Both SNAREs and Sey1p are required for efficient nuclear fusion during yeast mating. SNAREs appear to act at the step of nuclear envelope fusion, whereas Sey1p remodels the ER network to permit nuclear congression. In addition, SNARE sey1Δ double mutants reveal an Sey1p-independent, SNARE-mediated ER fusion pathway.
Abstract
During yeast mating, two haploid nuclei fuse membranes to form a single diploid nucleus. However, the known proteins required for nuclear fusion are unlikely to function as direct fusogens (i.e., they are unlikely to directly catalyze lipid bilayer fusion) based on their predicted structure and localization. Therefore we screened known fusogens from vesicle trafficking (soluble N-ethylmaleimide–sensitive factor attachment protein receptors [SNAREs]) and homotypic endoplasmic reticulum (ER) fusion (Sey1p) for additional roles in nuclear fusion. Here we demonstrate that the ER-localized SNAREs Sec20p, Ufe1p, Use1p, and Bos1p are required for efficient nuclear fusion. In contrast, Sey1p is required indirectly for nuclear fusion; sey1Δ zygotes accumulate ER at the zone of cell fusion, causing a block in nuclear congression. However, double mutants of Sey1p and Sec20p, Ufe1p, or Use1p, but not Bos1p, display extreme ER morphology defects, worse than either single mutant, suggesting that retrograde SNAREs fuse ER in the absence of Sey1p. Together these data demonstrate that SNAREs mediate nuclear fusion, ER fusion after cell fusion is necessary to complete nuclear congression, and there exists a SNARE-mediated, Sey1p-independent ER fusion pathway.
INTRODUCTION
During mating in the budding yeast Saccharomyces cerevisiae two haploid cells fuse to form a diploid. The fusion process requires a coordinated sequence of “shmooing” (polarized growth toward the mating partner), prezygote formation (adhesion and cell wall degradation), plasma membrane fusion, nuclear congression, and finally nuclear fusion (karyogamy; reviewed in Ydenberg and Rose, 2008). Nuclear fusion is subdivided into the sequential steps of outer membrane fusion, bridge expansion, inner membrane fusion, and spindle pole body fusion (Melloy et al., 2007). However, although many genes required for karyogamy are known, the mechanisms at each individual step of karyogamy are poorly characterized (Rose, 1996; Melloy et al., 2009).
Electron tomography of karyogamy-defective mutants revealed only one gene required specifically for outer membrane fusion: PRM3 (Melloy et al., 2009). However, Prm3p is unlikely to act as a fusogen (i.e., capable of fusing membranes directly) and more likely plays an accessory role. Prm3p is a small peripheral membrane protein (133 residues without a transmembrane domain), and only the ∼60 C-terminal residues are required for function, making it unlike other known fusogens (Shen et al., 2009; Kozlov et al., 2010). Because genetic screens have not yielded other outer membrane fusogen candidates, it is probable either that the true fusogen is essential for life or there are multiple redundant fusogens. We therefore screened for mating defects in known fusogens from other pathways.
Soluble N-ethylmaleimide–sensitive factor attachment protein receptors (SNAREs) are the most common intracellular fusogens and mediate all forms of vesicle trafficking. In the secretory pathway, proteins are translocated into the endoplasmic reticulum (ER) and transported in vesicles to the Golgi body. From the trans-Golgi network, proteins can be sent to either the plasma membrane or other compartments of the endomembrane system, including the vacuole. At each trafficking step, membrane fusion is mediated by SNARE proteins. SNARE proteins have a characteristic heptad-repeat SNARE motif (Sutton et al., 1998). Typically, four SNARE proteins each contribute a single SNARE motif to form a four-helix coiled-coil SNARE complex (Sutton et al., 1998; Burri and Lithgow, 2004). SNARE complex assembly is believed to supply the energy required for membrane fusion (Stein et al., 2009). The SNARE complex is zippered by multiple hydrophobic layers, but the central position or “0-layer” almost always consists of three glutamine (Q) residues and one arginine (R) residue (Fasshauer et al., 1998; Sutton et al., 1998; see Discussion for exceptions). SNARE proteins are designated R or Q based on the 0-layer residue.
In anterograde transport, COPII-coated vesicles move from the ER to the Golgi body (Figure 1; see Kienle et al., 2009, for a more detailed overview). The anterograde vesicles incorporate two SNAREs, Bos1p and Sec22p, from the ER membrane, which fuse with Bet1p and Sed5p on the cis-Golgi membrane (Newman et al., 1990; Sacher et al., 1997; McNew et al., 2000; Parlati et al., 2000; reviewed in Lee et al., 2004). In retrograde trafficking, COPI-coated vesicles move from the Golgi to the ER (Cosson and Letourneur, 1994; Letourneur et al., 1994; Lewis and Pelham, 1996). The SNAREs Sec20p, Ufe1p, and Use1p form a complex on the ER that binds incoming Sec22p on the vesicles (Lewis et al., 1997; Spang and Schekman, 1998; Burri et al., 2003; Dilcher et al., 2003). In both types of transport, Sec22p is the primary R-SNARE, although Ykt6p can function redundantly in its absence (Liu and Barlowe, 2002).
FIGURE 1:
Overview of SNARE-mediated vesicle trafficking. Subset of trafficking pathways is shown; intra-Golgi trafficking and intermediate steps of endosomal trafficking are not shown. The ER is shown in green. SNAREs required for nuclear fusion are listed in bold (see Results and Figure 3A). Note that the entire ER network is interconnected and continuous, but a single slice through the center, as drawn here, appears discontinuous. Inherited ER tubules at the bud tip are not drawn for clarity.
The ER network consists of interconnected sheets and tubules, which maintain their shape via reticulons (membrane-curvature–promoting proteins; Rtn1p, Rtn2p) and Yop1p (Voeltz et al., 2006; Hu et al., 2011; Chen et al., 2013). Homotypic ER fusion connects tubules and creates new three-way junctions. Of importance, homotypic ER fusion is mediated by a non-SNARE fusogen, Sey1p (called atlastin in mammals and Drosophila; Hu et al., 2009; Orso et al., 2009; Anwar et al., 2012). Sey1p is a dynamin-like GTPase that resides as an integral membrane protein in the ER and fuses membranes as a dimer (Hu et al., 2009; Anwar et al., 2012). Sey1p activity is antagonized by Lnp1p, as lnp1Δ cells exhibited a more highly reticulated ER network than did wild type, indicative of excess ER-fusion events (Chen et al., 2012). Of interest, loss of atlastin in mammals results in a severely unbranched, sheet-like ER network and in Drosophila causes ER fragmentation (Orso et al., 2009), whereas sey1Δ yeast cells have only a subtle change in ER morphology and no growth defect (Hu et al., 2009; Chen et al., 2012). This discrepancy likely exists because in yeast, but presumably not in mammals or Drosophila, there is a second, redundant homotypic ER fusion pathway mediated by the SNARE Ufe1p (Patel et al., 1998; Anwar et al., 2012).
To identify the fusogens for karyogamy, we screened SNARE mutants and sey1Δ and found that both were required for efficient karyogamy. We demonstrate that Sey1p is required early to remodel the ER network after cell fusion, which permits the completion of nuclear congression, whereas the ER-resident SNAREs Sec20p, Ufe1p, Use1p, and Bos1p are required for nuclear envelope (NE) fusion. We also show that the Sey1p-independent ER fusion pathway depends on at least three retrograde SNAREs—Sec20p, Ufe1p, and Use1p—but not Bos1p, Sec22p, or Ykt6p.
RESULTS
ER-bound SNAREs are required for efficient nuclear fusion
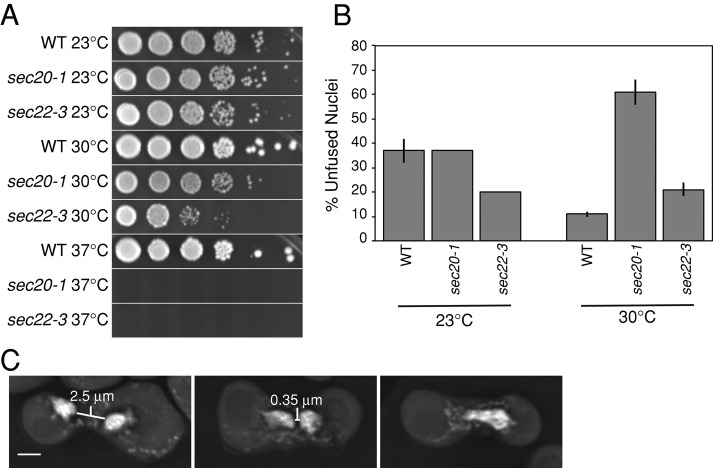
To determine whether the known fusogens from intracellular trafficking (SNAREs) mediate karyogamy (nuclear fusion during mating), we examined the efficiency of nuclear fusion in a collection of SNARE gene mutants (Table 1). Most SNAREs are essential for viability, and we therefore used temperature-sensitive alleles that permit growth at 23 but not 37°C. In addition to the temperature-sensitive use1-10AA allele, we included a use1-0layer allele, which has no growth defect at any temperature (Dilcher et al. 2003). Because wild-type cells mate poorly above 34°C (Grote, 2010) and an earlier step in mating—cell fusion—depends on SNARE-mediated secretion (Grote, 2010), we identified temperatures at which cells could mate but at which growth was clearly limited by a partial defect in SNARE function (Figure 2, A and B). To assay mating, we first grew cells at the permissive temperature (23°C for most strains) and then mated them at several semipermissive temperatures. Cells from the mating mixtures were fixed and stained with 4′,6′-diamidino-2-phenylindole (DAPI), and unbudded zygotes were identified and examined for the efficiency of nuclear fusion. Zygotes with two separate nuclei were scored as karyogamy defective (Figure 2C, left and middle). For most alleles, 30°C was chosen as the semipermissive temperature, with the exception of ufe1-1, which was mated at 33°C. By these means we were able to measure the efficiency of karyogamy under conditions in which cells are viable but SNARE function is greatly reduced.
