Emerin, a nuclear membrane protein, and myosin IIB contribute to nuclear movement by regulating the directionality of nuclear movement and dorsal actin cable flow. Emerin interacts with myosin IIB and is required for its perinuclear localization. The results show that the nuclear envelope actively organizes cytoplasmic polarity.
Abstract
In migrating fibroblasts, rearward movement of the nucleus orients the centrosome toward the leading edge. Nuclear movement results from coupling rearward-moving, dorsal actin cables to the nucleus by linear arrays of nesprin-2G and SUN2, termed transmembrane actin-associated nuclear (TAN) lines. A-type lamins anchor TAN lines, prompting us to test whether emerin, a nuclear membrane protein that interacts with lamins and TAN line proteins, contributes to nuclear movement. In fibroblasts depleted of emerin, nuclei moved nondirectionally or completely failed to move. Consistent with these nuclear movement defects, dorsal actin cable flow was nondirectional in cells lacking emerin. TAN lines formed normally in cells lacking emerin and were coordinated with the erratic nuclear movements, although in 20% of the cases, TAN lines slipped over immobile nuclei. Myosin II drives actin flow, and depletion of myosin IIB, but not myosin IIA, showed similar nondirectional nuclear movement and actin flow as in emerin-depleted cells. Myosin IIB specifically coimmunoprecipitated with emerin, and emerin depletion prevented myosin IIB localization near nuclei. These results show that emerin functions with myosin IIB to polarize actin flow and nuclear movement in fibroblasts, suggesting a novel function for the nuclear envelope in organizing directional actin flow and cytoplasmic polarity.
INTRODUCTION
Nuclear positioning is an active, regulated process that functions in cellular and developmental events, including fertilization and cell division, migration, and differentiation (Wilhelmsen et al., 2006; Starr, 2009; Starr and Fridolfsson, 2010; Gundersen and Worman, 2013). In migrating cells, the nucleus is usually positioned rearward of the cell centroid (Luxton and Gundersen, 2011; Gundersen and Worman, 2013). In many cases, this posterior positioning is achieved by active movement of the nucleus before forward migration of the cell (Gomes et al., 2005; Desai et al., 2009; Luxton et al., 2010; Dupin et al., 2011). Interfering with normal nuclear positioning reduces cell migration, suggesting that proper nuclear positioning is crucial for efficient cell migration.
Many nuclear movements are driven by the microtubule cytoskeleton, yet there is growing recognition that actin also powers nuclear movement (Starr and Fridolfsson, 2010; Dupin and Etienne-Manneville, 2011; Luxton et al., 2011; Gundersen and Worman, 2013). Actin-dependent nuclear movement occurs when moving actin cables couple to the nucleus via the linker of nucleus and cytoskeleton (LINC) complex, which consists of outer nuclear membrane nesprins and inner nuclear membrane SUNs anchored to the nuclear lamina (Crisp et al., 2006; Östlund et al., 2009; Starr and Fridolfsson, 2010; Folker et al., 2011). In fibroblasts polarizing for migration, nesprin-2G (the giant isoform) and SUN2 assemble into linear arrays termed transmembrane actin-associated nuclear (TAN) lines, which couple the nucleus to dorsal actin cables that move retrogradely (Luxton et al., 2010, 2011; Folker et al., 2011). SUN2 and A-type lamins (lamin A/C) anchor TAN lines, allowing force exerted by moving actin cables to move the nucleus (Folker et al., 2011). Nesprin-1 and nesprin-2 have also been shown to contribute to nuclear positioning events in muscle cells and neurons (Zhang et al., 2007b, 2009, 2010), and, at least in certain cases, actin has also been implicated in nuclear positioning events in these cells (Norden et al., 2009; Schenk et al., 2009; Solecki et al., 2009).
TAN lines resemble adhesive structures on the plasma membrane, such as integrin-based focal adhesions and cadherin-based cell–cell junctions, in that they are composed of integral membrane proteins and formed in response to actin filaments and myosin contractility. In addition to integral membrane proteins, focal adhesion and cell–cell junctions contain many accessory proteins that regulate the strength and dynamics of the adhesion and perform signaling roles. Similar to these other adhesive structures, TAN lines are likely to be composed of more than just the main structural proteins nesprin-2G and SUN2. Indeed, SAMP-1, an inner nuclear membrane protein that contributes to SUN1 and emerin nuclear envelope localization (Gudise et al., 2011), was recently localized to TAN lines and shown to be necessary for nuclear movement in fibroblasts (Borrego-Pinto et al., 2012).
Emerin interacts with SAMP1 and associates with LINC complex components nesprin-1, nesprin-2 (Mislow et al., 2002; Zhang et al., 2005), and SUN1/2 (Haque et al., 2010). In addition, emerin binds actin-pointed ends (Fairley et al., 1999; Holaska et al., 2004). Emerin was originally described as an integral inner nuclear membrane protein lacking in most cases of X-linked Emery–Dreifuss muscular dystrophy due to mutations in the EMD gene (Bione et al., 1994; Manilal et al., 1996; Nagano et al., 1996). Emerin was later shown to interact with lamin A/C (Fairley et al., 1999; Sullivan et al., 1999; Clements et al., 2000), which contribute to the anchoring of TAN lines (Folker et al., 2011). Whereas emerin concentrates in the inner nuclear membrane in cells containing lamin A/C, a pool of emerin is also localized in the outer nuclear membrane, where it has been implicated in controlling spacing between the nucleus and the centrosome (Salpingidou et al., 2007). On the basis of these findings, we hypothesized that emerin contributes to TAN line function and the rearward movement of nuclei that reorients the centrosome in 3T3 fibroblasts polarizing for migration. Surprisingly, we found that emerin does not play a major role in TAN line formation or function but instead affects nuclear movement by ensuring the directional flow of actin cables toward the rear of the cell.
RESULTS
Emerin functions in nuclear movement
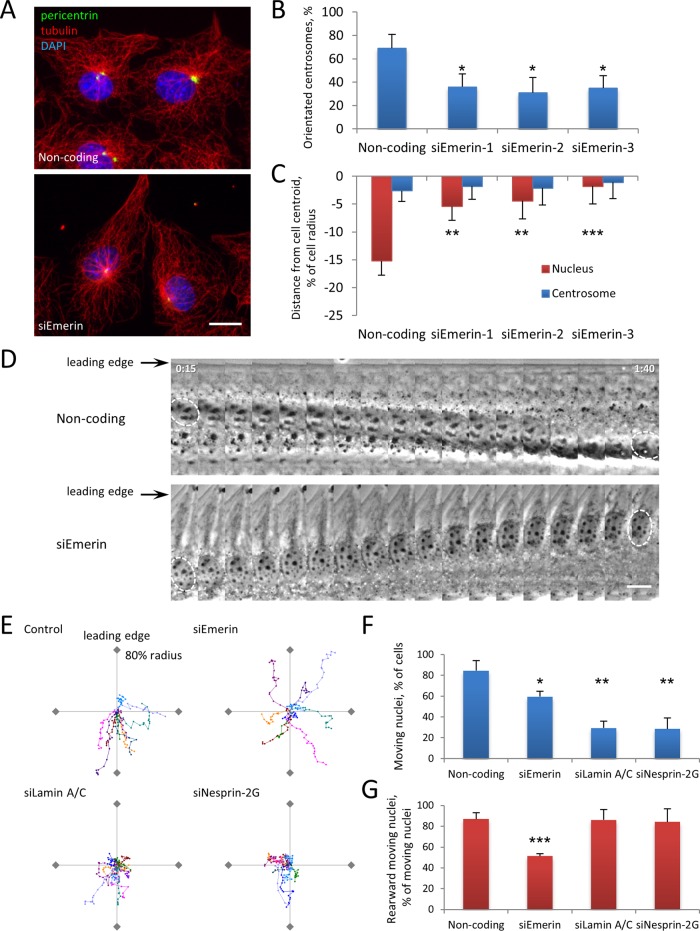
Lysophosphatidic acid (LPA) stimulation of serum-starved, wounded monolayers of NIH3T3 fibroblasts activates two Cdc42 pathways that together orient the centrosome to a position between the nucleus and leading edge: an actin- and myosin-dependent rearward movement of the nucleus and a microtubule- and dynein-dependent maintenance of the centrosome at the cell center (Palazzo et al., 2001; Gomes et al., 2005; Schmoranzer et al., 2009). To test the role of emerin in nuclear movement, we reduced its expression in NIH3T3 fibroblasts by using small interfering RNAs (siRNAs) and then stimulated cells with LPA. Multiple siRNA oligonucleotides targeting emerin efficiently reduced its expression (Supplemental Figure S1) and inhibited LPA-stimulated centrosome orientation compared with cells transfected with a noncoding siRNA (Figure 1, A and B). Lack of centrosome orientation in emerin-depleted cells was due to a failure of rearward nuclear positioning; no effect was observed on centrosome positioning at the cell centroid (Figure 1, A and C). Similar results were observed in emerin-null mouse embryo fibroblasts (MEFs; Supplemental Figure S2).
