Table 2. Repeat Composition of the Studied Genomes of Species of Orobanche, Phelipanche, Schwalbea, and Lindenbergia Deduced from the Individual Genome Screenings.
| GP (%) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| L. philippensis | S. americana | O. cumana | O. gracilis | O. crenata | O. pancicii | P. purpurea | P. lavandulacea | P. ramosa | |
| Retrotransposon | |||||||||
| Ty1/Copia | 17.21 | 8.09 | 16.01 | 18.41 | 21.42 | 18.82 | 10.67 | 20.53 | 22.83 |
| Ty3/Gypsy | 1.93 | 2.67 | 17.02 | 28.34 | 21.44 | 24.16 | 20.59 | 15.16 | 15.92 |
| Unclassified LTR | 0.02 | 1.88 | — | 1.71 | 0.13 | 0.69 | 0.04 | — | 0.25 |
| LINE/SINE | 0.67 | 0.37 | 0.41 | 0.47 | 0.56 | 1.04 | 0.21 | 0.13 | 0.17 |
| Transposon | |||||||||
| hAT | — | 0.29 | 0.06 | 0.11 | 0.12 | 0.24 | — | 0.04 | 0.07 |
| Mutator | 0.16 | 0.87 | 0.11 | 0.23 | 0.11 | 0.28 | 0.17 | 0.42 | 0.53 |
| RC/Helitron | — | 0.18 | 0.06 | — | 0.22 | 0.12 | 0.02 | 0.07 | 0.35 |
| En-Spm | 0.17 | 1.25 | 0.55 | 0.65 | 0.74 | 1.04 | 0.62 | 1.27 | 1.37 |
| PIF-Harbinger | — | — | — | 0.04 | — | 0.01 | — | — | — |
| Tc1-Mariner | — | 0.02 | — | 0.03 | — | — | — | 0.01 | 0.04 |
| rDNA | 1.81 | 1.56 | 0.67 | 1.36 | 1.74 | 1.34 | 0.06 | 0.48 | 0.76 |
| snRNA | — | — | — | — | — | — | — | 0.34 | — |
| Satellite | 3.61 | 2.95 | 3.10 | 5.08 | 3.88 | 2.28 | 1.75 | 2.36 | 2.59 |
| Unclassified | 4.06 | 4.63 | 7.57 | 3.71 | 4.57 | 6.06 | 3.82 | 1.22 | 2.15 |
| Total | 29.63 | 24.75 | 45.57 | 60.13 | 54.94 | 56.09 | 37.97 | 42.03 | 47.02 |
Note.—GP, genome proportion; snRNA, small noncoding RNA; —, not detected.
In each species, retroelements make up most of the repetitive DNA (estimates range from 13.01% to 48.93%). The majority of retroelements are Ty1/Copia and Ty3/Gypsy retrotransposons, with their respective genome proportions ranging from 8.09% to 22.83% and from 1.93% to 28.34%. The genomic proportion of Ty3/Gypsy elements, which are notably rare in L. philippensis and S. americana (1.93% and 2.67%, respectively), appears more variable than those of Ty1/Copia elements. Compared with LTR retrotransposons, non-LTR retrotransposons and DNA transposons were found relatively infrequently (0.13%–1.04% and 0.33%–2.36%, respectively).
Estimates of rDNA and satellite abundance in the nine genomes show that they make up a substantial fraction, rDNA representing between 0.48% and 1.81% of the genomes. Only in P. purpurea is their abundance much lower (0.06%). The abundance of satellites is also variable, ranging from 1.75% in L. philippensis to 5.08% in O. gracilis. The repetitive DNA fractions include 1.22% to 7.57% unclassified sequences, which might include additional repeat types.