TABLE 1:
SNARE alleles used in this study.
| SNARE allele | Mutation | Reference |
|---|---|---|
| sec20-1 | L234S | Lewis et al. (1997) |
| ufe1-1 | S282N, L295P | Lewis et al. (1997) |
| use1-0layer | D183G | Dilcher et al. (2003) |
| use1-10AA | Q18R, Q132R, E139D, Q156R, S168G, Q177R, D183G, Q185R, F220Y, F242S | Dilcher et al. (2003) |
| sec22-3 | R157G | Sacher et al. (1997) |
| bos1-1 | L190S | Stone et al. (1997) |
| bet1-1 | L72F | Stone et al. (1997) |
| sed5-1 | R255G | Banfield et al. (1995) |
| ykt6-ts | Y128H, D139G, T151A | Ben-Aroya et al. (2008) |
| vti1-1 | E145K, G148R | Fischer von Mollard and Stevens (1998) |
| vam7-167 | L134P, L287P | Sato et al. (1998) |
FIGURE 2:
Defining a semipermissive temperature. (A) A representative growth assay showing wild-type (MY2051), sec20-1 (MY2065), and sec22-3 (MY2069) cells grown on YEPD. Each spot from left to right is a 10-fold dilution. Plates were incubated for 2 d at 30 or 37°C and for 3 d at 23°C. (B) Nuclear fusion efficiencies (see Materials and Methods) for wild type (MY2051 × MY2050), sec20-1 (MY2065 × MY2064), and sec22-3 (MY2069 × MY2068) at the indicated temperatures. At 23°C, each strain was assayed for nuclear fusion in one trial, and wild type shows the average and range of the wild-type control from each trial. At 30°C, the data are the same as shown in Figure 3A. (C) Representative examples of unfused (left and middle) and fused nuclei (right). Nuclei were stained with DAPI and fixed with 3:1 methanol:acetic acid as described in Materials and Methods. Examples are from a sey1Δ × sey1Δ cross (MS8072 × MS8073). Scale bar, 2 μm.
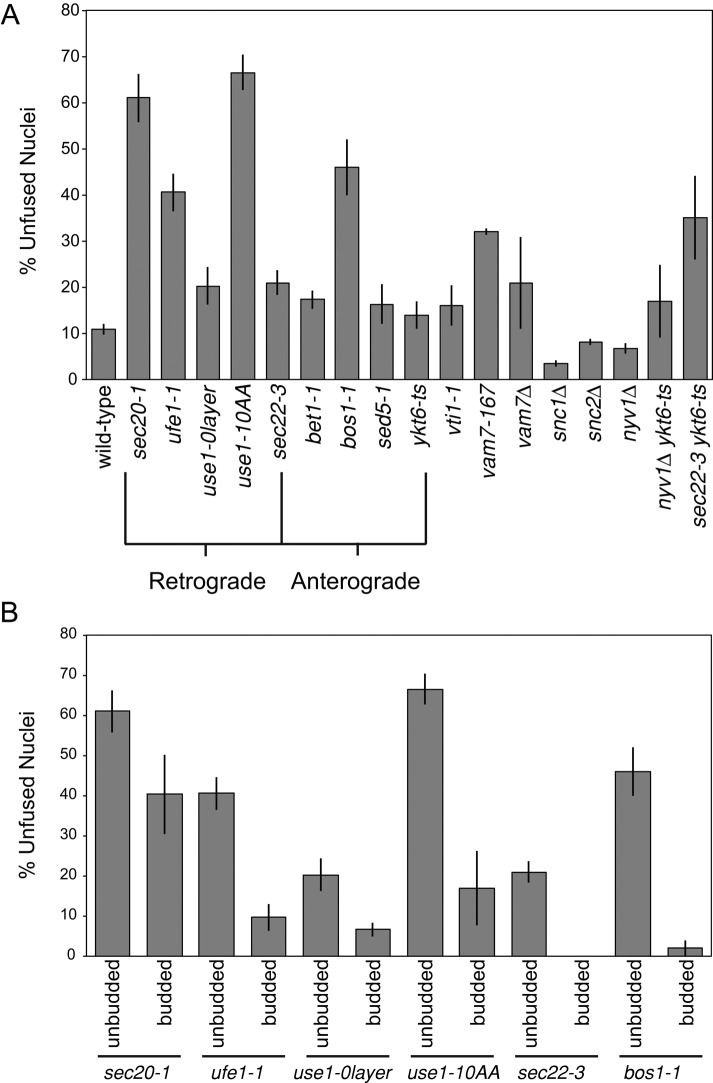
At the semipermissive temperature, sec20-1, ufe1-1, use1-10AA, and bos1-1 mutants all exhibited strong karyogamy defects (Figure 3A). Sec20p, Ufe1p, and Use1p mediate retrograde trafficking to the ER, whereas Bos1p mediates anterograde trafficking from the ER. Of note, these four SNAREs reside primarily in the ER/nuclear envelope (Figure 1). In contrast, mutations in sed5, sec22, bet1, and ykt6 had no or minor defects relative to wild type. These SNAREs also mediate ER/Golgi trafficking but are resident on the vesicles or Golgi. Mutations affecting SNAREs unrelated to ER–Golgi trafficking had either minor or no defects relative to wild type. Although the temperature-sensitive vam7-167 strain exhibited a moderate defect, the vam7 deletion exhibited no defect relative to wild type, suggesting that the Vam7-167 protein is interfering with other SNAREs. Thus we conclude that a subset of nuclear-envelope associated SNAREs, and not ones mediating the secretory pathway in general, is required for karyogamy.
FIGURE 3:
ER-bound SNAREs are required for efficient nuclear fusion. (A) Nuclear fusion efficiencies derived from quantitative matings. Each cross is the indicated genotype for both MATa and MATα (e.g., sec20-1 × sec20-1). Wild type was averaged across multiple isogenic controls for each SNARE mutant; no significant difference for wild-type fusion rates was seen across genotypes; see Table 2 for complete genotypes and strain names. Each strain was grown to log phase at 23°C and then mated at 30°C (except 33°C for ufe1-1). The sec22-3 ykt6-ts strain was grown at 18°C and mated at 23°C (mating was almost absent at 27°C). Only unbudded or small-budded zygotes were scored (see Materials and Methods). The means of multiple experiments were averaged (at least three independent experiments per genotype, with each individual experiment counting ∼50–100 zygotes). Errors bars show ± SEM. (B) Nuclear fusion efficiencies as in A, but budded zygotes (excluded in A) are also shown.
Karyogamy mutants are classified as either unilateral or bilateral; bilateral mutants have a defect only when mated against themselves, whereas unilateral mutants are defective even when mated against wild type. The SNARE mutants either had no or very little nuclear fusion defect when mated against wild-type cells, indicating that they are bilateral mutants (Supplemental Figure S1). It is possible that the wild-type SNAREs can transfer into both nuclear envelopes after cell fusion, either through new translation or diffusion within the ER. Alternatively, nuclear fusion may not require the same SNARE proteins to be resident in both nuclear envelopes.
Nuclear fusion is only partially blocked in the SNARE mutants. Possibly, there is still sufficient SNARE activity at the semipermissive temperature to support some fusion. In this model, the initial interaction between apposed nuclei might not support nuclear envelope fusion, but eventually, over longer time periods, a functional SNARE complex may form and complete membrane fusion. To test this hypothesis, we compared unbudded zygotes, which are early in the conjugation process, with zygotes that have already initiated budding and have therefore completed conjugation and reentered the mitotic pathway. Consistent with this hypothesis, we found that the nuclear fusion defect was readily apparent in unbudded zygotes but greatly reduced in budded zygotes (Figure 3B). Thus the temperature-sensitive SNARE mutations greatly delay nuclear fusion but do not abolish it, consistent with their partial loss of function. We conclude that the SNAREs are required for nuclear fusion. Because we cannot completely abolish SNARE activity in our in vivo assay, we cannot rule out the possibility that there is a redundant, but less efficient, fusion pathway that functions in the absence of SNARE activity.
