Abstract
Objectives
We evaluated whether pancreatic main duct fluid can provide protein biomarkers with prognostic value.
Methods
Mass spectrometry proteomics was applied to as little as 20 microliters of fluid collected at the time of tumor surgical resection. Biomarker proteins identified for 27 patients were correlated with clinical outcomes.
Results
Thirteen patients had pancreatic ductal adenocarcinomas (PDAC), 4 had IPMN with in situ adenocarcinoma, 5 had ampullary adenocarcinomas, 2 had IPMNs and 3 had benign diseases. In pathologic stage II or higher PDAC, moderate or high expression of S100A8 or S100A9 proteins was associated with a median disease recurrence-free survival of 5.8 months compared to 17.3 months in patients with low expression (p=0.002). Median overall survival was 12.6 versus 27 months for patients with moderate to high versus low S100A8 and A9 expression (p=0.02).
Conclusions
This analysis suggests distinct proteomic signatures for pancreatic cancer. Patients in our study with elevated levels of S100A8 or A9 in the ductal fluid, a near absence of pancreatic enzymes, and high levels of mucins, were found to have significantly worse prognosis. Although further validation is needed to corroborate these findings, analysis of pancreatic ductal fluid is a promising tool for identifying biomarkers of interest.
Keywords: PDAC, pancreatic duct fluid, cyst fluid, proteomic biomarkers, mucin, S100
Considerable effort has been made to identify biomarkers of interest in pancreatic cancer for diagnosis and prognosis. Pancreatic cancer is a deadly malignancy and the mainstay of treatment remains complete surgical resection. Unfortunately, most pancreatic cancers are diagnosed too late, with up to half of new diagnoses presenting with metastatic disease and an additional 20% with locally advanced disease.1 However, pancreatic lesions of unknown significance are increasingly being detected by imaging studies performed for other indications, and the management of these lesions can be controversial.2-4 The distinction between malignancy and benign disease is especially important in mucinous neoplasms, where the incidence of invasive carcinoma can range between 6% and 36%.5 The identification of a biomarker for prediction of invasive carcinoma may be useful for guiding management decisions.
The discovery of biomarkers for prognosis is also of interest for clinicians for identifying potential pathways as therapeutic targets in pancreatic cancer. Examples include the recognition of microRNA-21 and RAD51 expression as associated with 5-fluorouracil and gemcitabine resistance.6,7 Annexin II and S100A6 over-expression both relate to poor prognosis and shortened disease-free survival, via related mechanisms.8,9 The mapping of the interplay between protein expression and pathway up-regulation is key for our understanding of tumor biology and may lead us to novel therapeutic agents. For instance, triptolide, a diterpene triepoxide, has been shown to induce apoptosis in pancreatic cancer cells via targeting of heat shock protein 70, which is over-expressed in certain pancreatic cell lines.10 Investigations such as these are integral for the development of future chemotherapeutics against pancreatic cancer.
There are several sample sources employed in proteomic profiling of pancreatic cancer, including pancreatic cancer cell lines, xenograft models, human tumor blocks, serum, and pancreatic ductal fluid. We previously utilized SDS gel fractionation followed by digestion and HPLC-tandem mass spectrometry (GeLC-MS/MS) in identifying a proteomic protein panel in pancreatic cyst fluids that were aspirated via endoscopic ultrasound for diagnosis.11 This method, which was further validated by Western blotting, unveiled around 30 biomarker proteins, many of which were known to be related to pancreatic cancer. However, several novel homologs were discovered, such as mucin 5B and CEACAM 6 and 7, which were not previously associated with pancreatic cancer. We next applied GeLC-MS/MS to main pancreatic ductal fluids that were aspirated at the time of planned surgical resection for varying pancreatic pathologies. Here we present our results, with the identification of MUC homologs, amylase, and S100A8/A9 as biomarkers of particular interest.