To assess the effect of the different genomic coverage (table 1), we subsampled O. cumana reads such that 6% instead of 23% of its genome was covered; the resulting genomic proportions of repetitive DNA were 44.89% versus 45.57%. In small genomes, decreasing coverage results in a larger numbers of smaller, less well-annotated clusters (supplementary table 1). Fragmentation with lower proportions of repeats in small genomes may be a general phenomenon (Novak et al. 2010).
Repeat Family Distribution in Orobanche and Phelipanche
Exclusion of L. philippensis and S. americana for which fewer reads were sequenced left one combined data set of 2,450,000 reads of 300 bp length (350,000 reads per species) and a second in which each genome was sampled at the same coverage (2.43%; see Materials and Methods). In contrast to the subsampling of O. cumana, the proportionally sampled data set remains sufficiently large to not suffer from a less efficient classification (supplementary table 1). The repeat types found were essentially the same using both combined data sets as detected in the individual genome screening (supplementary table 1). Only one rare superfamily, PIF-Harbinger, remained undetected in either of the combined data sets. The combined data sets also tended to overestimate genomic repeat proportions compared with the individual genome screenings. For example, from the combined data sets, the O. crenata genome appeared to contain from 56.4% to 61.1% repetitive DNA, whereas this genome analyzed on its own contained 54.4% repetitive DNA (supplementary table 1). As expected, the combined data set allows detecting low-copy repeat families that remain undetected in the individual genome screenings. The only exception to this pattern was O. gracilis in which less repetitive DNA was detected in the proportionally sampled combined data set than in the individual genome screening (supplementary table 1). This could be due to a higher proportion of species-specific families in this species. Interestingly, the combined data set in which we used identical read numbers per species did not show this effect, and therefore, we focus on this data set.
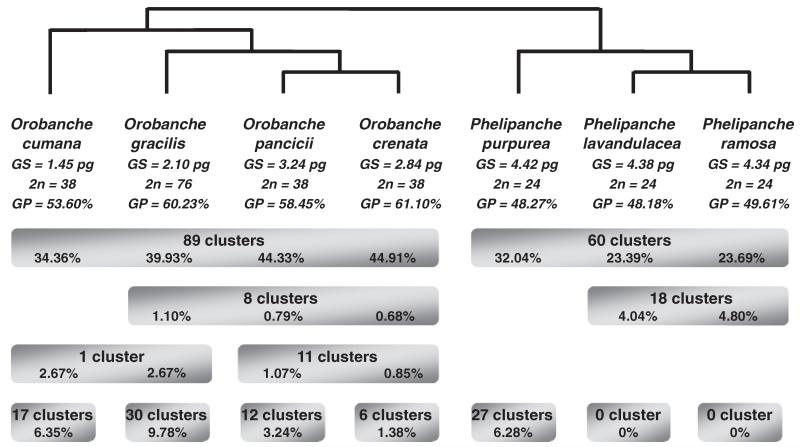
The distribution of most repeat families among the seven species is shown in figure 2. Species-specific clusters make up 1.38% of the O. crenata genome, 3.24% of the O. pancicii genome, 6.28% of the P. purpurea genome, 6.35% of the O. cumana genome, and 9.78% of the O. gracilis genome, whereas no cluster is exclusive to P. lavandulacea and P. ramosa. Genus-specific clusters make up most of the repetitive DNA fraction, comprising 34.36%–44.91% and 23.39%–32.04% of the Orobanche and Phelipanche genomes, respectively. The 44 remaining clusters are unevenly distributed among the seven genomes or absent (or undetected) in only one or two species. A peculiar feature in P. lavandulacea and P. ramosa is the overrepresentation of these widespread clusters. In these two species, they comprise around 21% of the genome, whereas they make up less than 13.3% of the other genomes. The identified families of satellite DNA are mostly species- or genus-specific (data not shown).
Fig. 2. Distribution of repetitive DNA clusters in seven species of Orobanche and Phelipanche, focusing on the 13 main distribution patterns and the corresponding numbers of clusters.