SNAREs facilitate membrane fusion by forming a four-helix bundle usually consisting of three Q-SNAREs and one R-SNARE (Fasshauer et al., 1998). In yeast, there are only five known R-SNAREs: Sec22p, Ykt6p, Nyv1p, Snc1p, and Snc2p. Because Sec20p, Ufe1p, Use1p, and Bos1p are all Q-SNAREs, we examined the set of R-SNAREs more extensively for a role in karyogamy. No single R-SNARE mutant had a nuclear fusion defect (Figure 3A). Because the R-SNAREs Sec22p and Ykt6p are believed to function redundantly in anterograde and retrograde trafficking (Liu and Barlowe, 2002), we created a sec22-3 ykt6-ts double mutant. The permissive temperature for growth of the double mutant was reduced from 23 to 18°C, and the permissive temperature for mating was reduced from 30 to 23°C. Nevertheless, it exhibited only a minor karyogamy defect (Figure 3A), similar to that seen for SNARE mutations affecting other steps in secretion. Furthermore, this defect may be exaggerated, as wild-type mating is less efficient at 23°C (Figure 2B). The sec22Δ mutant exhibited no karyogamy defect at any temperature tested and was inviable when combined with ykt6-ts, as previously reported (Liu and Barlowe, 2002; unpublished data). Therefore we tentatively conclude that a novel complex comprising four Q-SNAREs mediates nuclear fusion. It is possible, however, that more than two different R-SNAREs redundantly contribute to nuclear envelope fusion.
The ER fusogen, Sey1p, is also required for efficient nuclear fusion
SNAREs are not the only proteins that mediate membrane fusion in yeast. Mitochondrial fusion in yeast is controlled by Fzo1p, Ugo1p, and Mgm1 (Meeusen and Nunnari, 2005). Although these proteins localize to the mitochondria and have not been reported to reside on the ER/NE membrane, we nevertheless tested the fzo1Δ strain for defects in nuclear fusion. We observed no karyogamy defect (6% unfused nuclei). Homotypic ER fusion in yeast and other eukaryotes is mediated by the dynamin-like GTPase Sey1p, a transmembrane ER-protein (Hu et al., 2009; Anwar et al., 2012). We first confirmed reports that sey1Δ strains do not have a mitotic growth defect at any temperature (Hu et al., 2009). Nevertheless, sey1Δ cells exhibited an intermediate karyogamy defect at 30 and 33°C, similar to the SNARE mutants (Figure 4A). Of interest, the sey1Δ defect became significantly more severe when cells were mated at lower temperatures (Figure 4A). A decrease in the efficiency of nuclear fusion at low temperature was also observed for the wild type; this might reflect a change in membrane fluidity or inherent cold sensitivity of other mating proteins required after cell fusion. Like the SNARE mutants, the sey1Δ cells had no defect when mated against wild type (mean 4% defect). Single deletion of YOP1, an ER membrane protein involved in shaping, but not fusing, the ER network, caused a small, but not significant defect in nuclear fusion (p = 0.46, t test). Additional deletion of RTN1 and RTN2, two additional ER remodeling proteins (Voeltz et al., 2006), did not increase the magnitude of the yop1Δ defect, although it was significant relative to wild type (p = 0.04, t test; Figure 4A). Therefore, although proper ER morphology may play a contributing role, we conclude that nuclear fusion depends largely on the fusogen Sey1p and not on more general ER-shaping proteins. Of note, a recent report also implicated Sey1p in nuclear fusion (Chen et al., 2012).
FIGURE 4:
Sey1p is required for efficient nuclear fusion. (A) Nuclear fusion efficiencies as described in Figure 3A, except that budded zygotes were not excluded (zygotes that had initiated or completed nuclear division were still excluded). In most experiments, cells were grown to log phase at 23°C and then mated at the indicated temperatures. Values are aggregated from multiple sey1Δ strains in different background genotypes (Table 2); background genotype did not significantly affect nuclear fusion rates. The means of multiple experiments were averaged (at least three independent experiments per genotype). Errors bars show ± SEM. (B) Nuclear fusion efficiencies in unbudded and budded zygotes, as in Figure 3B. Of the 27 sey1Δ 30°C experiments, we analyzed a subset of five to quantify the unbudded and budded nuclear fusion rates shown here.
It is interesting that the sey1Δ mutant exhibited an intermediate karyogamy defect. In the SNARE mutants, this was explained by partial SNARE activity at the semipermissive temperature. This explanation seems unlikely for sey1Δ cells, as there is no residual Sey1p activity. In support of the idea that the sey1Δ mutant lacks residual Sey1p activity, we observed that, unlike the SNARE mutants, there was no increase in the frequency of nuclear fusion over time (budded zygotes showed the same frequency of fusion as unbudded zygotes; Figure 4B). These data suggest that either there is a redundant mechanism for nuclear fusion in the absence of Sey1p or Sey1p mediates a stochastic but time-independent process required for nuclear fusion.
sey1Δ mutants block nuclear congression; SNAREs fuse the nuclear envelope
It was surprising that two different membrane fusion systems, SNAREs and Sey1p, might both be required for nuclear envelope fusion. The severity of the nuclear fusion defects (>60%) for each seemed too high to be consistent with simple additive processes, suggesting that either the proteins interact as part of one complex or the two fusogens might act at different steps in karyogamy. Previous work (Kurihara et al., 1994; Shen et al., 2009) demonstrated that one hallmark of a nuclear envelope fusion defect (e.g., kar5, prm3) is that nuclei become very closely apposed (<0.3 μm separation). In contrast, mutations that abolish nuclear congression (e.g., kar1, kar3) result in widely separated nuclei. We therefore examined the nuclear separation distances in DAPI-stained mutant zygotes to determine whether the SNAREs or Sey1p might be required at an earlier step. As previously observed, kar1Δ15 zygotes did not complete nuclear congression (mean internuclear distance, 2.3 μm; Figure 5A), whereas kar5Δ mutant zygotes contained nuclei that were uniformly closely apposed (mean, 0.3 μm; Figure 5A). Similarly, the ufe1-1 mutant zygotes contained closely apposed nuclei (mean, 0.4 μm). In contrast, the unfused nuclei in sey1Δ mutant zygotes exhibited a broad range of internuclear distances (mean, 0.9 μm) and were significantly further apart than either the kar5Δ (p < 0.001) or the ufe1-1 mutant (p = 0.003) but closer together than the kar1Δ15 mutant (p < 0.001; Figure 5A). The highly variable internuclear distances in sey1Δ zygotes suggest that they initiate but do not complete nuclear congression. On the contrary, the closely apposed nuclei of the ufe1-1 zygotes suggest that Ufe1p may be specifically required for nuclear fusion but not congression. It is intriguing that the use1-10AA, bos1-1, and sec20-1 mutants showed an intermediate range of internuclear distances, suggesting that they may partially contribute to congression.
FIGURE 5:

sey1Δ zygotes accumulate ER and block nuclear congression. (A) Boxplots of the minimum distances, determined manually, between nuclei in zygotes of the indicated genotypes. Distances are pooled from multiple experiments, except for kar1Δ15 data, which are derived from a single experiment. Standard boxplots are used; black bars represent the median value, gray diamonds represent the mean (calculated excluding outliers), and outliers are shown as points beyond the 1.5*interquartile range (IQR) past the box. Notches represent an approximate 95% confidence interval for the median, calculated as ±1.58*IQR/sqrt(n). For clarity, a few outliers beyond 5 μm are not shown. p values vs. sey1Δ (two-sided, two-sample Kolmogorov–Smirnov test): sec20-1, 0.18; ufe1-1, 0.003; use1-10AA, 0.002; bos1-1, 0.10; kar5Δ, <0.001; sey1Δ kar5Δ, 0.10; kar1Δ15, <0.001. (B) Live microscopy of wild- type (MS5 × MS34) and (C) sey1Δ (MS8072 × MS8073); MATa cells express GFP-Prm3p (MR6362), and MATα cells express mCherry-HDEL (MR6474). Numbers indicate minutes elapsed. Scale bars, 2 μm. (D) Matings as in B, but zygotes were first fixed in formaldehyde, stained with DAPI, and then imaged on the same day (see Materials and Methods). NE, GFP-Prm3p (MR6362); DNA, DAPI; merge, NE + DNA + brightfield image. (E) Representative examples of zygotes mated as in D with (arrow, left) and without (right) accumulated ER. Examples zygotes are sey1Δ (MS8072 × MS8073). ER, mCherry-HDEL (MR6474); DNA, DAPI. Graph on the right, percentage of zygotes that had accumulated ER (scored simply as yes or no) between nuclei. Only zygotes containing unfused nuclei and clearly marked ER (some cells had a diffuse labeling) were scored (WT, n = 4; sey1Δ, n = 62; sec20-1, n = 11; ufe1-1, n = 17; bos1-1, n = 8). Error bars show ± SE for a binomial distribution. All scale bars, 2 μm.