FIGURE 1:
Emerin is required for centrosome orientation and nuclear movement. (A) Representative images of LPA-stimulated NIH3T3 cells transfected with noncoding or emerin siRNA (siEmerin) and immunostained for tubulin (red), the centrosomal marker pericentrin (green), and DAPI for nuclei (blue). (B) Quantification of LPA-stimulated centrosome orientation in NIH3T3 cells treated with noncoding siRNA or three different siRNAs against emerin as indicated. Centrosome orientation between the leading edge and nucleus was scored as described previously (Palazzo et al., 2001); random orientation is 33% by this measure. Error bars, SD from three experiments (N > 120 cells). (C) Quantification of nucleus and centrosome position in emerin-depleted NIH3T3 cells treated with noncoding siRNA or three different siRNAs against emerin as indicated. The cell centroid is defined as 0; positive values, toward the leading edge; negative, away. Error bars, SEM from three experiments (N ≥ 100 cells). Centrosome position was not affected by siEmerin (p > 0.9, ANOVA). (D) Kymographs from representative phase contrast movies of LPA-stimulated NIH3T3 cells treated with noncoding or emerin siRNAs. Nuclei are outlined in the first and last frames. Note the rearward-moving nucleus in noncoding siRNA control (top) and the forward-moving nucleus in emerin siRNA–treated cells (bottom). Time, hours:minutes after LPA stimulation; each panel represents 5 min. (E) Representative traces of LPA-stimulated nuclear movement in NIH3T3 cells treated with the indicated siRNAs. Ten traces are plotted with a common origin. Axes represent 80% of cell radius; leading edge is at the top of the y-axis. Points represent 5 min; total time, 90 min. (F, G) Quantification of moving nuclei (F) and percentage of moving nuclei that moved rearward (G) from phase contrast movies of LPA-stimulated NIH3T3 cells treated with indicated siRNA. Error bars, SD from three experiments (N ≥ 60 cells). In B,C, F, and G statistical significance is compared with noncoding siRNA. Bars, 10 μm (A, D).
Live-cell imaging revealed that LPA-induced nuclear movement was inhibited in many emerin-depleted cells, but in others nuclei moved in unusual directions, including toward the leading edge (Figure 1, D–F). Traces of nuclear paths showed that most nuclei moved rearward in control cells, whereas they either did not move or moved randomly relative to the front–back axis in emerin-depleted cells (Figure 1E). Quantification showed a significant decrease in the number of moving nuclei and the number of nuclei that moved rearward in emerin-depleted cells (Figure 1, F and G). Whereas emerin depletion inhibited rearward nuclear movement to a similar degree as lamin A/C or nesprin-2G depletion, more nuclei moved randomly in emerin-depleted cells (Figure 1, E–G). Consistent with the defects in centrosome orientation and nuclear movement, we observed that emerin depletion resulted in decreased migration into wounded monolayers (Supplemental Figure S3). Thus emerin functions in centrosome orientation and nuclear movement but may do so via a mechanism distinct from that of nesprin-2G or lamin A/C.
Actin flow is random in cells lacking emerin
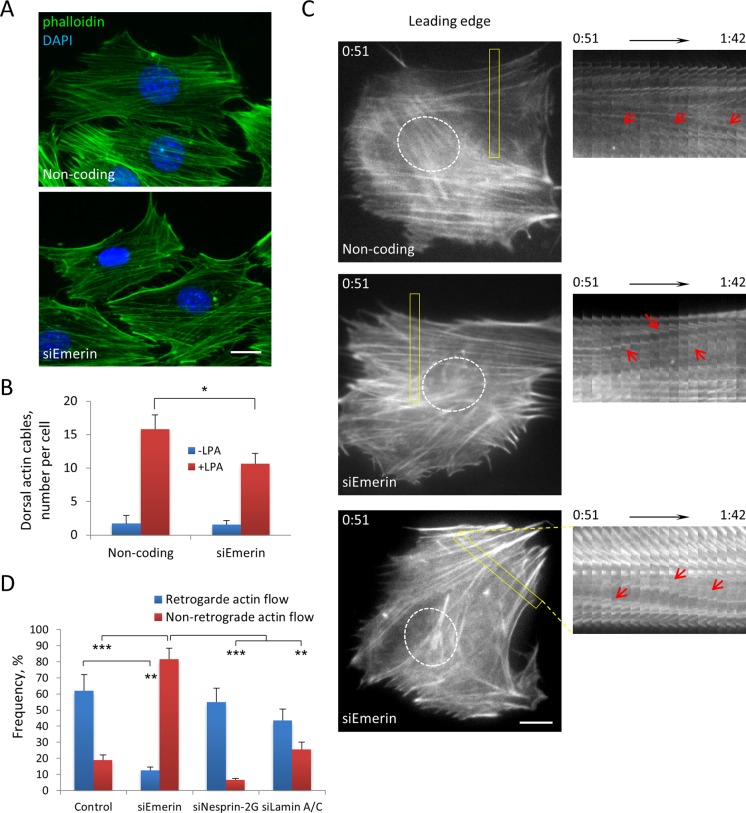
The abnormal movements of nuclei in emerin-depleted cells suggested that emerin might affect actin retrograde flow. Consistent with a previous study (Hale et al., 2008), we found that global F-actin organization appeared similar in LPA-stimulated control and emerin-depleted cells, although the number of dorsal actin cables observed over nuclei was reduced by about one-third (Figure 2, A and B). In contrast to the relatively modest changes in actin filament organization observed in fixed cells, live-cell imaging of actin with mCherry-LifeAct (Riedl et al., 2008) revealed dramatic alterations in the behavior of actin cables in emerin-depleted cells. In control cells, actin cables in the leading lamella moved retrogradely from near the leading edge toward the cell center (Figure 2C and Supplemental Movie S1). In contrast, in emerin-depleted cells, actin cables in the leading lamella moved in a manner that was not oriented along the front–back axis: they moved forward or obliquely and originated from sites other than the leading edge (Figure 2, C and D, and Supplemental Movies S2 and S3). These actin behaviors were rarely observed in control cells (Figure 2D). As we reported previously (Luxton et al., 2010), actin cables flowed retrogradely in nesprin-2G–depleted cells (Figure 2D), indicating that unoriented actin flow was not caused by inhibition of nuclear movement per se. Emerin and lamin A/C were recently found to regulate the MKL1-SRF transcriptional pathway by modulating actin dynamics (Ho et al., 2013). However, depletion of lamin A/C did not induce unoriented nuclear movement (Figure 1, E–G) and only increased random actin flow to a small extent that was not statistically significant (Figure 2D). This indicated that disrupting the MKL1-SRF pathway was not sufficient to cause the actin flow phenotypes caused by emerin depletion.