MATERIALS AND METHODS
Sample Acquisition
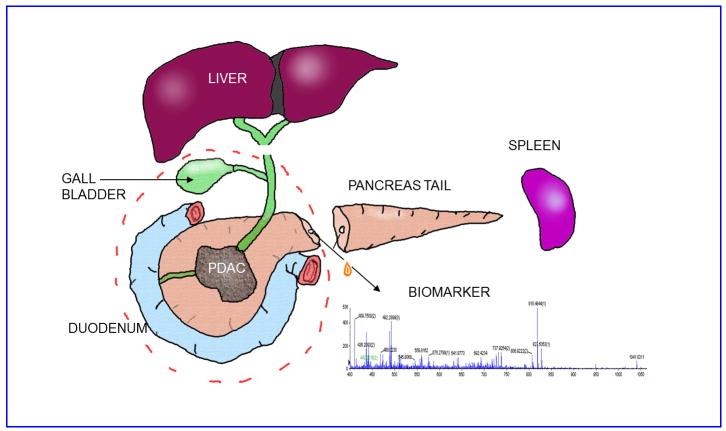
This study was approved by the Institutional Review Board of the Fox Chase Cancer Center. Patient information was blinded and de-identified prior to performing proteomics on the duct fluids. We prospectively aspirated pancreatic duct fluid at the time of surgical resection (Figure 1) performed for various pancreatic pathologies for 27 patients. All the patients were Caucasian. Briefly, the pancreatic neck or body was divided at time of resection, exposing the main pancreatic duct. A syringe with an attached plastic catheter was used to directly cannulate the duct to aspirate the fluid from either side of the cut, and the fluid was immediately put on ice. The volume of fluid obtained varied between 20 microliters to 5 mL. Each duct fluid sample was photographed, centrifuged for 10 min at 13,000 × g at 4 °C to remove cells and any insoluble materials, photographed again, and banked in aliquots at -80 °C. Patient information was deidentified prior to receipt by the proteomics laboratory.
FIGURE 1.
Diagram of pancreatic resection and main pancreatic duct fluid collection.
GeLC-MS/MS comparison of ductal protein expression
The mass spectrometer is effectively a high-resolution high-accuracy balance that can identify peptide fragments and sequences according to their unique masses. Individual protein concentration in a sample can be determined by mass spectrometry after resolution of the duct fluid proteins by SDS PAGE followed by trypsin digestion of each gel slice (GeLC-MS/MS). Reversed-phrase nanoHPLC provides the second round of fractionation so the tryptic peptides generated from each gel slice are ionized and introduced into a qTOF mass spectrometer wherein a third round of peptide resolution occurs. Counting of the number of tryptic peptides observed for a given protein is compared with the number of theoretical tryptic peptides that can be generated by that protein to lead to an approximate measure of protein quantity, a relative molar abundance value called emPAI score.12 This method is called GeLC-MS/MS as previously described in detail for cyst fluids.11 Fifteen microgram of duct fluid protein was used for each GeLC-MS/MS experiment. Protein determination was performed using the BioRad protein assay.13 The duct fluid protein was reduced with dithiothreitol (DTT) and alkylated with iodoacetamide (IAA) as previously described,13 then solubilized with lithium dodecylsulfate (LDS solution, Novex, Invitrogen) at 70°C for resolution by PAGE.11
Mass spectrometry was performed as follows: The peptide samples were acidified with 0.2% formic acid before being injected into a LC/MS/MS instrument QSTAR XL (Applied Biosystems/MDS Sciex, Foster City, CA). Agilent nano-HPLC (Agilent, Wilmington, DE) was equipped to interface the Q-TOF mass spectrometer. Samples were automatically loaded onto a C-18 trap column (ZORBAX 300SB-C18, 0.3 × 5 mm, 5 μm) and then eluted to a reversed-phase C-18 analytical column (ZORBAX 300SB-C18, 100 × 150 mm, 3.5 μm). The nanoflow was sprayed through a coated emitter (FS360-50-5-CE, New Objective Inc., Woburn, CA) at 300 nL/min. The spray voltage was set at +2200 V. The loading and washing steps took place within 4 min and then the system was switched to elution mode. The total analysis time for each injection was 115 min. Elution involved 5% of solution B at 0 to 4 min (A: 0.1 % formic acid. B: 0.1 % formic acid, 90 % acetonitrile), and then B was increased to 26 % at 8 min followed by 55 % at 80 min. In the next 5 min, B reached 100% and was maintained there for 10 min. Then the gradient was returned to starting position at 100 min. The system was equilibrated for 15 min at the end of the gradient.
The acquisition method of QSTAR was set at a 2 sec precursor “survey” scan followed by three 4 sec MS/MS scans. Parent ions with charge state of +2 or +3 and intensity above 15 counts were fragmented. The mass range for the “survey” scan was 400 to 1000 amu and was 100 to 2000 amu for MS/MS scan. For discovery of more proteins and peptides in each gel slice and to overcome the possibility of false-negatives due to under-sampling of co-eluting peptides, after the first LC/MS/MS run of a sample, an exclusion list composed of the peptides sequenced in the first LC/MS/MS run was assembled and used to direct another LC/MS/MS run to sequence only new peptides in the same sample. The two peak lists were combined for database searching for protein identification.