If Sey1p affects nuclear congression before nuclear fusion, then we would expect double mutants of sey1Δ and kar5Δ to be blocked mostly at the nuclear congression step and therefore have internuclear distances similar to the sey1Δ single mutant. Consistent with this hypothesis, we observed that unfused nuclei in the sey1Δ kar5Δ mutant zygotes had internuclear distances similar to the sey1Δ single mutant (Figure 5A).
All previous mutations found to cause nuclear congression defects have been in genes affecting cytoplasmic microtubules. DAPI staining of zygotes provides information about the positions of the nuclear masses but not about the nuclear envelopes. Therefore, to understand how an ER fusion protein could affect congression, we examined the ER and nuclear envelopes in the mutant zygotes. Specifically, the matings included markers to visualize the ER lumen (mCherry-HDEL) and outer nuclear envelope (green fluorescent protein [GFP]–Prm3p). In wild-type zygotes, very little peripheral ER was observed between the two nuclei; the nuclei moved together rapidly, and nuclear fusion ensued soon after cell fusion (<30 min; Figure 5B and Supplemental Movie S1). In contrast, in the sey1Δ zygotes, we frequently observed a large mass of ER at the site where cells had recently fused, most often between the two nuclei. Time-lapse microscopy revealed that the ER mass was present before cell fusion and persisted throughout mating, apparently blocking nuclear congression (Figure 5C and Supplemental Movie S2). In 22 time-lapse experiments, a mass of ER was observed between the nuclei in 15 zygotes (68%). In 12 of the 15 zygotes (80%) the nuclei failed to fuse. In three cases, the ER mass was observed but was not positioned between the nuclei (Supplemental Movie S3); in two of these three zygotes, the nuclei successfully fused. These data suggest that Sey1p is not directly involved in nuclear fusion but instead is required to maintain the dynamic ER network that permits the completion of nuclear congression.
In contrast to the sey1Δ mutants, in the SNARE mutant zygotes the nuclear envelopes were generally closely apposed. During nuclear fusion, Prm3p becomes enriched adjacent to the spindle pole body, where nuclear fusion occurs (Shen et al., 2009). In the SNARE mutant zygotes, the two nuclear envelopes appear to be connected at a point containing a bright GFP-Prm3p punctum (Figure 5D), indicating that the two nuclei have completed congression. This phenotype is identical to that seen for kar5 mutants. Furthermore, unlike the sey1Δ zygotes, the SNARE mutants only rarely accumulated ER between the unfused nuclei (Figures 5E and 6B). We therefore conclude that the SNAREs are required directly for nuclear envelope fusion, whereas Sey1p acts at an earlier step.
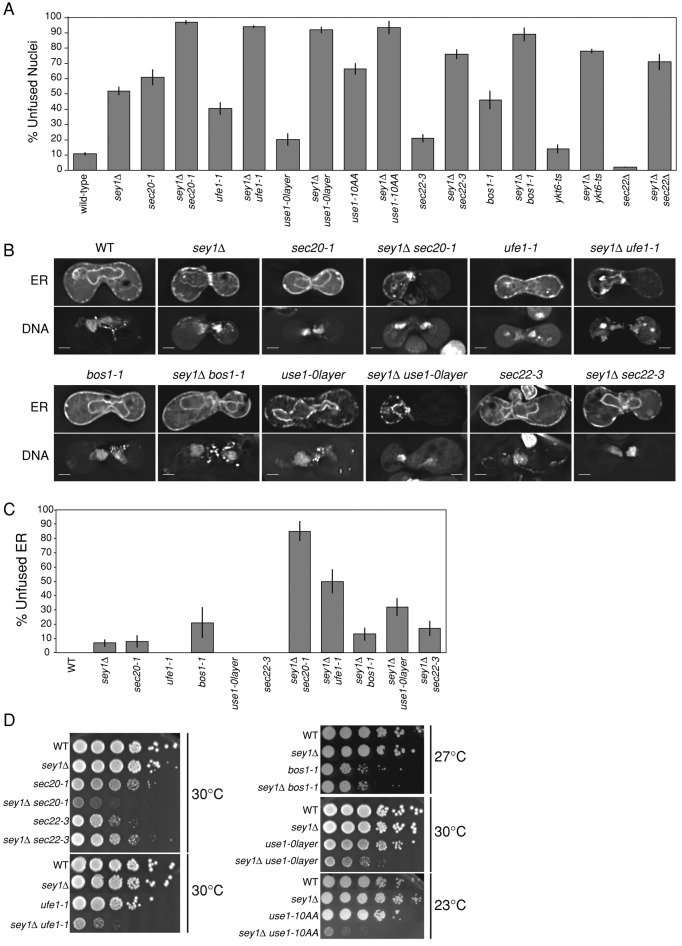
FIGURE 6:
sey1Δ SNARE double mutants exhibit synthetic ER defects. (A) Nuclear fusion efficiencies as in Figure 3A. (B) Example images of ER morphology. Cells mated and fixed as in Figure 5D. ER, mCherry-HDEL (MR6474); DNA, DAPI. Scale bars, 2 μm. (C) Percentage of zygotes with unfused or unequally distributed ER (mCherry-HDEL equilibrated rapidly in wild-type cells; cells more strongly labeled on one-half of the zygote were scored as unfused, such as in the example zygote for sey1Δ ufe1-1 in B). Only zygotes containing unfused nuclei and expressing the mCherry-HDEL marker were scored (but not excluded if ER labeling was somewhat diffuse, as ER fusion was still evident). Number of zygotes scored: WT, 7; sey1Δ, 90; sec20-1, 30; ufe1-1, 21; bos1-1, 14; use1-0layer, 5; sec22-3, 13; sey1Δ sec20-1, 26; sey1Δ ufe1-1, 34; sey1Δ bos1-1, 53; sey1Δ use1-0layer, 56; sey1Δ sec22-3, 47. Error bars show ± SE for a binomial distribution. (D) Growth assays as described in Figure 2A. Before diluting and plating, cultures were grown to saturation in YEPD at 23°C. Plates were grown at the indicated temperatures for 2 or 3 d. A variety of temperatures were tested, and the highest temperature permissive for robust growth is shown.
Sec20p, Ufe1p, and Use1p, but not Bos1p, redundantly fuse ER in sey1Δ cells
We hypothesize that Sey1p affects nuclear fusion indirectly by regulating the dynamic remodeling of the ER, which is required for efficient nuclear congression, whereas the SNAREs act directly at the step of nuclear envelope fusion. On the assumption that the two steps of ER fusion and NE fusion are sequential and occur independently, the fraction of double mutants that successfully complete karyogamy should be the product of the success rate of each single mutant. To test this hypothesis, we examined the frequency of nuclear fusion in matings between sey1Δ SNARE double mutants. In general, we found that most sey1Δ SNARE double-mutant matings exhibited high levels of karyogamy failure, which were more severe than the prediction based on the multiplicative model (Figure 6A). For example, in the sey1Δ single-mutant zygotes, NE fusion failed 53% (±3%) of the time, and in the sec20-1 mutant zygotes NE fusion failed 61% (±5%) of the time. In the multiplicative model we expect that nuclear fusion would fail 82% of the time. Instead, in the double-mutant zygotes we observed NE fusion failure at a rate of 97% (±2%), significantly greater than predicted (p = 0.01, one-sample t test). Similarly high, synergistic rates of NE fusion failure (>90%) were observed for the double mutants involving sey1Δ and ufe1-1 (94% failure observed vs. 72% expected, p = 0.002) and use1-0layer mutations (92% observed vs. 62% expected, p = 0.006). In contrast, the sey1Δ bos1-1, sey1Δ sec22-3, and sey1Δ sec22Δ double mutants were not significantly more defective than expected (p > 0.05), demonstrating that sey1Δ does not generally exacerbate all SNARE mutant phenotypes. We conclude from these results that, contrary to the initial assumptions, there is at least one step during karyogamy in which Sey1p and certain SNARE proteins function redundantly.
To determine which step of karyogamy might require both Sey1p and SNARE proteins, we examined ER and nuclear markers in the mutant zygotes as before. Remarkably, we found that some of the double-mutant strains exhibited more-extreme ER morphology defects (Figure 6B). Moreover, the ER-lumenal marker was frequently restricted to the membranes of one parent cell, even in some budded zygotes, indicating a complete block in ER fusion (Figure 6C). This differs from sey1Δ zygotes, in which the ER lumenal marker equilibrates soon after cell fusion, indicating that some ER fusion still occurs. Of note, even the sey1Δ use1-0layer double mutant exhibited a high karyogamy and ER fusion defect, despite the fact that the use1-0layer single mutant had no defects in karyogamy, ER morphology, or mitotic growth rate. These results corroborate previous studies on Ufe1p (Patel et al., 1998; Anwar et al., 2012) and demonstrate a role for Sec20p and Use1p in ER fusion.