Species relationships are shown based on figure 1. For each species, the genome size (GS), chromosome number, and total repeat genomic proportion (GP) are given and for each distribution pattern, the corresponding genomic proportion. Genomic proportions shown are deduced from the combined data set.
Discussion
Repetitive Sequences in Orobanchaceae Compared with Other Angiosperms
This study characterizes the repeat composition in genomes of nine Orobanchaceae using 454 GS FLX pyrosequencing, with between 6% and 23% genome coverage per species (table 1). Previous studies on Pisum sativum (Macas et al. 2007), Glycine max (Swaminathan et al. 2007), Hordeum vulgare (Wicker et al. 2009), Musa acuminata (Hribova et al. 2010), and an allotetraploid species of Nicotina and its progenitors (Renny-Byfield et al. 2011) found that a coverage of >0.5% allows the reconstruction of repeat units that have genomic proportion in excess of 0.01%. Thus, the genome coverage used here should allow robust estimates.
Most of the repetitive DNA found in the nine species (table 2) consists of transposable elements as is typical for flowering plants (Tenaillon et al. 2010). The most abundant types are Ty1/Copia and Ty3/Gypsy, which comprise 10.76% of the genome of L. philippensis, 19.14% of the S. americana genome, and more than 31.26% of the Phelipanche and Orobanche genomes. In terms of the overall abundance of repetitive DNA, Orobanchaceae appear to be in the mid-range of roughly similar-sized angiosperm genomes. For example, hundreds of families of transposable elements make up more than 85% of the H. vulgare and Zea mays genomes (Wicker et al. 2009; Schnable et al. 2009), with sizes of 5.55 and 2.73 pg, respectively, whereas M. acuminata has 30% highly or moderately repetitive DNA (Hribova et al. 2010), with a genome size of 0.63 pg, and P. sativum, 35%–55% highly or moderately repetitive DNA (Macas et al. 2007, Novak et al. 2010), with a genome size of 4.88 pg. Possibly, the large genome sizes of the seven nonphotosynthetic holoparasitic Orobanchaceae (compare with table 1) are due to whole genome or segmental duplications. At least one round of paleopolyploidzation has been suggested for Orobanche and Phelipanche based on the chromosome numbers (Schneeweiss, Palomeque et al. 2004).
Large Genomes in Parasitic Plants and within Orobanchaceae Genome Dynamics
The repetitive DNA proportions in the nine genomes, combined with the genome C-values, indicate that Orobanchaceae genomes are highly dynamic. The genomes of the autotrophic L. philippensis and the hemiparasite S. americana are much smaller than those of the holoparasitic Orobanche and Phelipanche, which fit the hypothesis of larger genome sizes in parasitic plants, possibly because they escape constraints imposed by root meristem growth rates in nonparasitic plants (Gruner et al. 2010). Although the C-values so far available for Orobanchaceae are insufficient to test the prediction that nonroot-developing plants have larger genomes, values from another parasitic plant family, the Convolvulaceae, fit the prediction (supplementary table 2; Mann–Whitney U test: P < 0.01). Interestingly, McNeal et al. (2007) reported that large genome sizes in holoparasites do not correlate with ploidy level.
Besides having large C-values, the holoparasites studied here also contain more repetitive DNA than do the photosynthetic Lindenbergia and Schwalbea (table 2). Conceivably, the evolution of (holo)parasitism in Orobanchaceae was accompanied by an increase in the fraction of repetitive DNA, an hypothesis that could be tested with a deeper sampling within Orobanchaceae, which contain several transitions from hemiparasitism (as in Schwalbea) to holoparasitism (Bennett and Mathews 2006). Lindenbergia philippensis and S. americana both contain fewer LTR retrotransposons than their parasitic relatives with only <3% Ty3/Gypsy elements, compared with 15.16%–28.34% in the parasites. They differ from each other in their Ty1/Copia element abundance (17.21 in L. philippensis vs. 8.09% in S. americana).