FIGURE 2:
Emerin affects retrograde flow. (A) Representative fluorescence images of F-actin (phalloidin) and nuclei (DAPI) in NIH3T3 cells transfected with noncoding or emerin siRNAs and stimulated with LPA. (B) Quantification of number of dorsal actin cables localized above the nucleus in cells treated with indicated siRNAs. Error bars, SD from three experiments with N ≥ 90 cells. C) Left, panels from movies of LifeAct-mCherry in NIH3T3 cells transfected with noncoding and emerin (two examples) siRNAs and stimulated with LPA. The position of the nucleus is shown by the dotted outline. Right, kymographs of boxed regions shown on the left. Red arrows, moving actin cables. Note that actin cables move retrogradely in noncoding siRNA control (top) and anterogradely (middle) or obliquely (bottom) in siEmerin. Time is in hours:minutes. D) Categorization of actin flow types (see Materials and Methods) from movies of Lifeact-mCherry–expressing cells treated with indicated siRNAs. Error bars, SD from three experiments (N ≥ 60 movies). Retrograde actin flow was significantly different from control in siEmerin but not in siNesprin-2 G or siLamin A/C. Nonretrograde actin flow was significantly different in siEmerin compared with control and siNesprin-2G and siLamin. Bars, 10 μm (A, C).
TAN lines form but infrequently move with nuclei in emerin-depleted cells
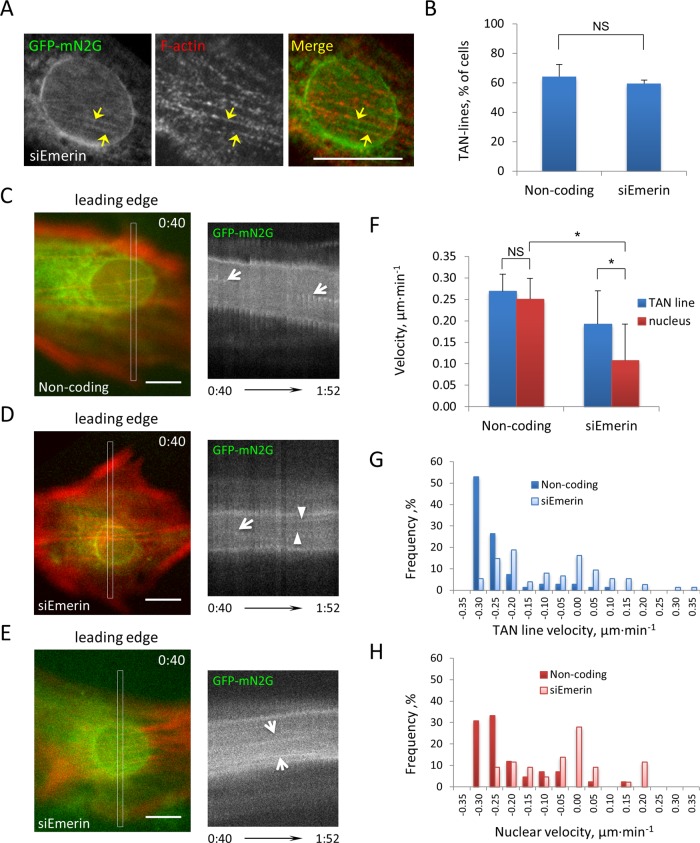
Given that some nuclei moved in emerin-depleted cells, we tested whether TAN lines still coupled actin to the nucleus upon emerin depletion. We expressed green fluorescent protein (GFP)-tagged mini-nesprin-2G (GFP-mN2G), a chimeric nesprin-2G construct containing the N-terminus actin-binding domain and the C-terminus KASH domain, in emerin-depleted cells to visualize TAN lines (Luxton et al., 2010). Depleting emerin did not affect LPA-induced TAN line formation (Figure 3, A and B). Localization of emerin with GFP-mN2G showed that emerin accumulated in TAN lines in only a small percentage of the cells (∼20%), and even in these cells emerin accumulated in only a subset of the TAN lines (Supplemental Figure S4). These results suggest that emerin is not a major structural component of TAN lines and does not directly contribute to their formation.
FIGURE 3:
TAN lines form in cells lacking emerin. (A) Representative fluorescence image of a nucleus in an LPA-stimulated NIH3T3 cell treated with emerin siRNA and expressing GFP-mN2G and stained for GFP, emerin, and F-actin. Arrows, TAN lines. (B) Quantification of TAN line formation in noncoding and emerin siRNA–treated NIH3T3 cells expressing GFP-mN2G. Error bars, SD from three experiments (N > 60 cells). siEmerin vs. noncoding, p > 0.3. (C–E) Left, panels from movies of LifeAct-mCherry and GFP-mN2G in NIH3T3 cells transfected with noncoding (C) and emerin (D, E) siRNAs and stimulated with LPA. Right, kymographs of GFP-mN2G from boxed regions shown on the left. Arrows, diagonal signals indicating retrograde (C, D) and anterograde (E) movement of TAN lines; arrowheads, horizontal signals (D) indicating immobile TAN lines. TAN lines moved with the moving nucleus in C and E but either slip (arrow) or are immobile (arrowheads) on the immobile nucleus in D. Time, hours:minutes (bottom). (F) Quantification of TAN line and nuclear velocities in cells treated with the indicated siRNAs. Error bars, SD from three experiments (N > 60 cells). (G, H) Frequency plots of TAN line and nuclear movement velocities. Positive velocity indicates forward movement, and negative velocity indicates rearward movement. Note that in emerin-depleted cells TAN line and nuclear velocities exhibited greater variation as well as positive values, indicating movement in a forward direction. Bars, 10 μm (A, C–E).
We next tested whether emerin affected TAN line coupling to move the nucleus. GFP-mN2G and mCherry-LifeAct were coexpressed to visualize TAN lines and actin cables, respectively. In control cells, as expected (Luxton et al., 2010; Folker et al., 2011), LPA-induced TAN lines moved with the nucleus (Figure 3C). In emerin-depleted cells, we observed distinct behaviors of the TAN lines depending whether or not the nucleus moved. In cells with nonmoving nuclei, some TAN lines failed to move, whereas others moved over an immobile nucleus, indicating slippage of TAN lines (Figure 3D and Supplemental Movie S4). In cells in which nuclei moved toward the leading edge, TAN lines moved forward with the nucleus (Figure 3E). Overall, TAN lines moved more slowly in emerin-depleted cells than in control cells (reflecting the many nonmoving nuclei in emerin-depleted cells) but faster than nuclei, reflecting the slippage of some TAN lines over nonmoving nuclei (Figure 3F). Although the maximal velocities were the same in emerin-depleted and control cells, both TAN line and nuclear movement exhibited larger variations in emerin-depleted cells, including forward movement, which was rarely seen in control cells (Figure 3, G and H). These results indicate that the lack of directional actin cable flow in emerin-depleted cells resulted in abnormal TAN line coupling to the nucleus and hence lack of efficient nuclear movement; in those cases in which nuclei moved (even if in an anterograde direction), TAN line coupling to the nucleus appeared normal.
A pool of emerin is localized in the outer nuclear membrane during nuclear movement
Emerin is primarily localized to the inner nuclear membrane, but a pool of emerin has been detected in the outer nuclear membrane (Salpingidou et al., 2007). To test whether there is a pool of emerin in the outer nuclear membrane during nuclear movement in NIH3T3 fibroblasts, we extracted cells with ice-cold digitonin before fixation and immunofluorescence. Cold digitonin extraction permeabilizes the plasma membrane but not the nuclear envelope membranes (Adam et al., 1990). Consistent with the earlier report (Salpingidou et al., 2007), we detected a pool of emerin in the outer nuclear membrane of 3T3 fibroblasts during nuclear movement (Supplemental Figure S5). Thus emerin may access cytoplasmic factors, including those that control actin cable movement.