Of interest, sey1Δ bos1-1 and sey1Δ sec22-3 double mutants had only slight ER-fusion defects and did not exhibit significantly worse karyogamy defects than expected. We next examined the growth rates of the single and double mutants. We found that the growth rates recapitulated the ER fusion data: sey1Δ combined with sec20-1, ufe1-1, use1-0layer, or use1-10AA resulted in strong synthetic growth defects, even at the permissive temperatures for the single mutants, whereas sey1Δ combined with bos1-1 or sec22-3 had no synthetic growth defect (Figure 6D). We conclude that Bos1p, while required for efficient nuclear fusion, has no role in peripheral ER fusion. These results demonstrate that SNARE-mediated ER fusion is distinct from NE fusion, as each process has distinct protein requirements.
DISCUSSION
In this study, we demonstrated that ER membrane fusion is required at two different steps during mating for yeast nuclear fusion. Four SNARE proteins—Sec20, Ufe1p, Use1p, and Bos1p—mediate a late step: fusion of the nuclear envelopes. These four SNAREs reside in the NE/ER, and mutations in these proteins block nuclear fusion regardless of whether they function in anterograde or retrograde trafficking. In contrast, other SNARE mutations affecting ER trafficking had no effect on nuclear fusion. Therefore the nuclear fusion defect is not due to a general ER-trafficking defect, suggesting that these SNAREs provide a novel karyogamy-specific function.
We also demonstrated that ER fusion and remodeling is required after cell fusion. In sey1Δ cells, ER membranes accumulate at the zone of cell fusion and block nuclear congression. Although some ER fusion still occurs in the sey1Δ mutants, it is insufficient to maintain normal ER-network morphology. In the absence of Sey1p, SNARE-mediated ER fusion provides a second, partially redundant pathway for ER remodeling, which is required for normal mitotic growth. The Sey1p-independent, SNARE-mediated ER fusion pathway requires at least the retrograde SNAREs Sec20p, Ufe1p, and Use1p, but not Sec22p, Ykt6p, or Bos1p.
Of importance, Bos1p is required for nuclear fusion but not peripheral ER fusion. This observation supports our model of two distinct membrane fusion steps in karyogamy, with Bos1p acting specifically in the later step. Furthermore, it argues that the NE fusion defect observed in the SNARE mutants is not simply a secondary consequence of an earlier ER fusion defect (Figure 7).
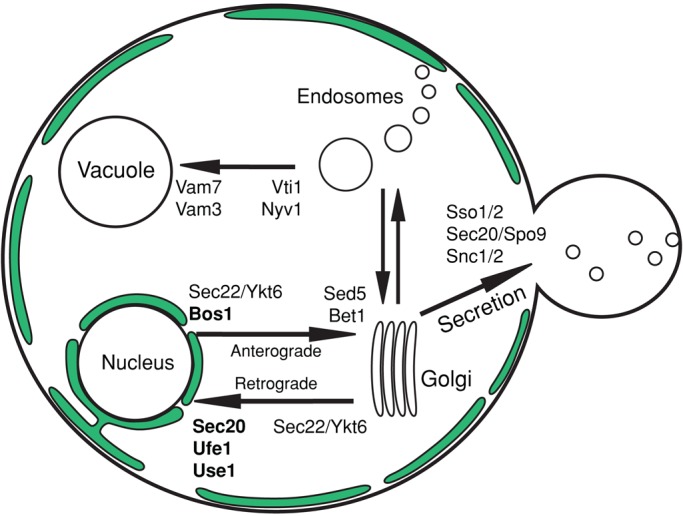
FIGURE 7:
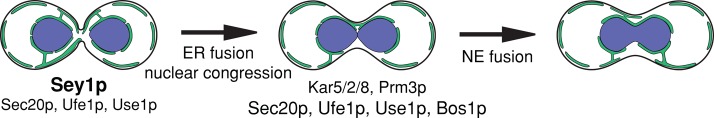
An updated pathway for ER and nuclear fusion. After cell fusion, nuclear congression and Sey1p-mediated ER fusion allow nuclei to become closely apposed. When Sey1p is absent, at least Sec20p, Ufe1p, and Use1p provide the remaining ER fusion activity. After nuclear apposition, outer nuclear envelope fusion requires the SNAREs Sec20p, Ufe1p, Use1p, and Bos1p, as well as the previously characterized proteins Prm3p and Kar5p (Melloy et al., 2009). Inner nuclear envelope fusion requires Kar5p, Kar2p, and Kar8p (Melloy et al., 2009). Black outline represents the plasma membrane, green lines represent the cortical ER and nuclear ER, with a few interconnecting tubules shown, and blue represents the nucleoplasm.
Usually, three Q-SNAREs and one R-SNARE comprise a SNARE complex. However, we found that karyogamy depends on four Q-SNAREs and not on the ER-related R-SNAREs Sec22p and Ykt6p. It remains possible that Sec22p and Ykt6p are involved but functionally redundant with another R-SNARE. The difficulty in constructing viable triple mutants makes this difficult to assess. Alternatively, there may be no R-SNARE required for homotypic ER/NE fusion. For example, the R-SNARE Snc2p can be mutated from R to Q with no apparent phenotype (Katz and Brennwald, 2000; Ossig et al., 2000), suggesting that at least some four–Q-SNARE complexes are functional in vivo.
Although we did not examine mutations in all SNAREs for karyogamy defects, our screen is likely to be exhaustive for the set of SNAREs that could mediate karyogamy. Proteins capable of fusing nuclei must reside in the ER/NE, and the only ER-bound SNAREs are Sec20, Ufe1p, Use1p, Sec22p, and Bos1p. The only SNAREs found to have roles in karyogamy comprise four of the five ER-bound SNAREs (the exception is Sec22p). The other SNAREs involved in ER trafficking are Golgi or vesicle bound and were not required for efficient nuclear fusion. We additionally sampled several vacuolar and endosome-associated SNAREs (Vti1p, Nyv1p, Vam7p) and found no karyogamy role. Four SNAREs—Ykt6p, Vam7p, Sec9p, and Spo20p—are soluble in the cytosol and bind peripherally to membranes and other SNAREs, and these could function similarly in nuclear fusion. Ykt6p is known to function redundantly with Sec22p (Liu and Barlowe, 2002). However, even in a sec22-3 ykt6-ts double mutant there was only a slight karyogamy defect. It seems likely that such mild defects arise as a secondary consequence of perturbed ER trafficking and recycling. Similarly, the vam7-167 mutant exhibited a mild defect, which was not observed for a vam7 deletion. Sec9p and Spo20p were not tested, as Sec9p is required solely for exocytosis, unlike Ykt6p and Vam7p, which function at multiple trafficking steps, and is thus unlikely to mediate nuclear fusion. Spo20p is expressed only during meiosis. Therefore we have assayed all of the known SNAREs that might mediate nuclear fusion.
Normal vesicle trafficking requires not only SNAREs, but also coat proteins, Rab-GTPases, associated GTPase-activating proteins and guanine nucleotide exchange factors, cytoskeletal proteins for movement, tethering complexes, and Sec1/Munc18 protein family members (Lee et al., 2004). In SNARE-mediated nuclear fusion, it is unlikely that vesicle formation and coat proteins are relevant. Similarly, because tethering complexes function partly by binding incoming coat proteins, they may not be important for nuclear fusion (reviewed in Cai et al., 2007). On the other hand, karyogamy-specific proteins may provide alternate functions, such as nuclear membrane tethering or modifying SNARE specificity. In support of this hypothesis, Prm3p, a key nuclear fusion protein, physically interacts with Sec20p in in vitro pull-down experiments (Shen, 2008). Furthermore, Prm3p overexpression partially suppresses the nuclear fusion defect observed for both sec20-1 and ufe1-1 mutants (Shen, 2008).
The role of Sey1p and ER morphology during karyogamy
Our data suggest that Sey1p is indirectly required for karyogamy and that aberrant ER morphology blocks nuclear congression. The observation that sey1Δ zygotes fuse nuclei in ∼50% of matings is likely due to heterogeneity in the amount and position of the accumulated ER before cell fusion (Figure 5E). Unlike SNARE mutant zygotes, the percentage of fused nuclei did not increase over time (i.e., in unbudded vs. budded zygotes; Figure 4B), implying that if sufficient ER accumulates to block fusion, then it is essentially irreversible. Although unfused nuclei may eventually interact, nuclei are competent to fuse only during a brief time period after cell fusion but before reentry into mitosis (Rose, 1991).