The repeats found in Orobanche and Phelipanche differ greatly (fig. 3), with as many as 50% of the repeat clusters being genus specific (fig. 2); disregarding autapomorphic clusters, this proportion increases to 65%. These results are in line with analyses in Oryza (Zuccolo et al. 2007), in which the pool of LTR retrotransposons is essentially conserved throughout the genus. The high proportion of genus-specific clusters (fig. 2) supports the hypothesis of differential genome dynamics in Orobanche and Phelipanche (Schneeweiss, Palomeque et al. 2004; Weiss-Schneeweiss et al. 2006; Park et al. 2007) and the emergence of new repeat families early in the radiation of the two genera. For example, both the two most abundant Ty3/Gypsy families and the second most abundant Ty1/Copia family from the combined data set are genus specific (fig. 3). Alternatively, differences between the two genera could result from differential amplification of repeat families present in the Orobanche–Phelipanche ancestor. An observation fitting with this is that the most abundant Ty1/Copia family (from the combined data set) is present in both genera and makes up 4.4% to 13.4% of the Phelipanche genomes but only 1% to 2.5% of the Orobanche genomes. The Phelipanche genomes also are more stable in terms of the genomic proportions composed of repetitive DNA (from 48% to 50%; supplementary table 1). There are no species-specific repeat families in the sister taxa P. lavandulacea and P. ramosa (fig. 2), so probably no recent bursts of transposition, although there is at least differential repeat amplification, for example, in P. purpurea, P. ramosa, and P. lavandulacea (figs. 2 and 3).
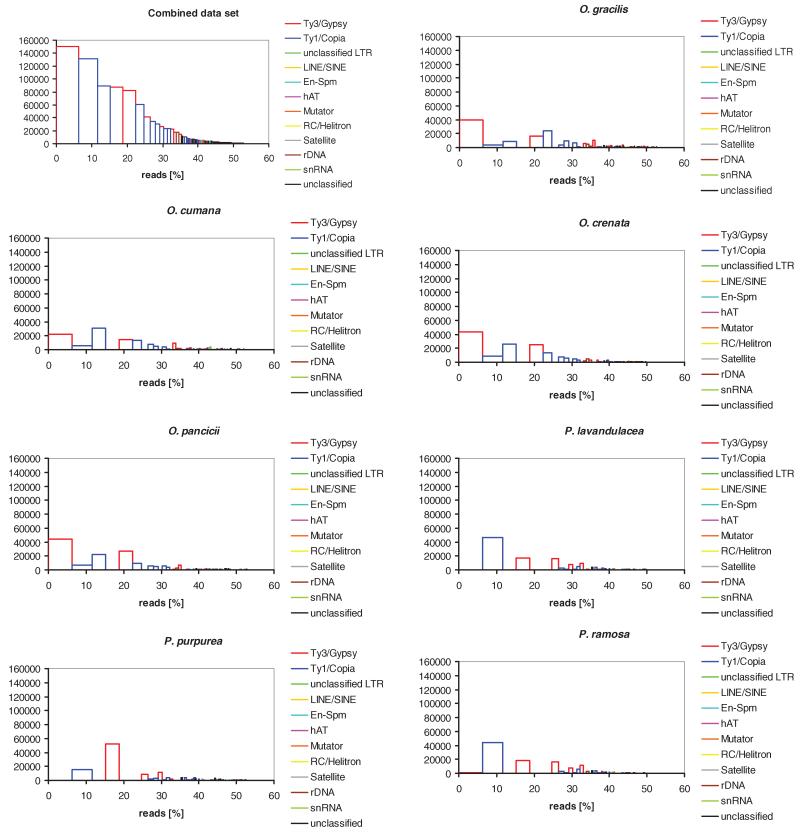
Fig. 3. Cluster size distributions in each species related to the master histogram from the combined data set clustering.