Emerin actin binding is not required for centrosome orientation and nuclear movement
Emerin interacts with the pointed end of actin filaments (Holaska et al., 2004). To test whether an emerin–actin interaction was required for nuclear movement, we expressed emerin mutants m151 and m175, which have two and three point mutations in the nucleocytoplasmic domain, respectively, and completely abolish actin binding (Holaska et al., 2004). Expression of wild-type emerin or either of the actin-binding mutants of emerin in emerin-depleted NIH3T3 fibroblasts rescued centrosome orientation (Supplemental Figure S6). In addition, all three constructs rescued rearward nuclear position, although the degree of rearward nuclear position was not as great as for wild-type emerin with the m151 mutant (Supplemental Figure S6). These results suggest that the emerin–actin interaction is not required for nuclear movement.
Myosin IIB knockdown leads to similar nuclear movement and actin flow phenotypes as emerin knockdown
Given that emerin's actin-binding activity was not required for nuclear movement, we next explored the role of myosin II. Isoforms of myosin II are major factors that participate in the organization of actin cables and their movement in migrating cells (Giuliano et al., 1992; Lin et al., 1996; Salmon et al., 2002; Gomes et al., 2005; Cai et al., 2006). The two major isoforms in fibroblasts, myosin IIA and myosin IIB, have unique and overlapping biophysical properties and cellular functions (Vicente-Manzanares et al., 2009). Myosin IIA is localized on stress fibers and other actin cables and tends to associate with peripheral actin structures; in contrast, myosin IIB preferentially associates with actin structures in the rear and middle of the cell, frequently exhibiting a perinuclear distribution (Kolega, 1998; Lo et al., 2004; Cai et al., 2006; Vicente-Manzanares et al., 2007). Myosin IIA has been implicated in retrograde actin movements, accounting for at least 70% of the retrograde movement of a marker of actin flow (Cai et al., 2006). No study has explored whether myosin II isoforms differentially contribute to actomyosin-dependent nuclear movement, although knockdown of myosin IIB causes nuclei to spin in migrating fibroblasts (Vicente-Manzanares et al., 2007).
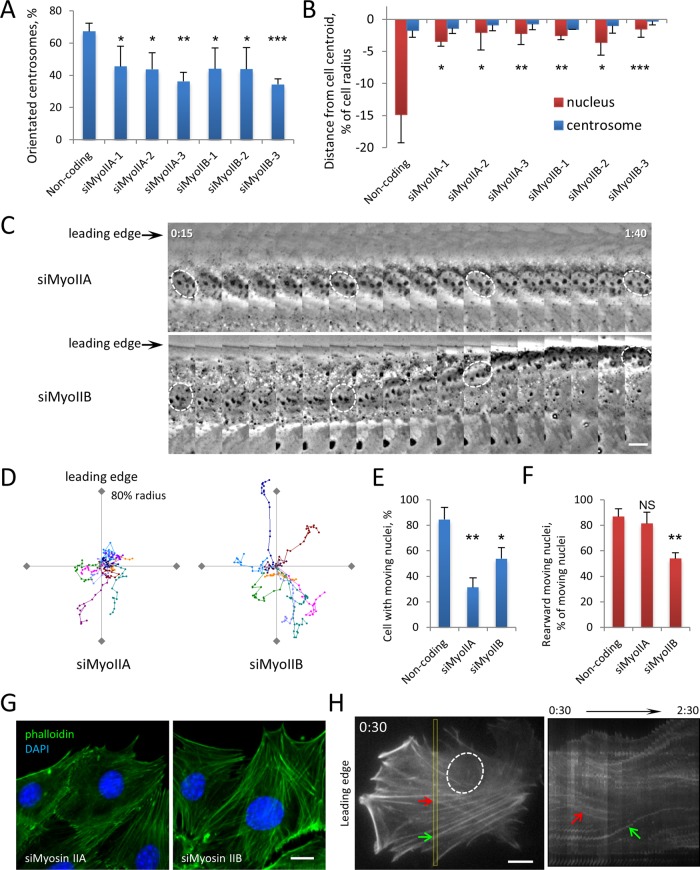
To test the role of individual myosin II A and B isoforms in nuclear movement, we decreased their expression using multiple siRNAs (Supplemental Figure S7). Depleting either isoform inhibited both LPA-stimulated centrosome orientation and rearward nuclear positioning (Figure 4, A and B). Live-cell imaging revealed that the two isoforms contributed differently to nuclear movement. Most nuclei in myosin IIA–depleted cells remained stationary after LPA stimulation, similar to those in nesprin-2G– and lamin A/C-depleted cells (Figure 4, C–F; compare to Figure 1, E and F). In contrast, although some nuclei in myosin IIB–depleted cells did not move, in ∼25% of the cases they moved nondirectionally as in emerin-depleted cells (Figure 4, C–F).
FIGURE 4:
Myosin IIB depletion causes nuclear movement and actin retrograde flow phenotypes similar to those of emerin depletion. (A) Quantification of centrosome reorientation in LPA-stimulated NIH3T3 cells treated with indicated siRNAs. Error bars, SD from three experiments (N ≥ 100 cells). (B) Nucleus and centrosome position in cells treated as in A. Error bars, SEM from three experiments (N ≥ 100 cells). None of the myosin siRNAs showed a difference in centrosome position compared with noncoding siRNA (p > 0.4, ANOVA). (C) Kymographs of nuclear movement from phase contrast movies of LPA-stimulated cells depleted of myosin IIA or myosin IIB. Nuclei are outlined in several frames. Time, hours:minutes after LPA stimulation; each panel represents 5 min. (D) Representative traces of LPA-stimulated nuclear movement in NIH3T3 cells depleted of myosin IIA or myosin IIB. Traces are plotted as in Figure 1E legend. (E, F) Quantification of moving nuclei (E) and percentage of moving nuclei that moved rearward (F) from phase contrast movies of LPA-stimulated NIH3T3 cells treated with indicated siRNA. Error bars, SD from three experiments (N ≥ 60 movies). (G) Representative fluorescence images of F-actin (phalloidin) and nuclei (DAPI) in myosin IIA– and myosin IIB–depleted NIH3T3 cells stimulated with LPA (see Figure 2A for noncoding control). (H) Initial panel (left, oval indicates nucleus) and kymograph of insert region (right) from movie of LifeAct-mCherry in myosin IIB–depleted NIH3T3 cells stimulated with LPA. Arrows indicate actin cables moving obliquely relative to the leading edge. Time, hours:minutes after LPA. Bars, 10 μm (C, G, H).
We next examined the effect of depleting myosin II isoforms on LPA-stimulated actin cable movement in NIH3T3 fibroblasts. Consistent with earlier reports (Cai et al., 2006; Vicente-Manzanares et al., 2007), actin cables were dramatically reduced in myosin IIA–depleted cells (Figure 4G), accounting for the lack of nuclear movement in these cells. Formation of actin cables was not dramatically affected in cells depleted of myosin IIB (Figure 4G), and dorsal actin cables still moved in about half of the cells after LPA stimulation. However, their directionality was lost: actin cables moved inward or in multiple directions with frequent changes in direction, similar to actin cable movement in emerin-depleted cells (Figure 4H and Supplemental Movie S5).