Data Analysis
The mass spectrometry data files were used to search the SwissProt Database Release 2010_09 using Mascot 2.2 (Matrix Science, London, U.K.), analyzing the MS/MS sequencing spectra of the +2 and +3 ions.11 Fixed modification of carbamidomethylcysteine, variable oxidation of methionine, and one trypsin miss-cleavage were allowed for protein identification but the latter two were disallowed for calculating the emPAI scores. Protein identification required at least two peptides with confidence over 95% of which one is not found in any other protein of the proteome (Mascot “BOLD RED”). Peptide mass tolerance was +/-150 ppm and fragment mass tolerance was 0.5 Da. False discovery rate due to coincidence in database was less than 3.5% for individual peptides as judged by hits at a decoy database containing randomized sequences in each entry. Data visualization was assisted by Scaffold 3 Q+ (Proteome Software).14
Patient information was subsequently unblinded and clinical results were analyzed to identify important questions for correlating with the proteomic data. In particular, pathology reports were reviewed by a surgeon and a pathologist. Patients were re-staged in accordance with the American Joint Commission on Cancer staging guidelines, 7th edition. Biomarker concentrations observed in the study were tested in correlation with various clinical parameters of interest. For our study, we evaluated overall survival (OS) and disease-free survival (DFS) in patients with S100A8 or S100A9 expression. Patients with a moderate expression of either S100A8 or S100A9 were considered positive for these biomarkers (i.e., emPAI score > 1) while patients with no expression or emPAI <1 were considered negative. Kaplan-Meier curves were computed for OS and DFS for the positive and negative sub-groups of patients identified as described above (Figure 2). The log-rank test was used to compare OS and DFS between these patient subgroups. A Type I Error of 5% was used to determine statistical significance.
FIGURE 2.
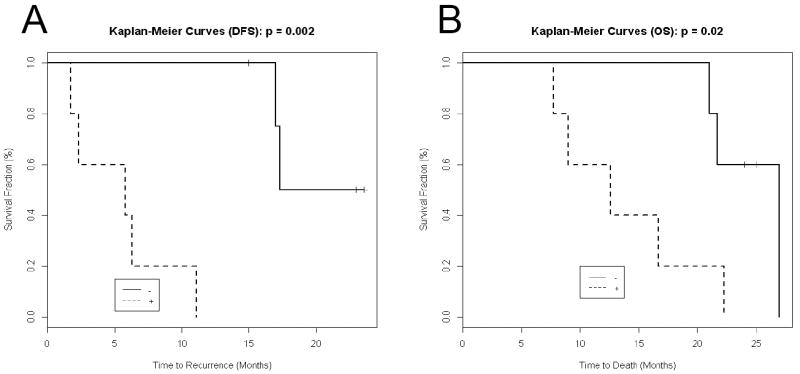
Survival of patients after pancreatic resection for tumors of grade T2 or above. A. Disease-free survival. B. Overall survival. Dotted lines denote patients #8-12 with high S100A8+A9 (combined emPAI score over 2), no amylase (except when IPMN was present) but high mucins (Table 1). Solid lines were patients #1-5 with low S100A8+A9 (combined emPAI score < 2), have amylase but no mucins (Table 1).
Concentrations of mucins and S100s
The S100A8 and A9 proteins form a heterodimer in vivo most of the time. Hence, we simplified expressing their relative concentrations in Table 1 as the sum of the emPAI scores of the two proteins. The same was done for the mucins 1, 2, 5AC, 5B, 6 and 13, collectively call mucins as the sum of their emPAI scores detected under our experimental conditions.
Table 1.
Patient information and biomarker levels.
| Duct fluid # | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Pathology Report | IPMN + ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Adeno-squamous CA |
| Stage | T2N1 | T2N1 | T3N0 | T3N1 | T3N1 | T3N1 | T3N0 |
| [protein] mg/mL | 0.5 | 3.4 | 1.7 | 1 | 1.5 | 0.5 | 0.7 |
| Serum albumin* | 4.3 | 5.5 | 7.9 | 6.2 | 3.1 | 6.6 | 7.9 |
| Amylase* | 2.9 | 5.0 | 2.2 | 2.5 | 2.5 | 2.2 | - |
| Mucins * | 1 | 0 | 0 | 4 | 0 | 7 | 30 |
| S100A8+A9* | - | 0.7 | - | - | - | - | - |
| Duct fluid # | 8 | 9 | 10 | 11 | 12 | 13 |
| Pathology Report | IPMN + ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA | Ductal adeno CA |
| Stage | T3N1 | yT1N0 | T3N1 | yT3N1 | T3N1 | yT0N0 |
| [protein] mg/mL | 1.3 | 1.2 | 2.8 | 4.6 | 1.4 | 6.4 |
| Serum albumin* | 7.9 | 8.8 | 6.6 | 5.9 | 9.3 | 7.9 |
| Amylase* | 2.1 | - | - | - | - | - |
| Mucins * | 11 | 71 | 10 | 10 | 18 | 12 |
| S100A8+A9* | 2.1 | 1.9 | 8.2 | 10.6 | 12.7 | 8.3 |
denotes numbers as emPAI score roughly proportional to molar protein concentration.