Clusters are sorted by decreasing size. x axis, genomic proportion (expressed as the read percentage); y axis, read number.
The “dynamics” of transposable elements is a complex concept involving factors such as transposition control mechanisms, removal rates of repeats, and environmental and genomic stresses. For example, transposable element elimination in Arabidopsis thaliana is more effective than in Arabidopsis lyrata (Hu et al. 2011), which provides an example of differential transposable element dynamics. Our study shows that although diploid Phelipanche species have 1.3–3× larger genomes than Orobanche species, they have lower proportions of high-copy repetitive DNA (table 2). This mostly results from an accumulation of Ty3/Gypsy retrotransposons in Orobanche that appears to be unrelated to any transposition “bursts” (table 2, supplementary fig. 2).
Genome Downsizing in the Tetraploid O. gracilis
Polyploidy in Orobanche is restricted to a few lineages and species, including the normally tetraploid O. gracilis and its relatives, the species of Orobanchaceae with the smallest known chromosomes (Schneeweiss, Palomeque et al. 2004). Previous studies have found that polyploidy can be associated with either selective amplification or loss of repetitive DNA (Parisod et al. 2010 for review), but an analysis of 1C values in over 3,000 diploid and polyploid angiosperm species suggested that genome downsizing in polyploids may be the rule (Leitch and Bennett 2004; Leitch and Leitch 2008). The O. gracilis monoploid genome size (Cx value), 1.05 pg, fits with genome downsizing in this species compared with the three diploid Orobanche species in this study (1.45, 2.84, and 3.24 pg for O. cumana, O. crenata, and O. pancicii, respectively; table 1).
As expected under the assumption of a stable proportion of repetitive DNA per diploid genome, O. gracilis has more highly or moderately repetitive DNA than any of the diploid species (table 2). The tetraploidization event, which occurred at an unknown time in the past, appears to have mostly involved accumulation of Ty3/Gypsy elements, which comprise 28.34% in O. gracilis but <24.16% in the other species. Orobanche gracilis also possesses a higher number of exclusive clusters (30 clusters; fig. 3; the same was found with the combined data set having the same coverage for each species; data not shown), suggesting a diversification of repeat families within its genome or that of the ancestor of the entire clade (fig. 1).
A loss of repetitive sequences due to polyploidization and genome downsizing has been reported from the young allopolyploid species Nicotiana tabacum (Renny-Byfield et al. 2011). These authors found that sequence loss affected most repeat types, but especially paternally derived repeats. In the tetraploid O. gracilis, we do not know whether the genome doubling arose from allopolyploidy or autopolyploidy. Perhaps, autopolyploidy is more likely, given that there are tetraploid and hexaploid populations, and even within-population variation in ploidy level (Greilhuber and Weber 1975; Schneeweiss, Palomeque et al. 2004). The analysis of the combined data sets revealed that >25% of the 89 clusters otherwise common in Orobanche are absent or extremely rare in O. gracilis. This proportion seems too high to be due to only stochastic losses. As observed in N. tabacum (Renny-Byfield et al. 2011), the polyploidization and subsequent genome downsizing in O. gracilis affected all repeat types and transposable elements (Ty1/Copia, Ty3/Gyspy, unclassified retrotransposons, RC/Helitron, satellites, and unclassified clusters) not only specific classes.