Emerin interacts with myosin IIB to couple the nucleus to sites of myosin IIB accumulation
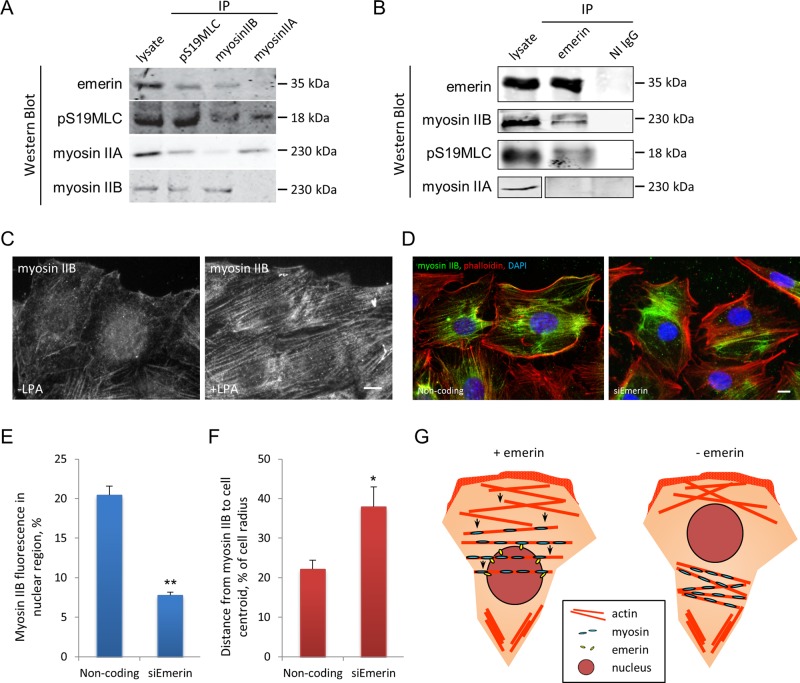
Given the similarity of nuclear and actin cable movement phenotypes in emerin-depleted and myosin IIB–depleted cells, we hypothesized that emerin either interacts with myosin IIB or affects its behavior. To test for an interaction, we immunoprecipitated myosin IIA, myosin IIB, and the phosphorylated myosin regulatory light chain pS19MLC, which binds to both isoforms and is an indicator of activated myosin II. Emerin coimmunoprecipitated with myosin IIB and pS19MLC but not with myosin IIA (Figure 5A). Conversely, myosin IIB and pS19MLC, but not myosin IIA, coimmunoprecipitated with emerin (Figure 5B). These data show that emerin associates specifically with myosin IIB and that some of the emerin-associated myosin IIB is active. When we immunoprecipitated myosin IIB and immunoblotted with antibodies against pS19MLC, we did not detect a difference in pS19MLC levels between controls and emerin-depleted cells (Supplemental Figure S8), indicating that emerin is not required for myosin IIB activation.
FIGURE 5:
Emerin interacts specifically with myosin IIB. (A) Western blots of indicated proteins immunoprecipitated by pSer19MLC and myosin II antibodies from NIH3T3 cell lysates. Lysate, 2% of input. (B) Western blots of indicated proteins immunoprecipitated by emerin antibody or nonimmune immunoglobulin G (NI IgG) from NIH3T3 cell lysates. Lysate, 25% of input. (C) Representative immunofluorescence micrographs showing myosin IIB localization before and 30 min after LPA stimulation of NIH3T3 cells. (D) Myosin IIB and actin localization relative to the nucleus (DAPI) in LPA-stimulated NIH 3T3 cells treated with noncoding or emerin siRNAs. Note the reduced perinuclear myosin IIB in cells treated with emerin siRNA compared with control cells treated with noncoding siRNA. (E) Quantification of myosin IIB immunofluorescence signals localized to the nuclear region in cells treated with indicated siRNAs. (F) Quantification of distance from centroid of myosin IIB immunofluorescence signal to cell centroid (as percentage of cell radius) in cells treated with the indicated siRNAs. In E and F, error bars are SD of three experiments with N > 90. (G) Model depicting emerin coupling of myosin IIB to the nucleus to control the directionality of actin cable flow in cells polarizing for migration. In cells that lack emerin, the displacement of myosin IIB from the perinuclear area alters the central “contractility sink” and prevents the establishment of normal retrograde actin flow. Bars, 10 μm (C, D).
Because emerin did not appear to participate in myosin IIB activation, we next tested whether it affected myosin IIB localization. LPA activates myosin II (Gomes et al., 2005), and we found that it induced the accumulation of myosin IIB on actin cables and in perinuclear regions (Figure 5C and Supplemental Figure S9). Myosin IIB was not obviously colocalized on the nuclear envelope with emerin, yet myosin IIB was localized on dorsal actin cables over the nucleus and in the adjacent perinuclear region (Figure 5D). In cells depleted of emerin, myosin IIB still accumulated near the center of the cell, although its distribution no longer overlapped with the nuclear envelope where emerin would normally be localized (Figure 5D). We quantified the myosin IIB signal that overlapped with the nucleus and found that in control cells, >20% of the total myosin IIB signal was within the nuclear area, whereas in emerin-depleted cells <8% of the myosin IIB signal was within the nuclear area (Figure 5E). This suggests that emerin is necessary to couple the localization of myosin IIB to the nucleus. To determine whether this coupling affected overall myosin IIB distribution in the cell, we determined the centroid of the myosin IIB signal relative to the cell centroid. The distance from the centroid of the myosin IIB signal to the cell center was significantly greater in emerin-depleted cells than in control cells treated with noncoding siRNA (Figure 5F). These results show that without emerin the central localization of myosin IIB is not maintained.
DISCUSSION
Our results support a model in which emerin acts to couple myosin IIB to the nucleus to control the directionality of actin cable flow in cells polarizing for migration (Figure 5G). According to this model, in the presence of emerin, myosin IIB is maintained near the nucleus, resulting in a high level of actomyosin contractility in the center of the cell near the nucleus. In the absence of emerin, myosin IIB is not constrained to the perinuclear area, resulting in an uncoupling of actomyosin contractility from the nucleus and unoriented actin flow. In support of this model, we found that both emerin and myosin IIB depletion caused similar nondirectional nuclear movement and actin cable flow and that emerin depletion disrupted the normal coupling of the nucleus with perinuclear myosin IIB. In addition, emerin specifically interacted with myosin IIB but not myosin IIA. This emerin “pathway” is distinct from the nuclear movement pathway involving nesprin-2G, SUN2, and A-type lamins, as shown by the distinct nuclear movement and actin cable flow phenotypes exhibited by emerin deficiency compared with deficiencies in these other proteins. Emerin may also localize in the endoplasmic reticulum, especially at high expression levels, and we cannot rule out the possibility that this pool of emerin also contributes to organizing actin retrograde flow. Nonetheless, we favor the model in which outer nuclear membrane emerin is the principal pool involved because depletion of emerin strongly affected myosin IIB localization around the nucleus.
A question raised by the model is how emerin localization of myosin IIB acts to polarize actin cable flow. One possibility is that emerin acts as a diffusion trap to maintain a pool of active myosin IIB in high concentration near the nucleus and this pool of myosin IIB serves as a “contractility sink” for the inward movement of actin filaments. Myosin IIB has a lower rate of ATP hydrolysis and a higher duty cycle than myosin IIA (Kovacs et al., 2003; Wang et al., 2003), and these properties would contribute to maintaining a tonic state of contraction centered on the nucleus and near the middle of the cell. Without emerin in the outer nuclear membrane, myosin IIB may concentrate but becomes unconstrained from the nucleus and localizes elsewhere in the cell, which disrupts the positioning of the contractile sink in the cell center necessary to maintain the directionality of actin flow. We note that this model can account for only one element of the cellular system controlling actin flow in migrating cells; for example, polymerization of actin filaments near the leading edge and their movement by myosin II must also contribute (Ponti et al., 2004). In addition, whereas emerin and myosin IIB have similar phenotypes with respect to actin flow in cultured cells, they are likely to perform additional functions, given that myosin IIB–knockout mice are early embryonic lethal with defects in heart and brain (Tullio et al., 1997, 2001), whereas emerin-knockout mice are viable, with only subtle motor and cardiac muscle phenotypes (Melcon et al., 2006; Ozawa et al., 2006).