Amylase = Alpha-amylase 2B. DFS = disease-free survival in months, OS = overall survival.
Values of mucins were emPAI score x 100. Values of S100A8+A9 equal the sum of the emPAI scores for both proteins. “-“ = none detected.
RESULTS
Around 503 proteins were identified with confidence among the duct fluids in this study (Supplemental Digital Content). Similar to cyst fluids, duct fluids contain blood contamination and pancreatic digestive enzymes. In Tables 1 and 2, we found we can use serum albumin levels as a reasonable representative reporter for blood contamination (151 blood proteins were quantified in this study. See Supplemental Digital Content, rows in red color). Similarly, amylase is a good representative indicator for the level of pancreatic digestive enzymes present (30 digestive enzymes were quantified in this study. See Supplemental Digital Content, rows in green color). Mucin1, mucin 2, mucin 5AC, mucin 5B, mucin 6, and mucin 13 were quantified in this study (Tables 1 and 2). Complete proteome for each duct fluid is shown in Supplemental Digital Content.
Table 2.
Non-PDAC patients, relationship of patient information and biomarker levels.
| Duct fluid # | 14 | 15 | 16 | 17 | 18 | 19 | 20 |
|---|---|---|---|---|---|---|---|
| Pathology Report | Ampullary adeno CA | Ampullary adeno CA | Ampullary adeno CA | Ampullary adeno CA | Ampullary adeno CA | Serous cystadenoma | Serous cystadenoma |
| Stage | T2N0 | T2N0 | T3N0 | T3N0 | T3N1 | NA | NA |
| [protein] mg/mL | 4.6 | 2.5 | 3.9 | 2.5 | 11 | 13.3 | 23.1 |
| Serum albumin* | 7.9 | 7.0 | 2.1 | 4.3 | 3.1 | 1.1 | |
| Amylase* | 2.9 | 2.7 | 2.1 | 2.9 | 3.2 | 5.0 | 2.9 |
| Mucins * | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| S100A8+A9* | - | - | 1.2 | 0.7 | - | - | - |
| Duct fluid # | 21 | 22 | 23 | 24 | 25 | 26 | 27 |
| Pathology Report | Chronic pancreatitis | IPMN | IPMN | IPMN + in situ adeno CA | IPMN + foci adeno CA | IPMN + in situ adeno CA | IPMN + in situ adeno CA |
| Stage | NA | NA | NA | T0N0 | T1N0 | TisN0 | yTisN0 |
| [protein] mg/mL | 4.6 | 2.18 | 3.5 | 0.6 | 1.4 | 0. 8 | 1.7 |
| Serum albumin* | 2.6 | 5.9 | 7.9 | 8.8 | 7.9 | 7.9 | 5.5 |
| Amylase* | 1.4 | 3.2 | - | 3.4 | - | 1.9 | 3.4 |
| Mucins * | 0 | 54 | 43 | 5 | 24 | 3 | 13 |
| S100A8+A9* | 0.3 | 1.2 | 2.8 | 0.7 | 2.3 | 0.4 | - |
denotes numbers as emPAI score roughly proportional to molar protein concentration.
Amylase = Alpha-amylase 2B. DFS = disease-free survival in months, OS = overall survival.
Values of mucins were emPAI score x 100. Values of S100A8+A9 equal the sum of the emPAI scores for both proteins. “-“ = none detected.
The pathology diagnoses of the 27 patients are shown in Tables 1 and 2. Of note, nine patients had discordant pre-operative and post-operative diagnoses (data not shown), suggesting the urgency of discovering diagnostic and prognosis biomarkers.
Upon analysis of biomarker expression, we noted a distinct lack of abundance of biomarker proteins in the extrapancreatic pathologies (ampullary neoplasms) and benign pathologies (chronic pancreatitis and serous cystadenomas). Of this aggregate, the median number of biomarkers expressed is 0.0 (Supplemental Digital Content, rows in blue color in pages 4 and 5, Table 1). Conversely, the median number of biomarkers expressed by remaining pathologies (IPMN, pancreatic carcinomas) is 7. Two biomarker families were represented most prominently among the samples: mucins (MUC) and S100s.
When mucin homologs MUC1, MUC5AC and MUC5B were taken into consideration, we found that only 1 of 7 patients (14%) with extrapancreatic neoplasms and benign pancreatic disease expressed any MUC, versus 17 of 21 patients (81%) with pancreatic malignancies and IPMN (Tables 1 and 2). Of note, the one patient with ampullary adenocarcinoma with MUC5AC expression was found to have signet cells with mucinous features on pathology. Interestingly, all four patients that lacked MUC expression had PDAC. These four patients also fell into the cohort of PDAC patients who had improved overall survival, as seen below.