Conclusion
Our study constitutes the first analysis that maps changes in the abundance of all major repeats in a plant genome against a species phylogeny of a relatively densely sampled clade. It is also the first comparative genomic analysis of any parasitic angiosperm clade. The results reveal that highly or moderately repetitive DNA, mostly LTR retrotransposons, make up between 24.75% and 60.13% of the genomes, and repetitive DNAs appear to be the major contributors to genome size variation among the nine species. The genomic proportions consisting of repetitive DNA do not strictly correlate with genome size. Rather, the polyploid species studied here, O. gracilis, has one of the smallest genomes and one of the highest proportions of Ty3/Gypsy elements, yet underwent genome downsizing leading to the loss of numerous types of repeat clusters. The accumulation of Ty3/Gypsy retrotransposons in general appears to be related to a higher diversity of repetitive DNA types (families), rather than bursts of transposition as had been hypothesized (Park et al. 2007). Finally, the larger genomes of the obligatorily parasitic Orobanchaceae, compared with the autotrophic and hemiparasitic species in this family, fits a hypothesis of larger genome sizes in parasitic plants (Gruner et al. 2010), perhaps because of relaxed selection on root meristem growth rates or genomic “economy,” given that parasites obtain all their resources from the host.
Supplementary Material
Acknowledgments
The authors thank Aretuza Sousa for counting the chromosomes of Lindenbergia philippensis. This work was supported by German Science Foundation (RE 603/9-1), sequencing of the Lindenbergia and Schwalbea genomes was supported by the Austrian Science Foundation (FWF) grant P19404-B03 to G.M.S., and development of bioinformatics tools was supported by the grant AVOZ50510513 to J.M.
Footnotes
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J, Jean M, Belzile F. The allotetraploid Arabidopsis thaliana-Arabidopsis lyrata subsp. petraea as an alternative model system for the study of polyploidy in plants. Mol Genet Genomics. 2009;281:421–435. doi: 10.1007/s00438-008-0421-7. [DOI] [PubMed] [Google Scholar]
- Bennett JR, Mathews S. Phylogeny of the parasitic plant family Orobanchaceae inferred from phytochrome A. Am J Bot. 2006;93:1039–1051. doi: 10.3732/ajb.93.7.1039. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 2005;95:127–132. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Greilhuber J, Lucretti S, Meister A, Lysak MA, Nardi L, Obermayer R. Plant genome size estimation by flow cytometry: inter-laboratory comparison. Ann Bot. 1998;82(Suppl A):17–26. [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Flavell RB, Bennett MD, Smith JB, Smith DB. Genome size and proportion of repeated nucleotide-sequence DNA in plants. Biochem Genet. 1974;12:257–269. doi: 10.1007/BF00485947. [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E. Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science. 1983;220:1049–1051. doi: 10.1126/science.220.4601.1049. [DOI] [PubMed] [Google Scholar]
- Greilhuber J. “Self-tanning”—a new and important source of stoichiometric error in cytophotometric determination of nuclear DNA content in plants. Plant Syst Evol. 1988;158:87–96. [Google Scholar]
- Greilhuber J, Borsch T, Müller K, Worberg A, Porembski S, Barthlott W. Smallest angiosperm genomes found in Lentibulariaceae, with chromosomes of bacterial size. Plant Biol. 2006;8:770–777. doi: 10.1055/s-2006-924101. [DOI] [PubMed] [Google Scholar]
- Greilhuber J, Weber A. Aneusomaty in Orobanche gracilis. Plant Syst Evol. 1975;124:67–77. [Google Scholar]
- Gruner A, Hoverter N, Smith T, Knight CA. Genome size is a strong predictor of root growth meristem rate. J Bot. 2010 2010. [Google Scholar]
- Hawkins JS, Grover CE, Wendel JF. Repeated big bangs and the expanding universe: directionality in plant genome size evolution. Plant Sci. 2008;174:557–562. [Google Scholar]