We attribute most of the abnormal behaviors of the TAN lines (immobility, nonoriented movements) observed in emerin-depleted cells to the abnormal directionality of actin cable movements. Nonetheless, emerin also localizes to the inner nuclear membrane and may contribute to TAN line anchoring, as we observed occasional slippage of TAN lines on immobile nuclei, a phenotype reminiscent of that in lamin A/C-deficient cells (Folker et al., 2011). Other inner nuclear membrane proteins, such as SAMP-1, may also be required for TAN line anchoring (Borrego-Pinto et al., 2012).
Emerin interacts directly with actin and modulates actin polymerization (Holaska et al., 2004; Ho et al., 2013), yet our results show that these activities of emerin are not required for nuclear movement. Instead, emerin's actin activity seems to be required to regulate intranuclear actin and the G-actin responsive MKL1-SRF transcriptional pathway (Ho et al., 2013). Given that the point mutations in emerin-m175 and m151 also disrupt its interaction with GCL, Btf, YT521-B, and Lmo7 (Berk et al., 2013), it is likely that these interactions are also not required for emerin's function in nuclear movement.
Our results on emerin add to the growing number of cases in which proteins of the nuclear envelope or lamina contribute to the organization of the cytoplasmic cytoskeleton. Defects in the LINC complex were shown to affect the steady-state distribution of the actin cytoskeleton and focal adhesions (Hale et al., 2008; Khatau et al., 2009), although the effects were subtler than those we detected, perhaps because we examined actin dynamics directly and used a polarized cell model. The nucleus also acts directly as a site to nucleate and organize microtubules in some cells (e.g., muscle cells), although the nuclear envelope proteins involved have not been identified. Given the number of nuclear envelope constituents that can interact with cytoskeletal elements, it will be important to explore further how the nucleus contributes to cytoskeletal organization. Lamin A variants expressed in autosomal dominant Emery–Dreifuss muscular dystrophy and depletion of SUNs and nesprins, which have also been implicated in the disease (Zhang et al., 2007a; Puckelwartz et al., 2009; Haque et al., 2010), all result in defective nuclear positioning during fibroblast cell migration (Luxton et al., 2010; Folker et al., 2011). We have now found that emerin, which is absent in most cases of X-linked Emery–Dreifuss muscular dystrophy, is also involved in nuclear positioning. Our results show that although gross nuclear movement defects occur from alterations in A-type lamins and emerin, the underlying mechanisms differ. Alterations in A-type lamins cause defective anchoring of TAN lines, whereas loss of emerin primarily causes mislocalization of myosin IIB and disorganization of cytoplasmic actin flow. Abnormal positioning of nuclei in the center of muscle fibers is characteristic of muscular dystrophies, including Emery–Dreifuss muscular dystrophy. It will be interesting to test whether the roles that emerin, nesprins, and lamin A/C play in nuclear positioning in simple systems such as fibroblasts also are required to properly position nuclei in muscle cells.
MATERIALS AND METHODS
Reagents
LPA was from Avanti Polar Lipids (Alabaster, AL). Alexa 647–phalloidin was from Invitrogen (Carlsbad, CA). Unless noted, all other chemicals were from Sigma-Aldrich (St. Louis, MO). Lifeact-mCherry (Riedl et al., 2008) was from ibidi (Munich, Germany). GFP-mN2G was previously described (Luxton et al., 2010). pS19MLC mouse monoclonal antibody (mAb; #3675), myosin IIA (#3403), myosin IIB (#3404), and pS19MLC (#3671) rabbit polyclonal antibodies were from Cell Signaling (Danvers, MA). Emerin rabbit antibody (sc-15378) was from Santa Cruz Biotechnology (Santa Cruz, CA). Emerin mouse mAb (VP-E602) was from Vector Laboratories (Burlingame, CA). Pericentrin mouse antibody (#611814) was from BD Transduction Laboratories (San Jose, CA). Pericentrin rabbit antibody (PRB-432C) was from Covance (Princeton, NJ). Tyrosinated α-tubulin rat mAb (YL1/2) was from the European Collection of Animal Cell Cultures (Salisbury, United Kingdom). GFP (AB16901) chicken antibody and actin (clone C4, MAB1501) mouse mAbs were from Millipore (Billerica, MA). Lamin A/C mouse mAb (MANLAC1) was from the MDA Monoclonal Antibody Resource at the Wolfson Center for Inherited Neuromuscular Disease (Oswestry, United Kingdom).
Cell culture, microinjection, and siRNA transfection
NIH3T3 fibroblasts were cultured in DMEM (Life Technologies, Grand Island, NY) with 10% calf serum (Gemini, West Sacramento, CA). Emerin knockout and wild-type MEFs were isolated as previously described (Melcon et al., 2006) and cultured in DMEM containing 15% fetal bovine serum (Gemini, West Sacramento, CA). 4-(2-hydroxyethyl)-1-Piperazineethanesulfonic acid (HEPES; 10 mM, pH 7.4), penicillin, and streptomycin (Thermo Scientific, Waltham, MA) were added to DMEM. For analysis of nuclear movement and centrosome orientation, confluent monolayers of serum-starved NIH3T3 cells were wounded and stimulated with 10 μM LPA for 2 h as previously described (Gomes et al., 2005; Gomes and Gundersen, 2006). For live-cell imaging, wounded NIH3T3 cell monolayers were maintained in serum-free medium, and 20 μM LPA was added to initiate nuclear movement. To assay migration, monolayers were starved for 1 d, wounded, and stimulated with 2.5% calf serum. For microinjection, 10–50 μg/ml purified plasmid DNA in 10 mM HEPES, 140 mM KCl (pH 7.4), was pressure microinjected into nuclei of cells at the wounded edge using glass micropipettes and allowed to express for 1 h. GFP-emerin, GFP-emerin-m151, and GFP-emerin-m175 were generously provided by James Holaska (University of Chicago, Chicago, IL).
For siRNA knockdown, NIH3T3 fibroblasts were transfected with 40 nM siRNA using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Two days after siRNA transfection, cells were serum starved for 2 d before conducting experiments. siRNA oligonucleotides were from Shanghai GenePharma (Shanghai, China). siRNAs against nesprin-2G and lamin A/C were previously described (Luxton et al., 2010; Folker et al., 2011). Other siRNA sequences used were as follows: emerin, 5′-GAGCAAGGACUAUAAUGAUTT-3′, 5′-CUUUGUUUACUAUUCCAUATT-3′, and 5′-CCAGGUGCGUGAUGACAUUTT-3′; myosin IIA, 5′-AGCACCAGGCAAUGAUCACCG-3′, 5′-AAGACAGAGUAGCUGAAUUCA-5′, and 5′-GGACCUGGAGGCACACAUUG-3′; and myosin IIB, 5′-CAAGAAUAAUGCACUUAAATT-3′, 5′-GACCAGAAUUCCAAAUUUATT-3′, and 5′-CGUUCACCGCAUCAAUAAATT-3′. Protein levels were analyzed by immunoblotting using antibodies against myosin IIA, pS19MLC at 1:500, and myosin IIB, actin, and emerin at 1:1000.