A recent study also implicated SEY1 in nuclear fusion (Chen et al., 2012). However, in that study, only budded zygotes were examined, and nuclei appeared to have congressed but not fused, suggesting that Sey1p mediates nuclear fusion directly. However, budded zygotes usually contain nuclei that have oriented and moved toward the zygotic bud, masking any original congression defect (Kurihara et al., 1994). Looking at unbudded sey1Δ zygotes, we found instead that congression is defective and propose that Sey1p mediates nuclear fusion indirectly.
SNARE-mediated ER fusion
Ufe1p was previously implicated in ER fusion (Patel et al., 1998). A recent study of sey1Δ ufe1-1 double mutants showed greatly decreased ER fusion rates, beyond either single mutant (Anwar et al., 2012). The authors hypothesized that there are two pathways for ER fusion and that the SNARE-mediated pathway is secondary to Sey1p-mediated ER fusion. They further identified negative genetic interactions (slow growth) between sey1Δ and sec20-1 and use1-10AA and a lack of interaction with sec22Δ. Our results confirmed that at least Sec20p, Ufe1p, and Use1p mediate Sey1p-independent ER fusion and excluded a role for Sec22p, Ykt6p, and Bos1p. However, as with nuclear fusion, Sec22p/Ykt6p redundancy could mask a role in ER fusion.
The sey1Δ SNARE double mutants exhibited slow growth even at temperatures at which the single SNARE mutants had normal growth and retrograde trafficking (23°C). This suggests either that SNAREs function somewhat differently in ER fusion and vesicle trafficking or vesicle trafficking is less sensitive to minor loss of SNARE activity. Of interest, sey1Δ use1-0layer cells had a strong ER defect, whereas the use1-0layer single mutant had no ER, karyogamy, or growth phenotype at any temperature tested. Therefore the use1-0layer mutation may be a separation-of-function allele that could aid future studies of the SNARE-mediated ER-fusion pathway.
Together our results demonstrate roles for SNAREs and Sey1p in nuclear fusion and a novel role for SNAREs in ER fusion. Future studies should address whether the proteins required in vesicle trafficking are also required in NE and ER fusion and whether there are SNARE-interacting proteins specific to each pathway that do not function in vesicle trafficking.
MATERIALS AND METHODS
Strains and yeast methods
All strains and plasmids used are described in Table 2. MS strains are isogenic to S288C; MY strains have various backgrounds. The mutations associated with the temperature-sensitive alleles are listed in Table 1. General methods, including cell culture, media, and transformations, have been described previously (Rose et al., 1990).
TABLE 2:
Strains and plasmids.
| Strain | Genotype | Source |
|---|---|---|
| MS5 | MATa ura3-52 leu2-3112 | Rose laboratory |
| MS34 | MATα ura3-52 leu2-3112 | Rose laboratory |
| MS1270 | MATa ura3-52 leu2-3112 ade2-101 trp1Δ1 cyh-r kar1Δ15 | Rose laboratory |
| MS1271 | MATα ura3-52 leu2-3112 trp1Δ1 kar1Δ15 | Rose laboratory |
| MS7658 | MATa ufe1Δ::HIS3 ura3-52 leu2-3112 his3Δ200 trp1Δ1 [ufe1-1 LEU2] | Rose laboratory |
| MS7659 | MATα ufe1Δ::HIS3 ura3-52 leu2-3112 his3Δ200 ade2-101 [ufe1-1 LEU2] | Rose laboratory |
| MS8072 | MATa ura3-52 leu2-3112 sey1Δ::KanMX | This study |
| MS8073 | MATα ura3-52 leu2-3112 sey1Δ::KanMX | This study |
| MS8278 | MATa ura3-52 leu2-3112 HIS3+ ufe1Δ::HIS3 sey1Δ::KanMX [ufe1-1 LEU2] | This study |
| MS8279 | MATα ura3-52 leu2-3112 HIS3+ ufe1Δ::HIS3 sey1Δ::KanMX [ufe1-1 LEU2] | This study |
| MS7311 | MATα ade2-101 his3Δ200 kar5Δ::HIS3 trp1Δ1 ura3-52 | Rose laboratory |
| MS7312 | MATa ade2-101 his3Δ200 kar5Δ::HIS3 leu2-3112 ura3-52 | Rose laboratory |
| MS8281 | MATa ura3-52 leu2-3112 trp1Δ1 HIS3+ kar5Δ::HIS3 sey1Δ::KanMX | This study |
| MS8282 | MATα ura3-52 leu2-3112 HIS3+ kar5ΔHIS3 sey1Δ::KanMX | This study |
| MS8299 | MATa ura3-52 trp1Δ1 | This study |
| MS8300 | MATα ura3-52 leu2-3112 | This study |
| MS8301 | MATa ura3-52 sey1Δ::KanMX leu2-3112 | This study |
| MS8302 | MATα ura3-52 sey1Δ::KanMX leu2-3112 | This study |
| MS8303 | MATa ura3-52 sec22Δ::KanMX leu2-3112 trp1Δ1 | This study |
| MS8304 | MATα ura3-52 sec22Δ::KanMX trp1Δ1 | This study |
| MS8305 | MATa ura3-52 sec22Δ::KanMX sey1Δ::KanMX trp1Δ1 | This study |
| MS8306 | MATα ura3-52 sec22Δ::KanMX sey1Δ::KanMX leu2-3112 trp1Δ1 | This study |
| MY2050 | MATα ura3-52 leu2-3112 | Rose laboratory |
| MY2051 | MATa ura3-52 leu2-3112 | Rose laboratory |
| MY2064 | MATα sec20-1 his4-619 ura3-52 | Rose laboratory |
| MY2065 | MATa sec20-1 his4-619 ura3-52 | Rose laboratory |
| MY12582 | MATa ura3-52 leu2-3112 sey1Δ::KanMX | This study |
| MY13359 | MATα ura3-52 his4-619 sey1Δ::KanMX | This study |
| MY13368 | MATa ura3-52 leu2-3112 sey1Δ::KanMX sec20-1 | This study |
| MY12606 | MATα his4-619 ura3-52 sey1Δ::KanMX sec20-1 | This study |
| MY2068 | MATα sec22-3 his4-619 ura3-52 | Rose laboratory |
| MY2069 | MATa sec22-3 ura3-52 | Rose laboratory |
| MY12584 | MATα sec22-3 his4-619 ura3-52 sey1Δ::KanMX | This study |
| MY12585 | MATa sec22-3 ura3-52 sey1Δ::KanMX | This study |
| MY9941 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- | G. Fischer von Mollard (Universität Göttingen, Göttingen, Germany) |
| MY9942 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- | G. Fischer von Mollard |
| MY9943 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-0layer LEU2] | G. Fischer von Mollard |
| MY9944 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-0layer LEU2] | This study |
| MY9945 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-10AA HIS3] | G. Fischer von Mollard |
| MY9946 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-10AA HIS3] | G. Fischer von Mollard |
| MY12607 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 sey1Δ::KanMX [use1-0layer LEU2] | This study |
| MY12608 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 sey1Δ::KanMX [use1-0layer LEU2] | This study |
| MY13590 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- | This study |
| MY13591 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- | This study |
| MY13592 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 suc2Δ9 mel- sey1Δ::KanMX | This study |
| MY13593 | MATα leu2-3112 ura3-52 his3Δ200 trp1-Δ901 suc2-Δ9 mel- sey1Δ::KanMX | This study |
| MY13594 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-10AA HIS3] | This study |
| MY13596 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- use1Δ::TRP1 [use1-10AA HIS3] | This study |
| MY13807 | MATa leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- sey1Δ::KanMX use1Δ::TRP1 [use1-10AA HIS3] | This study |
| MY13808 | MATα leu2-3112 ura3-52 his3Δ200 trp1Δ901 lys2-801 suc2Δ9 mel- sey1Δ::KanMX use1Δ::TRP1 [use1-10AA HIS3] | This study |
| MY9997 | MATa bos1-1 lys2-801 leu2-3112 | H. Riezman (University of Geneva, Geneva, Switzerland) |
| MY9998 | MATα bos1-1 ura3 trp1 his4 lys2 leu2 | H. Riezman |
| MY9999 | MATa ura3 trp1 his4 lys2 leu2 | H. Riezman |
| MY10000 | MATα trp1 leu2 ura3 his4 lys2 | H. Riezman |
| MY12588 | MATa bos1-1 lys2-801 leu2-3112 sey1Δ::KanMX | This study |
| MY12589 | MATα bos1-1 ura3 trp1 his4 lys2 leu2 sey1Δ::KanMX | This study |
| MY12590 | MATa ura3 trp1 his4 lys2 leu2 sey1Δ::KanMX | This study |
| MY12591 | MATα trp1 leu2 ura3 his4 lys2 sey1Δ::KanMX | This study |
| MY9839 | MATα ura3-52 his4-619 bet1-1 | S. Ferro-Novick (University of California, San Diego, La Jolla, CA) |