- Hribova E, Neumann P, Matsumoto T, Roux N, Macas J, Dolezel J. Repetitive part of the banana (Musa acuminata) genome investigated by low-depth 454 sequencing. BMC Plant Biol. 2010;10:204. doi: 10.1186/1471-2229-10-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. 30 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH. Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics. 2004;166:1437–1450. doi: 10.1534/genetics.166.3.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LJ, Leitch IJ. Exploring giant plant genomes with next-generation sequencing technology. Chromosome Res. 2011;19:939–953. doi: 10.1007/s10577-011-9246-z. [DOI] [PubMed] [Google Scholar]
- Kondo K, Segawa L, Musselman J, Mann WF. Comparative ecological study of the chromosome races in certain root parasitic plants of the southeastern USA. Bol Soc Brot, sér. 1981;2:793–807. [Google Scholar]
- Leitch AR, Leitch IJ. Perspective—genomic plasticity and the diversity of polyploid plants. Science. 2008;320:481–483. doi: 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Leitch IJ, Bennett MD. Genome downsizing in polyploid plants. Biol J Linn Soc. 2004;82:651–663. [Google Scholar]
- Leitch IJ, Hanson L, Lim KY, Kovarik A, Chase MW, Clarkson JJ, Leitch AR. The ups and downs of genome size evolution in polyploid species of Nicotiana (Solanaceae) Ann Bot. 2008;101:805–814. doi: 10.1093/aob/mcm326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macas J, Neumann P, Navratilova A. Repetitive DNA in the pea (Pisum sativum L.) genome: comprehensive characterization using 454 sequencing and comparison to soybean and Medicago truncatula. BMC Genomics. 2007;8:427. doi: 10.1186/1471-2164-8-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manen JF, Habashi C, Jeanmonod D, Park JM, Schneeweiss GM. Phylogeny and intraspecific variability of holoparasitic Orobanche (Orobanchaceae) inferred from plastid rbcL sequences. Mol Phylogenet Evol. 2004;33:482–500. doi: 10.1016/j.ympev.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Manetti ME, Rossi M, Costa APP, Clausen AM, van Sluys MA. Radiation of the TntI retrotransposon superfamily in three Solanaceae genera. BMC Evol Biol. 2007;7:34. doi: 10.1186/1471-2148-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeal J, Arumugunathan K, Kuehl J, Boore J, dePamphilis C. Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae) BMC Biol. 2007;5:55. doi: 10.1186/1741-7007-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray MG, Peters DL, Thompson WF. Ancient repeated sequences in the pea and mung bean genomes and implications for genome evolution. J Mol Evol. 1981;17:31–42. [Google Scholar]
- Neumann P, Koblizkova A, Navrátilová A, Macas J. Significant expansion of Vicia pannonica genome size mediated by amplification of a single type of giant retroelement. Genetics. 2006;173:1047–1056. doi: 10.1534/genetics.106.056259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden AH, Kirkman LK. Factors controlling the fire-induced flowering response of the federally endangered Schwalbea americana L. (Scrophulariaceae) J Torrey Bot Soc. 2004;131:16–22. [Google Scholar]
- Novak P, Neumann P, Macas J. Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics. 2010;11:378. doi: 10.1186/1471-2105-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Oldiges H, Gohde W, Jain VK. Flow cytometric measurement of nuclear DNA content variations as a potential in vivo mutagenicity test. Cytometry. 1981;2:189–191. doi: 10.1002/cyto.990020311. [DOI] [PubMed] [Google Scholar]
- Parisod C, Alix K, Just J, Petit M, Sarilar V, Mhiri C, Ainouche M, Chalhoub B, Grandbastien MA. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Park JM, Manen JF, Colwell AE, Schneeweiss GM. A plastid gene phylogeny of the non-photosynthetic parasitic Orobanche (Orobanchaceae) and related genera. J Plant Res. 2008;121:365–376. doi: 10.1007/s10265-008-0169-5. [DOI] [PubMed] [Google Scholar]