Microscopy
For immunofluorescence microscopy, cells on coverslips were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) and permeabilized with 0.3% Triton X-100 in phosphate-buffered saline (PBS) containing 5% normal goat serum. For digitonin permeabilization, fixed cells were treated with 0.02% digitonin on ice for 5 min and blocked with 5% normal goat serum. Cells were stained with primary antibodies for 1 h, washed three times with PBS, and stained with 4′,6-diamidino-2-phenylindole (DAPI), secondary antibodies, and/or phalloidin for 1 h. Coverslips were then washed three times with PBS and mounted with Fluoromount-G (Southern Biotech, Birmingham, AL). Images were acquired with either a 40× PlanApo objective (numerical aperture [NA] 1.0) or a 60× PlanApo objective (NA 1.4) and a CoolSNAP HQ charge-coupled device (CCD) camera (Photometrics, Tucson, AZ) on a Nikon TE300 inverted microscope (Nikon, Melville, NY) controlled by MetaMorph (Molecular Devices, Sunnyvale, CA) and processed with ImageJ (National Institutes of Health, Bethesda, MD).
Phase contrast live-cell movies were acquired with a 20× (NA 0.45) or 40× (NA 0.6) ELWD Plan objective and a CoolSNAP HQ CCD camera on a Nikon TE300 microscope with temperature controller (37°C) and motorized xyz stage to acquire multiple movies simultaneously using MetaMorph's multidimensional acquisition application. Acquisition rate was 5 min/frame. AVI files and montage images were generated using ImageJ. Fluorescence live-cell movies of TAN lines and actin cables were acquired at 37°C (3 or 5 min/frame) with a 60× PlanApo objective (NA 1.49) and an iXon X3 CCD camera (Andor, Belfast, United Kingdom) on a Nikon Eclipse Ti microscope controlled by Nikon's NIS-Elements software. AVI files and montage images were generated using either ImageJ or NIS-Elements.
Image and data analysis
Centrosome orientation to a position between the nucleus and the leading edge was analyzed as previously described using cells immunofluorescently stained for pericentrin, tyrosinated α-tubulin, and nuclei (Palazzo et al., 2001; Gomes and Gundersen, 2006). Nuclear and centrosomal positions were determined from images of cells immunofluorescently stained for the centrosome (pericentrin), cell boundaries (actin or microtubules), and nuclei (DAPI). Images were uploaded into custom software that identifies the positions of the nuclear centroid and the centrosome, cell boundaries, and the wound direction. Software determinations of cell boundaries were inspected and corrected manually where necessary using the software to adjust computer drawn boundaries. The x, y positions (x, parallel to wound edge; y, perpendicular) of both the nucleus centroid and centrosome were calculated and normalized to the average cell radius calculated by the software. We used the same software to determine the proportion of the whole-cell myosin IIB that colocalized with the nucleus. The software summed thresholded background-subtracted myosin IIB fluorescence signals for the whole cell and for the nucleus (defined by DAPI staining), and the ratio was calculated to be the percentage of nuclear localization of myosin IIB. The centroids of myosin IIB fluorescence were calculated for each cell as the signal-weighted centroid of thresholded images, and their distances to cell centroid were calculated and normalized to cell radius. Further details on the software are available upon request. Data were exported to Excel (Microsoft, Redmond, WA) for statistical analysis and plotting.
Nuclear positions in live-cell movies were tracked using custom software that calculates the nuclear centroid after manually outlining the nucleus. Nuclear centroid positions in individual frames were then combined and successive positions connected by lines to generate traces representing the paths of the nuclei. Ten representative traces were combined on an X, Y plot with the starting position at 0, 0 (e.g., see Figure 1E). Nuclear movement paths were grouped into three categories: 1) no movement—nuclei that moved <35% of the cell radius (approximately the diameter of the nucleus) in 90 min; 2) rearward movement—nuclei that moved >35% of cell radius in 90 min and within 35° of the front–back axis of the cell; and 3) nonoriented movement—nuclei that moved >35% of cell radius but at an angle >35% of the front–back axis of the cell.
Movies of actin cable flows were used to categorize the flow into three types: 1) retrograde—dorsal actin cables moved from the leading edge of the cell toward the trailing edge of the cell; 2) random—actin cables moved either from all directions toward the nucleus or in a direction that was not perpendicular to the leading edge and/or changed their direction; and 3) nondetectable movement. The percentages of category 3 were low (≤20%), and there was no significant difference between noncoding or emerin siRNA-treated cells, so this category was not included in Figure 2D.
For measuring velocity of TAN lines and nuclear movement, kymographs were generated using NIS-Elements and exported to ImageJ. The slopes of individual TAN lines and the leading edge of the nucleus were measured in ImageJ and exported to Excel to calculate the velocity.
Cell migration velocity in wound-healing assays was analyzed by tracing the advance of the wound edge at different time points using ImageJ. We determined the area that the wound advanced by comparing traces at two times. We then divided this area by the length of the wound to determine the advancement of the cells.
Traces of nuclear movement were plotted with custom software. All other plots were generated using Excel.
Statistical analysis was performed using Excel. Unless noted, p values were calculated with unpaired two-tailed Student's t test comparing experimental to controls. One-way analysis of variance (ANOVA) was used to test whether multiple groups were statistically similar to each. In the figures, statistical significance is represented as follows: *p < 0.05, **p < 0.01, ***p < 0.001, or NS (not significant).
Immunoprecipitation
NIH3T3 fibroblasts at ∼70% confluency were placed on ice and lysed for 30 min in 1% Triton X-100 in 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, 1 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethanesulfonyl fluoride, and a mixture of protease inhibitors. Lysates were clarified by centrifugation at 13,000 × g for 30 min at 4°C. The lysate was then divided equally (∼ 5 mg lysate/immunoprecipitate) and incubated for 4 h with 5 μl of the specified antibody at 4°C. For emerin and pS19MLC the rabbit polyclonal antibodies were used. Immunoprecipitates were collected with 50 μl of protein G beads (Invitrogen, Carlsbad, CA), washed three times in lysate buffer, and then eluted with SDS sample buffer, boiled, separated by SDS–PAGE, transferred to nitrocellulose strips, and immunoblotted.
Supplementary Material
Acknowledgments
We thank Glenn E. Morris (Wolfson Center for Inherited Neuromuscular Disease, Oswestry, United Kingdom) for MANLAC1 antibody and James M. Holaska for emerin constructs. This study was supported by National Institutes of Health Grants NS059352 and GM099481. E.S.F. was supported in part by an American Heart Association postdoctoral fellowship.
Abbreviations used:
- DAPI
4′,6-diamidino-2-phenylindole
- GFP-mN2G
green fluorescent protein–mini-nesprin-2G
- LINC
linker of nucleoskeleton and cytoskeleton
- LPA
lysophosphatidic acid
- TAN
transmembrane actin-associated nuclear
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-06-0307) on October 23, 2013.
*Present address: Department of Biology, Boston College, Chestnut Hill, MA 02467.