| MY9840 | MATa ura3-52 his4-619 bet1-1 | S. Ferro-Novick |
| MY9841 | MATa ura3-52 | S. Ferro-Novick |
| MY9842 | MATα sed5-1 ura3-52 leu2-3112 his3Δ200 trp1Δ901 lys801 suc2Δ9 | S. Ferro-Novick |
| MY10004 | MATα lys2-801 trp1Δ901 ura3-52 his3Δ200 sed5-1 | This study |
| MY10005 | MATa leu2-3112 trp1Δ901 ura3-52 sed5-1 | This study |
| MY10006 | MATα ura3-52 his3Δ200 | This study |
| MY10007 | MATa leu2-3112 ura3-52 lys-801 | This study |
| MY11614 | MATa ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 can1Δ::LEU2-MFA1pr::HIS3 ykt6-ts::URA3 | P. Hieter (University of British Columbia, Vancouver, Canada) |
| MY11615 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 can1Δ::LEU2-MFA1pr::HIS3 ykt6-ts::URA3 | P. Hieter |
| MY10191 | MATa leu2-3112 ura3-52 his3Δ200 ade2-101 trp1Δ901 suc2Δ9 vti1-1 | G. Fischer von Mollard |
| MY10265 | MATα ura3-52 ade2-101 cyh-R trp1Δ901 lys2-801 vti1-1 | This study |
| MY11926 | MATa leu2-3112 ura3-52 trp1Δ vti1-1 | This study |
| MY11927 | MATα leu2-3112 ura3-52 trp1Δ vti1-1 | This study |
| MY11617 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 snc1Δ::KanMX | Open Biosystems (Huntsville, AL) deletion collection |
| MY11618 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 snc1Δ::KanMX | Open Biosystems deletion collection |
| MY11619 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 snc2Δ::KanMX | Open Biosystems deletion collection |
| MY11620 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 snc2Δ::KanMX | Open Biosystems deletion collection |
| MY11621 | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 nyv1Δ::KanMX | Open Biosystems deletion collection |
| MY11622 | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 nyv1Δ::KanMX | Open Biosystems deletion collection |
| MY10193 | MATα leu2-3112 ura3-52 his3Δ200 ade2-101 trp1Δ901 lys2-901 suc2Δ9 [vam7-167 HIS3] | S. Emr (Cornell University, Ithaca, NY) |
| MY10266 | MATa leu2-3112 ura3-52 trp1Δ901 lys2-901 [vam7-167 HIS3] | This study |
| MY11928 | MATa nyv1Δ::KanMX ykt6-ts::URA3 his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 | This study |
| MY11929 | MATα nyv1Δ::KanMX ykt6-ts::URA3 his3Δ1 leu2Δ0 lys2Δ0 met15Δ0 | This study |
| MY13602 | MATa ura3- leu2Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13603 | MATα ura3- his3Δ1 leu2Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13604 | MATa ykt6-ts::URA3 ura3- lys2Δ0 his3Δ1 leu2Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13605 | MATα ykt6-ts::URA3 ura3- lys2-Δ0 leu2Δ0 | This study |
| MY13606 | MATa sec22-3 ura3- lys2Δ0 his3Δ1 leu2Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13607 | MATα sec22-3 ura3- his3-Δ1 leu2-Δ0 | This study |
| MY13608 | MATa sec22-3 ykt6-ts::URA3 ura3- | This study |
| MY13613 | MATα sec22-3 ykt6-ts::URA3 ura3- lys2Δ0 leu2-Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13632 | MATa ura3- leu2- lys2Δ0 | This study |
| MY13633 | MATα ura3- leu2- his3Δ1 can1Δ::MFA1pr::HIS3 | This study |
| MY13634 | MATa ura3- leu2- ykt6-ts::URA3 his3Δ1 lys2Δ0 can1Δ::MFA1pr::HIS3 | This study |
| MY13635 | MATα ura3- leu2- ykt6-ts::URA3 | This study |
| MY13636 | MATa ura3- leu2- sey1Δ::KanMX | This study |
| MY13637 | MATα ura3- leu2- sey1Δ::KanMX can1Δ::MFA1pr::HIS3 | This study |
| MY13638 | MATa ura3- leu2- sey1Δ::KanMX ykt6-ts::URA3 his3Δ1 can1Δ::MFA1pr::HIS3 | This study |
| MY13639 | MATα ura3- leu2- sey1Δ::KanMX ykt6-ts::URA3 lys2Δ0 | This study |
| MY14358 | MATa ura3- leu2- his3- | This study |
| MY14359 | MATα ura3- leu2- his3- met15Δ0 | This study |
| MY14360 | MATa ura3- leu2- his3- yop1Δ::URA3 rtn1Δ::KanMX rtn2Δ::URA3 | This study |
| MY14361 | MATα ura3- leu2- his3- yop1Δ::URA3 rtn1Δ::KanMX rtn2Δ::URA3 SEC61-GFP::LEU2 | This study |
| MY14362 | MATα ura3- leu2- his3- yop1Δ::URA3 rtn1Δ::KanMX rtn2Δ::URA3 SEC61-GFP::LEU2 | This study |
| N/A | MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0 yop1Δ::KanMX | Open Biosystems deletion collection |
| N/A | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 yop1Δ::KanMX | Open Biosystems deletion collection |
| Plasmid | Relevant markers | Source |
| pMR6362 | PRM3pr-GFP::PRM3 LEU2 CEN4 amp-r | This study |
| pMR6474 | ADH1pr-mCherry::HDEL LEU2 CEN4 amp-r | This study |
Growth assays
To assay growth rate on plates, we first grew cultures to saturation in yeast extract/peptone/dextrose (YEPD) at 23°C. Then 0.2 OD unit of cells was removed, pelleted, and resuspended in 200 μl of distilled H2O. Five 10-fold serial dilutions were made in a 96-well plate, spotted on YEPD plates, and grown at the temperature and time indicated in the figure legends.
Nuclear fusion assays
Nuclear fusion assays were performed as previously described (Gammie and Rose, 2002). Briefly, mating mixtures were grown to log phase, and 0.5 OD unit of cells of each mating type were mixed. If any temperature-sensitive strains were to be tested, all strains were grown at 23°C. Otherwise, strains were grown at 30°C. The mating mixture was vacuum filtered onto a 0.45-μm nitrocellulose filter (EMD Millipore, Billerica, MA) and incubated on a YEPD plate at 30 or 33°C for 2.5 h, 27°C for 3.0 h, or 23°C for 3.25 h. For slow-growing strains, mating was periodically checked every 15 min until a reasonable amount of mating had occurred (>∼1% of cells) and there was a mixture of budded and unbudded zygotes. If the mixture never reached ∼1% zygotes, it was designated as incapable of efficient mating and not quantified. Finally, strains were fixed in 3:1 methanol:acetic acid and stained with 1 μg/ml DAPI.
Usually both budded and unbudded zygotes were imaged and scored. Cells that appeared to have entered mitosis (began dividing their nuclei) were excluded from analysis. In addition, in the single-mutant SNARE strains, as explained in the text and figure legends, the budded zygotes were excluded from analysis. To objectively classify zygotes, the area of the zygote and the area of the bud were measured, and those zygotes with a bud area >10% of the area of the zygote were classified as “budded.” Zygotes with small buds (bud area <10% of the zygote area) were included with the unbudded zygotes.
Live- and fixed-cell microscopy
For live-cell microscopy, cells were grown at 30°C and mated on filter disks, as described for the nuclear fusion assay, for 1.5 h at 23°C. During this period, a 0.17-mm DeltaT4 Culture Dish (Bioptechs, Butler, PA) was coated with 25 μl of concanavalin A (0.1 mg/ml in 20 mM Na acetate, pH 5.8) for 30 min and then washed twice with 20 mM Na acetate, pH 5.8. Cells were then washed into 1 ml of room-temperature, 1× phosphate-buffered saline (PBS), and 15 μl of cells was added to the concanavalin A–treated DeltaT4 dish and allowed to settle for 10 min. Cells were washed once with 100 μl of appropriate selective medium (usually synthetic complete medium lacking uracil and leucine), and then 1 ml of medium was added to the dish. Cells were imaged at room temperature over the course of several hours.
For fixed-cell microscopy, cells were washed from the filter disk to 900 μl of 1× PBS, and 100 μl of 20% paraformaldehyde in distilled H2O was added. Cells were fixed at room temperature for 15 min and then washed once in 1 ml of 1× PBS, stained with 2 μg/ml DAPI in PBS for 15 min, washed twice again in 1 ml of 1× PBS, resuspended in 100 μl of 1× PBS, and then imaged on the same day.