- Park JM, Schneeweiss GM, Weiss-Schneeweiss H. Diversity and evolution of Ty1-copia and Ty3-gypsy retroelements in the non-photosynthetic flowering plants Orobanche and Phelipanche (Orobanchaceae) Gene. 2007;387:75–86. doi: 10.1016/j.gene.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Pertea G, Huang X, Liang F, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST data sets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. 12 co-authors. [DOI] [PubMed] [Google Scholar]
- Raskina O, Barber JC, Nevo E, Belyayev A. Repetitive DNA and chromosomal rearrangements: speciation-related events in plant genomes. Cytogenet Genome Res. 2008;120:351–357. doi: 10.1159/000121084. [DOI] [PubMed] [Google Scholar]
- Renny-Byfield S, Chester M, Kovařík A, et al. Next-generation sequencing reveals genome downsizing in allotetraploid Nicotiana tabacum, predominantly through the elimination of paternally derived repetitive DNAs. Mol Biol Evol. 2011;28:2843–2854. doi: 10.1093/molbev/msr112. 11 co-authors. [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. 156 co-authors. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM. Correlated evolution of life history and host range in the nonphotosynthetic parasitic flowering plants Orobanche and Phelipanche (Orobanchaceae) J Evol Biol. 2007;20:471–478. doi: 10.1111/j.1420-9101.2006.01273.x. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM, Colwell AE, Park JM, Jang CG, Stuessy TF. Phylogeny of holoparasitic Orobanche (Orobanchaceae) inferred from nuclear ITS-sequences. Mol Phylogenet Evol. 2004;30:465–478. doi: 10.1016/s1055-7903(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Schneeweiss GM, Palomeque T, Colwell AE, Weiss-Schneeweiss H. Chromosome numbers and karyotype evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2004;91:439–448. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Smit A, Hubley R, Green P. RepeatMasker Open-3.0. 1996 Cited 2011 Feb. Available from: http://www.repeatmasker.org.
- Spannagl M, Noubibou O, Haase D, Yang L, Gundlach H, Hindemitt T, Klee K, Haberer G, Schoof H, Mayer KF. MIPSPlantsDB—plant database resource for integrative and comparative plant genome research. Nucleic Acids Res. 2007;35(Database issue):D834–D840. doi: 10.1093/nar/gkl945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swaminathan K, Varala K, Hudson ME. Global repeat discovery and estimation of genomic copy number in a large, complex genome using a high-throughput 454 sequence survey. BMC Genomics. 2007;8:132. doi: 10.1186/1471-2164-8-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temsch EM, Greilhuber J, Krisai R. Genome size in liverworts. Preslia. 2010;82:63–80. [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 2010;15:471–478. doi: 10.1016/j.tplants.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Teryokhin ES. Weed Broomrapes. Aufstieg-Verlag; Landshut (Germany): 1997. [Google Scholar]
- Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- Weiss-Schneeweiss H, Greilhuber J, Schneeweiss GM. Genome size evolution in holoparasitic Orobanche (Orobanchaceae) and related genera. Am J Bot. 2006;93:148–156. doi: 10.3732/ajb.91.3.439. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. 13 co-authors. [DOI] [PubMed] [Google Scholar]
- Wicker T, Taudien S, Houben A, Keller B, Graner A, Platzer M, Stein N. A whole-genome snapshot of 454 sequences exposes the composition of the barley genome and provides evidence for parallel evolution of genome size in wheat and barley. Plant J. 2009;59:712–722. doi: 10.1111/j.1365-313X.2009.03911.x. [DOI] [PubMed] [Google Scholar]
- Wolfe AD, Randle CP, Liu L, Steiner KE. Phylogeny and biogeography of Orobanchaceae. Folia Geobot. 2005;40:115–134. [Google Scholar]
- Young ND, Steiner KE, dePamphilis CW. The evolution of parasitism in Scrophulariaceae/Orobanchaceae: plastid gene sequences refute an evolutionary transition series. Ann Miss Bot Gard. 1999;86:876–893. [Google Scholar]
- Zuccolo A, Sebastian A, Talag J, Yu Y, Kim H, Collura K, Kudrna D, Wing RA. Transposable element distribution, abundance and role in genome size variation in the genus Oryza. BMC Evol Biol. 2007;7:152. doi: 10.1186/1471-2148-7-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.