When analyzing IPMN and PDAC patients, two distinct groups emerged: those with moderate to high S100A8 or S100A9 expression (Table 1 #8-12) and those with low or no expression (Table 1 duct fluid #1-5). We found that in patients with pathologic stage II or above PDAC, expression of either S100A8 or S100A9 predicted a worse disease-free and overall survival. Two patients with in situ pancreatic adenocarcinoma (TisN0) arising from IPMN (Table 2 duct fluids #26 and 27) and one patient with no residual invasive adenocarcinoma seen in histologic sections following neoadjuvant chemoradiation (Table 1 duct fluid #13) were excluded from Kaplan-Meier analysis (Figure 2). Furthermore, one patient who died of post-operative complications was also excluded (Table 1 duct fluid #6). In the remaining patients, we found that moderate to high expression of S100A8 or S100A9 resulted in a median recurrence-free survival of 5.8 months compared to 17.3 months in those patients with low expression (p=0.002). Overall, median overall survival in patients with and without moderate to high expression of S100A8 or S100A9 was 12.6 months compared to 27 months (p=0.02) in patients with low expression of these biomarkers.
Pancreatic enzymes and mucins in duct fluids
Following the segregation of two groups of PDAC patients with different prognosis (Table 1), we noted that duct fluids #1-6 versus #8-13 also differed in the expression of pancreatic enzymes, represented by amylase, and in the opposite direction, mucins. Thus the duct fluid of one group (duct fluids #8-13) has high S100A8 and S100A9, a high level of mucins and an absence of pancreatic enzymes, represented by amylase, and has a less favorable prognosis. The second group of PDAC (duct fluids #1-7) has the opposite findings. Taken together, the biochemical evidence and prognostic data suggest that there may be two major groups of PDAC with different properties and prognosis.
DISCUSSION
The field of proteomic profiling for biomarker discovery for pancreatic cancer and other cancers has exploded in the past decade. The hope is to identify pathways that are up-regulated in cancer and target these pathways for diagnostic or therapeutic purposes. Previous profiling of pancreatic duct fluids have focused on validation of methods but have yielded hundreds of potentially interesting and cancer-associated biomarkers.15-17 The challenge remains to identify a biomarker, or a panel of biomarkers, with sufficient sensitivity and specificity to be of clinical value. We have previously described the utility of GeLC-MS/MS in proteomic profiling of pancreatic cyst fluids to unveil new homologs of biomarkers associated with pancreatic cancer.11 Here applying the same technique in pancreatic ductal fluid drawn directly at the time of surgery, we established a clinical correlation for the biomarkers identified. The increased expression of two families of biomarkers stood out: mucins for differentiating pancreatic malignancies and IPMN from other benign and extra-pancreatic neoplasms, and S100s for prognosticating overall and disease-free survival. Amylase is normally not thought of as a biomarker of PDAC. Surprisingly, loss of amylase, as a representative of the loss of pancreatic enzymes in the ductal fluids, is also associated with the worsening of the PDAC prognosis.
The expression of MUC5AC and MUC5B in the pancreas can be variable. IHC studies have demonstrated that MUC5B is expressed in normal pancreatic tissue, while both MUC5AC and MUC5B are expressed in pancreatic cancer.18,19 However, the significance of expression is controversial. Some studies suggested that MUC5AC-positive IPMN and PDAC patients have improved prognosis, while others suggested that MUC5AC expression enhances tumor cell invasive properties.20-22 MUC1, which has some prognostic value in IPMN, did not have a significant clinical role in our patients. Interestingly, although mucins are expressed in extrapancreatic malignancies (cholangiocarcinoma and ampullary carcinomas),23,24 only one of five ampullary adenocarcinomas in our study elaborated mucin (it is often difficult to distinguish the site of origin of cancers near the ampulla of Vater, and so this cancer could have well arisen from the pancreas). As expected, MUC4, which is specific to pancreatic cancer in IHC studies,25 was not identified in our duct fluid analysis, given that MUC4 is a membrane-bound mucin. This highlights the fact that tumor biomarker expression may be distinct from ductal fluid expression which may be secondary to the limits of detection or to different patterns of expression and secretion.
One initial observation in this report was the lack of digestive enzymes noted in some duct fluid samples (e.g.: duct fluids #9-13). Because all samples in this study were collected from the main pancreatic duct, we expected that all samples would contain digestive enzymes. We considered whether there was technical error in performing GeLC-MS/MS. However, for samples #9-13 which showed almost no digestive enzymes, represented by amylase, mucins were present and at levels as high as in other duct fluids that contained amylase (duct fluids #1-8), suggesting that GeLC-MS/MS was performed properly and that some samples truly had enzymes at levels below our limit of detection.