REFERENCES
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk JM, Tifft KE, Wilson KL. The nuclear envelope LEM-domain protein emerin. Nucleus. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat Genet. 1994;8:323–327. doi: 10.1038/ng1294-323. [DOI] [PubMed] [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- Cai Y, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91:3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements L, Manilal S, Love DR, Morris GE. Direct interaction between emerin and lamin A. Biochem Biophys Res Commun. 2000;267:709–714. doi: 10.1006/bbrc.1999.2023. [DOI] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai RA, Gao L, Raghavan S, Liu WF, Chen CS. Cell polarity triggered by cell-cell adhesion via E-cadherin. J Cell Sci. 2009;122:905–911. doi: 10.1242/jcs.028183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin I, Etienne-Manneville S. Nuclear positioning: mechanisms and functions. Int J Biochem Cell Biol. 2011;43:1698–1707. doi: 10.1016/j.biocel.2011.09.004. [DOI] [PubMed] [Google Scholar]
- Dupin I, Sakamoto Y, Etienne-Manneville S. Cytoplasmic intermediate filaments mediate actin-driven positioning of the nucleus. J Cell Sci. 2011;124:865–872. doi: 10.1242/jcs.076356. [DOI] [PubMed] [Google Scholar]
- Fairley EA, Kendrick-Jones J, Ellis JA. The Emery-Dreifuss muscular dystrophy phenotype arises from aberrant targeting and binding of emerin at the inner nuclear membrane. J Cell Sci. 1999;112:2571–2582. doi: 10.1242/jcs.112.15.2571. [DOI] [PubMed] [Google Scholar]
- Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano KA, Kolega J, DeBiasio RL, Taylor DL. Myosin II phosphorylation and the dynamics of stress fibers in serum-deprived and stimulated fibroblasts. Mol Biol Cell. 1992;3:1037–1048. doi: 10.1091/mbc.3.9.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes ER, Gundersen GG. Real-time centrosome reorientation during fibroblast migration. Methods Enzymol. 2006;406:579–592. doi: 10.1016/S0076-6879(06)06045-9. [DOI] [PubMed] [Google Scholar]
- Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Gudise S, Figueroa RA, Lindberg R, Larsson V, Hallberg E. Samp1 is functionally associated with the LINC complex and A-type lamina networks. J Cell Sci. 2011;124:2077–2085. doi: 10.1242/jcs.078923. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional connections between the nucleus and the actin and microtubule networks in laminopathic models. Biophys J. 2008;95:5462–5475. doi: 10.1529/biophysj.108.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature. 2013;497:507–511. doi: 10.1038/nature12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holaska JM, Kowalski AK, Wilson KL. Emerin caps the pointed end of actin filaments: evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004;2:E231. doi: 10.1371/journal.pbio.0020231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolega J. Cytoplasmic dynamics of myosin IIA and IIB: spatial “sorting” of isoforms in locomoting cells. J Cell Sci. 1998;111(Pt 15):2085–2095. doi: 10.1242/jcs.111.15.2085. [DOI] [PubMed] [Google Scholar]
- Kovacs M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II: kinetic characterization of the non-muscle IIA isoform. J Biol Chem. 2003;278:38132–38140. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- Lin CH, Espreafico EM, Mooseker MS, Forscher P. Myosin drives retrograde F-actin flow in neuronal growth cones. Neuron. 1996;16:769–782. doi: 10.1016/s0896-6273(00)80097-5. [DOI] [PubMed] [Google Scholar]
- Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15:982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Worman HJ, Gundersen GG. TAN lines: a novel nuclear envelope structure involved in nuclear positioning. Nucleus. 2011;2:173–181. doi: 10.4161/nucl.2.3.16243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gundersen GG. Orientation and function of the nuclear-centrosomal axis during cell migration. Curr Opin Cell Biol. 2011;23:579–588. doi: 10.1016/j.ceb.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manilal S, Nguyen TM, Sewry CA, Morris GE. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum Mol Genet. 1996;5:801–808. doi: 10.1093/hmg/5.6.801. [DOI] [PubMed] [Google Scholar]
- Melcon G, et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum Mol Genet. 2006;15:637–651. doi: 10.1093/hmg/ddi479. [DOI] [PubMed] [Google Scholar]
- Mislow JM, Holaska JM, Kim MS, Lee KK, Segura-Totten M, Wilson KL, McNally EM. Nesprin-1alpha self-associates and binds directly to emerin and lamin A in vitro. FEBS Lett. 2002;525:135–140. doi: 10.1016/s0014-5793(02)03105-8. [DOI] [PubMed] [Google Scholar]
- Nagano A, Koga R, Ogawa M, Kurano Y, Kawada J, Okada R, Hayashi YK, Tsukahara T, Arahata K. Emerin deficiency at the nuclear membrane in patients with Emery-Dreifuss muscular dystrophy. Nat Genet. 1996;12:254–259. doi: 10.1038/ng0396-254. [DOI] [PubMed] [Google Scholar]
- Norden C, Young S, Link BA, Harris WA. Actomyosin is the main driver of interkinetic nuclear migration in the retina. Cell. 2009;138:1195–1208. doi: 10.1016/j.cell.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa R, Hayashi YK, Ogawa M, Kurokawa R, Matsumoto H, Noguchi S, Nonaka I, Nishino I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am J Pathol. 2006;168:907–917. doi: 10.2353/ajpath.2006.050564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo AF, Joseph HL, Chen YJ, Dujardin DL, Alberts AS, Pfister KK, Vallee RB, Gundersen GG. Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr Biol. 2001;11:1536–1541. doi: 10.1016/s0960-9822(01)00475-4. [DOI] [PubMed] [Google Scholar]
- Ponti A, Machacek M, Gupton SL, Waterman-Storer CM, Danuser G. Two distinct actin networks drive the protrusion of migrating cells. Science. 2004;305:1782–1786. doi: 10.1126/science.1100533. [DOI] [PubMed] [Google Scholar]
- Puckelwartz MJ, et al. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet. 2009;18:607–620. doi: 10.1093/hmg/ddn386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon WC, Adams MC, Waterman-Storer CM. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ. A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol. 2007;178:897–904. doi: 10.1083/jcb.200702026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk J, Wilsch-Brauninger M, Calegari F, Huttner WB. Myosin II is required for interkinetic nuclear migration of neural progenitors. Proc Natl Acad Sci USA. 2009;106:16487–16492. doi: 10.1073/pnas.0908928106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmoranzer J, Fawcett JP, Segura M, Tan S, Vallee RB, Pawson T, Gundersen GG. Par3 and dynein associate to regulate local microtubule dynamics and centrosome orientation during migration. Curr Biol. 2009;19:1065–1074. doi: 10.1016/j.cub.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solecki DJ, Trivedi N, Govek EE, Kerekes RA, Gleason SS, Hatten ME. Myosin II motors and F-actin dynamics drive the coordinated movement of the centrosome and soma during CNS glial-guided neuronal migration. Neuron. 2009;63:63–80. doi: 10.1016/j.neuron.2009.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. A nuclear-envelope bridge positions nuclei and moves chromosomes. J Cell Sci. 2009;122:577–586. doi: 10.1242/jcs.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan T, Escalante-Alcalde D, Bhatt H, Anver M, Bhat N, Nagashima K, Stewart CL, Burke B. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio AN, Accili D, Ferrans VJ, Yu ZX, Takeda K, Grinberg A, Westphal H, Preston YA, Adelstein RS. Nonmuscle myosin II-B is required for normal development of the mouse heart. Proc Natl Acad Sci USA. 1997;94:12407–12412. doi: 10.1073/pnas.94.23.12407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullio AN, Bridgman PC, Tresser NJ, Chan CC, Conti MA, Adelstein RS, Hara Y. Structural abnormalities develop in the brain after ablation of the gene encoding nonmuscle myosin II-B heavy chain. J Comp Neurol. 2001;433:62–74. doi: 10.1002/cne.1125. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176:573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Kovacs M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem. 2003;278:27439–27448. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen K, Ketema M, Truong H, Sonnenberg A. KASH-domain proteins in nuclear migration, anchorage and other processes. J Cell Sci. 2006;119:5021–5029. doi: 10.1242/jcs.03295. [DOI] [PubMed] [Google Scholar]
- Zhang J, et al. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet. 2010;19:329–341. doi: 10.1093/hmg/ddp499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, et al. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet. 2007a;16:2816–2833. doi: 10.1093/hmg/ddm238. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Ragnauth CD, Skepper JN, Worth NF, Warren DT, Roberts RG, Weissberg PL, Ellis JA, Shanahan CM. Nesprin-2 is a multi-isomeric protein that binds lamin and emerin at the nuclear envelope and forms a subcellular network in skeletal muscle. J Cell Sci. 2005;118:673–687. doi: 10.1242/jcs.01642. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, Xu T, Zhuang Y, Han M. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007b;134:901–908. doi: 10.1242/dev.02783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.