Image acquisition and analysis
Samples were imaged on a DeltaVision deconvolution microscope (Applied Precision, Issaquah, WA), based on a Nikon TE200 (Melville, NY), using a 100×/numerical aperture 1.4 objective, a 50-W mercury lamp, and a Photometrics Cool Snap HQ charge-coupled device camera (Photometrics, Tucson, AZ). In all images, pixel width and height correspond to 49.2 nm. For nuclear fusion assays, we typically acquired z-stacks of 19 slices separated by 0.2 μm. For fixed-cell imaging of GFP or mCherry markers, fewer slices were used to minimize photobleaching, typically approximately nine slices separated by 0.3 μm, with an exposure time of 0.5–1.0 s. For time-course imaging of live cells, photobleaching and phototoxicity necessitated imaging a single slice at each time interval. All images were deconvolved using Applied Precision SoftWoRx imaging software.
Supplementary Material
Acknowledgments
We thank members of the Rose and Gammie labs for helpful support and discussion and Becky Davis for technical assistance and contributions to the quantitative mating assays. We also thank G. Fischer von Mollard, H. Riezman, S. Ferro-Novick, P. Hieter, and S. Emr for strains. This work was supported by National Institutes of Health Grant GM-37739. J.V.R. was supported by National Institutes of Health Training Grant 5 T32 GM 7388-35.
Abbreviations used:
- ER
endoplasmic reticulum
- NE
nuclear envelope
- SNARE
soluble N-ethylmaleimide–sensitive factor attachment protein receptor
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-08-0441) on October 23, 2013.
REFERENCES
- Anwar K, Klemm RW, Condon A, Severin KN, Zhang M, Ghirlando R, Hu J, Rapoport Ta, Prinz Wa. The dynamin-like GTPase Sey1p mediates homotypic ER fusion in S. cerevisiae. J Cell Biol. 2012;197:209–217. doi: 10.1083/jcb.201111115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK, Lewis MJ, Pelham HR. A SNARE-like protein required for traffic through the Golgi complex. Nature. 1995;375:806–809. doi: 10.1038/375806a0. [DOI] [PubMed] [Google Scholar]
- Ben-Aroya S, Coombes C, Kwok T, O'Donnell Ka, Boeke JD, Hieter P. Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol Cell. 2008;30:248–258. doi: 10.1016/j.molcel.2008.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burri L, Lithgow T. A complete set of SNAREs in yeast. Traffic. 2004;5:45–52. doi: 10.1046/j.1600-0854.2003.00151.x. [DOI] [PubMed] [Google Scholar]
- Burri L, Varlamov O, Doege Ca, Hofmann K, Beilharz T, Rothman JE, Söllner TH, Lithgow T. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc Natl Acad Sci USA. 2003;100:9873–9877. doi: 10.1073/pnas.1734000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S. ER network formation requires a balance of the dynamin-like GTPase Sey1p and the Lunapark family member Lnp1p. Nat Cell Biol. 2012;14:707–716. doi: 10.1038/ncb2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Novick P, Ferro-Novick S. ER structure and function. Curr Opin Cell Biol. 2013;25:1–6. doi: 10.1016/j.ceb.2013.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Letourneur F. Coatomer interaction with di-lysine endoplasmic reticulum retention motifs. Science. 1994;263:1629–1631. doi: 10.1126/science.8128252. [DOI] [PubMed] [Google Scholar]
- Dilcher M, Veith B, Chidambaram S, Hartmann E, Schmitt HD, Fischer von Mollard G. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003;22:3664–3674. doi: 10.1093/emboj/cdg339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G, Stevens TH. A human homolog can functionally replace the yeast vesicle-associated SNARE Vti1p in two vesicle transport pathways. J Biol Chem. 1998;273:2624–2630. doi: 10.1074/jbc.273.5.2624. [DOI] [PubMed] [Google Scholar]
- Gammie AE, Rose MD. Assays of cell and nuclear fusion. Methods Enzymol. 2002;351:477–498. doi: 10.1016/s0076-6879(02)51866-8. [DOI] [PubMed] [Google Scholar]
- Grote E. Secretion is required for late events in the cell-fusion pathway of mating yeast. J Cell Sci. 2010;123:1902–1912. doi: 10.1242/jcs.066662. [DOI] [PubMed] [Google Scholar]
- Hu J, Prinz WA, Rapoport TA. Weaving the web of ER tubules. Cell. 2011;147:1226–1231. doi: 10.1016/j.cell.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Shibata Y, Zhu PP, Voss C, Rismanchi N, Prinz Wa, Rapoport Ta, Blackstone C. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–561. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L, Brennwald P. Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol Biol Cell. 2000;11:3849–3858. doi: 10.1091/mbc.11.11.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienle N, Kloepper TH, Fasshauer D. Phylogeny of the SNARE vesicle fusion machinery yields insights into the conservation of the secretory pathway in fungi. BMC Evol Biol. 2009;9:19. doi: 10.1186/1471-2148-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov MM, McMahon HT, Chernomordik LV. Protein-driven membrane stresses in fusion and fission. Trends Biochem Sci. 2010;35:699–706. doi: 10.1016/j.tibs.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Beh CT, Latterich M, Schekman R, Rose MD. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J Cell Biol. 1994;126:911–923. doi: 10.1083/jcb.126.4.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MCS, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Letourneur F, Gaynor EC, Hennecke S, Démollière C, Duden R, Emr SD, Riezman H, Cosson P. Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell. 1994;79:1199–1207. doi: 10.1016/0092-8674(94)90011-6. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Pelham HR. SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- Lewis MJ, Rayner JC, Pelham HR. A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J. 1997;16:3017–3024. doi: 10.1093/emboj/16.11.3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Barlowe C. Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol Biol Cell. 2002;13:3314–3324. doi: 10.1091/mbc.E02-04-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Parlati F, Fukuda R, Johnston RJ, Paz K, Paumet F, Söllner TH, Rothman JE. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–159. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- Meeusen SL, Nunnari J. How mitochondria fuse. Curr Opin Cell Biol. 2005;17:389–394. doi: 10.1016/j.ceb.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, McIntosh JR, Rose MD. Nuclear fusion during yeast mating occurs by a three-step pathway. J Cell Biol. 2007;179:659–670. doi: 10.1083/jcb.200706151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy P, Shen S, White E, Rose MD. Distinct roles for key karyogamy proteins during yeast nuclear fusion. Mol Biol Cell. 2009;20:3773–3782. doi: 10.1091/mbc.E09-02-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AP, Shim J, Ferro-Novick S. BET1, BOS1, and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol Cell Biol. 1990;10:3405–3414. doi: 10.1128/mcb.10.7.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase atlastin. Nature. 2009;460:978–983. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- Ossig R, Schmitt HD, De Groot B, Riedel D, Keränen S, Ronne H, Grubmüller H, Jahn R. Exocytosis requires asymmetry in the central layer of the SNARE complex. EMBO J. 2000;19:6000–6010. doi: 10.1093/emboj/19.22.6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F, McNew JA, Fukuda R, Miller R, Söllner TH, Rothman JE. Topological restriction of SNARE-dependent membrane fusion. Nature. 2000;407:194–198. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- Patel SK, Indig FE, Olivieri N, Levine ND, Latterich M. Organelle membrane fusion: a novel function for the syntaxin homolog Ufe1p in ER membrane fusion. Cell. 1998;92:611–620. doi: 10.1016/s0092-8674(00)81129-0. [DOI] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in yeast. Annu Rev Microbiol. 1991;45:539–567. doi: 10.1146/annurev.mi.45.100191.002543. [DOI] [PubMed] [Google Scholar]
- Rose MD. Nuclear fusion in the yeast Saccharomyces cerevisiae. Annu Rev Cell Dev Biol. 1996;12:663–695. doi: 10.1146/annurev.cellbio.12.1.663. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston FM, Hieter P. Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sacher M, Stone S, Ferro-Novick S. The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J Biol Chem. 1997;272:17134–17138. doi: 10.1074/jbc.272.27.17134. [DOI] [PubMed] [Google Scholar]
- Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-like molecule, and Vam3p, a syntaxin homolog, function together in yeast vacuolar protein trafficking. Mol Cell Biol. 1998;18:5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S. Nuclear Envelope Fusion in the Yeast Saccharomyces cerevisiae PhD Thesis. Princeton, NJ: Princeton University; 2008. [Google Scholar]
- Shen S, Tobery CE, Rose MD. Prm3p is a pheromone-induced peripheral nuclear envelope protein required for yeast nuclear fusion. Mol Biol Cell. 2009;20:2438–2450. doi: 10.1091/mbc.E08-10-0987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Schekman R. Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 1998;143:589–599. doi: 10.1083/jcb.143.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525–528. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S, Sacher M, Mao Y, Carr C, Lyons P, Quinn AM, Ferro-Novick S. Bet1p activates the v-SNARE Bos1p. Mol Biol Cell. 1997;8:1175–1181. doi: 10.1091/mbc.8.7.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Ydenberg CA, Rose MD. Yeast mating: a model system for studying cell and nuclear fusion. Methods Mol Biol. 2008;475:3–20. doi: 10.1007/978-1-59745-250-2_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.