Pancreatic enzyme secretion is regulated by hormonal and nutrient signals26 from low basal levels to high levels after meals. The first 90 minutes of pancreatic secretion after food intake in humans appears to be via the release of storage zymogens followed by steady enzyme synthesis.27 In one report, infusion of 100 mM phenylalanine into the duodenum stimulated the secretion of trypsin, amylase and lipase. Further infusion of trypsin lowered this induction while inhibition of the trypsin in the duodenum returned the secretion of amylase and lipase to very low basal levels.28 The regulation of pancreatic enzyme release can also be altered by pancreatic diseases. In pancreatitis, the levels of amylase in the blood can be very high.29 For the PDAC cases with poor prognosis, it is possible that a reason for the low enzymes might be that the tumors had blocked the ducts such that no functioning acini were connected to the ducts from which the fluid was drawn, S100A8 and A9 had become elevated and mucins could still be secreted into the duct. Whether this explanation is sufficient or whether there are gene expression programs changed in the formation of this class of PDAC will be further studied.
We considered that in cases with chemoradiotherapy, the treatment can produce pancreatic atrophy. However, the chemoradiation treatment had no obvious effect on the biomarkers observed in the ductal fluids. This observation by itself may be interesting but its elucidation may require us to understand the origin of the biomarker proteins.
It is interesting that S100A8 and S100A9 were found to have some bearing on pancreatic cancer outcomes given that these two biomarkers are probably not expressed in pancreatic ductal cells but rather in stromal cells. The S100 family is postulated to be involved in inflammatory processes, including autoimmune diseases and a number of malignancies. This suggests that these inflammatory mediators may either enhance the cross-talk between tumor environment and tumor, or else may be sequelae of more aggressive disease. Although the one study examining IHC expression of S100A8 and S100A9 in pancreatic cancer tissue did not find a correlation with improved survival, no such study has been performed in ductal fluids to our knowledge. It is of interest to note that in colorectal cancer, S100A8 and S100A9 have been proposed as serologic biomarkers for diagnosis.30 S100A8 and S100A9 may also be involved in activating other downstream pathways via the multi-ligand RAGE receptor, including JAK/STAT and Ras, which are implicated in a number of cancers.31 As an aside, we considered the possibility that potential infection may lead to inflammation and rise in the levels of S100A8/A9. We thus reviewed the bile cultures of our PDAC cohort, which are obtained at the time of surgery, but there was no evident correlation with S100A8/A9 expression (data not shown).
For most clinicians, proteomic profiling of pancreatic ductal fluid remains of academic interest. There has been little information regarding correlation of ductal fluid biomarker expression and tumor IHC expression, the sensitivity or specificity of biomarker detection for disease or prognosis, or the relevance of biomarker expression. However, despite our small cohort of patients, we were able to identify two families of biomarkers of particular interest, mucin and S100. Together with whether pancreatic enzymes were absent in the duct fluids or not, our data suggested the possibility that there are two major groups of PDAC worth noting for patient treatment. We do believe that S100A8 and S100A9, in particular, deserve further study, given their ubiquity in other cancers and their potential as therapeutic targets. Future directions of interest may include the development of a serologic test for S100A8 and S100A9 in the setting of pancreatic cancer, analyzing other serologic markers of inflammation, and potentially evaluating the alteration of ductal fluid biomarker expression before and following neoadjuvant chemoradiation. That there may be at least two major groups of PDAC with different biomarkers and prognosis has many implications. The observations of the presence or not of amylase and mucins may each point to different cell types of origin of the cancer cells or changes in cell programming. Whether there are further differences in the genomes, in the locations of the two PDAC groups in the pancreas, whether the determinants lie in the cancer cells or in the stroma, are also topics of considerable interest for future studies.
Supplementary Material
Acknowledgments
Support by the Fox Chase Cancer Center Biosample Repository Facility, the Histopathology Facility and the Bioinformatics Facility are gratefully acknowledged. This study was supported by the Driskill Foundation, National Cancer Institute CA119242, Institutional Core Grant P30CA06927, and the Kresge Foundation.
The abbreviations used are
- GeLC-MS/MS
gel electrophoresis-HPLC-tandem mass spectrometry
- Amu
atomic mass unit
- PDAC
Pancreatic Ductal Adenocarcinoma
- IPMN
Intraductal papillary mucinous neoplasm
- MCN
mucinous cystic neoplasm
Footnotes
Conflict of interest: The authors declare that they have no conflict of interest
-
•Supplemental Digital Content. Table of the complete proteome of each ductal fluid. The composite proteome of all duct fluids and the complete proteome of each ductal fluid sample are detailed in this table.
Contributor Information
Kathryn T. Chen, Department of Surgical Oncology, Fox Chase Cancer Center, Philadelphia, PA 19111.
Phillip D. Kim, Developmental Therapeutics, Fox Chase Cancer Center, Philadelphia, PA 19111.
Kelly A. Jones, Developmental Therapeutics, Fox Chase Cancer Center, Philadelphia, PA 19111.
Karthik Devarajan, Department of Biostatistics, Fox Chase Cancer Center, Philadelphia, PA 19111.
Bhavinkumar B. Patel, Developmental Therapeutics, Fox Chase Cancer Center, Philadelphia, PA 19111.
John P. Hoffman, Department of Surgical Oncology, Fox Chase Cancer Center, Philadelphia, PA 19111.
Hormoz Ehya, Department of Pathology, Fox Chase Cancer Center, Philadelphia, PA 19111.
Min Huang, Department of Pathology, Fox Chase Cancer Center, Philadelphia, PA 19111.
James C. Watson, Department of Surgical Oncology, Fox Chase Cancer Center, Philadelphia, PA 19111.
Jeffrey L. Tokar, Department of Gastroenterology, Fox Chase Cancer Center, Philadelphia, PA 19111.
References
- 1.Wray CJ, Ahmad SA, Matthews JB, et al. Surgery for pancreatic cancer: recent controversies and current practice. Gastroenterology. 2005;128:1626–1641. doi: 10.1053/j.gastro.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 2.Handrich SJ, Hough DM, Fletcher JG, et al. The natural history of the incidentally discovered small simple pancreatic cyst: long-term follow-up and clinical implications. AJR Am J Roentgenol. 2005;184:20–23. doi: 10.2214/ajr.184.1.01840020. [DOI] [PubMed] [Google Scholar]
- 3.Hellstrom M, Svensson MH, Lasson A. Extracolonic and incidental findings on CT colonography (virtual colonoscopy) AJR Am J Roentgenol. 2004;182:631–638. doi: 10.2214/ajr.182.3.1820631. [DOI] [PubMed] [Google Scholar]
- 4.Sahani DV, Kadavigere R, Saokar A, et al. Cystic pancreatic lesions: a simple imaging-based classification system for guiding management. Radiographics. 2005;25:1471–1484. doi: 10.1148/rg.256045161. [DOI] [PubMed] [Google Scholar]
- 5.Testini M, Gurrado A, Lissidini G, et al. Management of mucinous cystic neoplasms of the pancreas. World J Gastroenterol. 2010;16:5682–5692. doi: 10.3748/wjg.v16.i45.5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hwang JH, Voortman J, Giovannetti E, et al. Identification of microRNA-21 as a biomarker for chemoresistance and clinical outcome following adjuvant therapy in resectable pancreatic cancer. PloS One. 2010;5:e10630. doi: 10.1371/journal.pone.0010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagathihalli NS, Nagaraju G. RAD51 as a potential biomarker and therapeutic target for pancreatic cancer. Biochim Biophys Acta. 2011;1816:209–218. doi: 10.1016/j.bbcan.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Takano S, Togawa A, Yoshitomi H, et al. Annexin II overexpression predicts rapid recurrence after surgery in pancreatic cancer patients undergoing gemcitabine-adjuvant chemotherapy. Ann Surg Oncol. 2008;15:3157–3168. doi: 10.1245/s10434-008-0061-5. [DOI] [PubMed] [Google Scholar]
- 9.Nedjadi T, Kitteringham N, Campbell F, et al. S100A6 binds to annexin 2 in pancreatic cancer cells and promotes pancreatic cancer cell motility. Br J Cancer. 2009;101:1145–1154. doi: 10.1038/sj.bjc.6605289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips PA, Dudeja V, McCarroll JA, et al. Triptolide induces pancreatic cancer cell death via inhibition of heat shock protein 70. Cancer Res. 2007;67:9407–9416. doi: 10.1158/0008-5472.CAN-07-1077. [DOI] [PubMed] [Google Scholar]
- 11.Ke E, Patel BB, Liu T, et al. Proteomic analyses of pancreatic cyst fluids. Pancreas. 2009;38:e33–42. doi: 10.1097/MPA.0b013e318193a08f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishihama Y, Oda Y, Tabata T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Li XM, Patel BB, Blagoi EL, et al. Analyzing alkaline proteins in human colon crypt proteome. J Proteome Res. 2004;3:821–833. doi: 10.1021/pr049942j. [DOI] [PubMed] [Google Scholar]
- 14.Searle BC. Scaffold: a bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 2010;10:1265–1269. doi: 10.1002/pmic.200900437. [DOI] [PubMed] [Google Scholar]
- 15.Chen R, Pan S, Yi EC, et al. Quantitative proteomic profiling of pancreatic cancer juice. Proteomics. 2006;6:3871–3879. doi: 10.1002/pmic.200500702. [DOI] [PubMed] [Google Scholar]
- 16.Gronborg M, Bunkenborg J, Kristiansen TZ, et al. Comprehensive proteomic analysis of human pancreatic juice. J Proteome Res. 2004;3:1042–1055. doi: 10.1021/pr0499085. [DOI] [PubMed] [Google Scholar]
- 17.Tian M, Cui YZ, Song GH, et al. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer. 2008;8:241. doi: 10.1186/1471-2407-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vincent A, Perrais M, Desseyn JL, et al. Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene. 2007;26:6566–6576. doi: 10.1038/sj.onc.1210479. [DOI] [PubMed] [Google Scholar]
- 19.Balague C, Audie JP, Porchet N, et al. In situ hybridization shows distinct patterns of mucin gene expression in normal, benign, and malignant pancreas tissues. Gastroenterology. 1995;109:953–964. doi: 10.1016/0016-5085(95)90406-9. [DOI] [PubMed] [Google Scholar]
- 20.Jinfeng M, Kimura W, Hirai I, et al. Expression of MUC5AC and MUC6 in invasive ductal carcinoma of the pancreas and relationship with prognosis. Int J Gastrointest Cancer. 2003;34:9–18. doi: 10.1385/IJGC:34:1:09. [DOI] [PubMed] [Google Scholar]
- 21.Yamazoe S, Tanaka H, Sawada T, et al. RNA interference suppression of mucin 5AC (MUC5AC) reduces the adhesive and invasive capacity of human pancreatic cancer cells. J Exp Clin Cancer Res. 2010;29:53. doi: 10.1186/1756-9966-29-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoshi H, Sawada T, Uchida M, et al. Tumor-associated MUC5AC stimulates in vivo tumorigenicity of human pancreatic cancer. Int J Oncol. 2011;38:619–627. doi: 10.3892/ijo.2011.911. [DOI] [PubMed] [Google Scholar]
- 23.Mall AS, Tyler MG, Ho SB, et al. The expression of MUC mucin in cholangiocarcinoma. Pathol Res Pract. 2010;206:805–809. doi: 10.1016/j.prp.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Van Heek NT, Maitra A, Koopmann J, et al. Gene expression profiling identifies markers of ampullary adenocarcinoma. Cancer Biol Ther. 2004;3:651–656. doi: 10.4161/cbt.3.7.919. [DOI] [PubMed] [Google Scholar]
- 25.Andrianifahanana M, Moniaux N, Schmied BM, et al. Mucin (MUC) gene expression in human pancreatic adenocarcinoma and chronic pancreatitis: a potential role of MUC4 as a tumor marker of diagnostic significance. Clin Cancer Res. 2001;7:4033–4040. [PubMed] [Google Scholar]
- 26.Liddle RA. Regulation of cholecystokinin secretion by intraluminal releasing factors. Am J Physiol. 1995;269(3 Pt 1):G319–327. doi: 10.1152/ajpgi.1995.269.3.G319. [DOI] [PubMed] [Google Scholar]
- 27.O’Keefe SJ, Bennet WM, Zinsmeister AR, et al. Pancreatic enzyme synthesis and turnover in human subjects. Am J Physiol. 1994;266(5 Pt 1):G816–821. doi: 10.1152/ajpgi.1994.266.5.G816. [DOI] [PubMed] [Google Scholar]
- 28.Dlugosz J, Folsch UR, Czajkowski A, et al. Feedback regulation of stimulated pancreatic enzyme secretion during intraduodenal perfusion of trypsin in man. Eur J Clin Invest. 1988;18:267–272. doi: 10.1111/j.1365-2362.1988.tb01257.x. [DOI] [PubMed] [Google Scholar]
- 29.Sunamura M, Lozonschi L, Takeda K, et al. Criteria for diagnosis of acute pancreatitis in Japan and clinical implications. Pancreas. 1998;16:243–249. doi: 10.1097/00006676-199804000-00007. [DOI] [PubMed] [Google Scholar]
- 30.Kim HJ, Kang HJ, Lee H, et al. Identification of S100A8 and S100A9 as serological markers for colorectal cancer. J Proteome Res. 2009;8:1368–1379. doi: 10.1021/pr8007573. [DOI] [PubMed] [Google Scholar]
- 31.Gebhardt C, Nemeth J, Angel P, et al. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.