Background: The membrane-associated tBid promotes Bax membrane insertion and activation.
Results: tBid adopts an extended structure in 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LPPG) micelles with its six helices including the BH3 domain interacting with the micelles.
Conclusion: An “on the membrane” binding mode was suggested for tBid interaction with Bax.
Significance: Revealing tBid structure on the membrane is key to the understanding of tBid-mediated Bax activation.
Keywords: Apoptosis, Bcl-2, NMR, Protein Dynamics, Protein Structure, LPPG Micelles, tBid
Abstract
The Bcl-2 family proteins regulate mitochondria-mediated apoptosis through intricate molecular mechanisms. One of the pro-apoptotic proteins, tBid, can induce apoptosis by promoting Bax activation, Bax homo-oligomerization, and mitochondrial outer membrane permeabilization. Association of tBid on the mitochondrial outer membrane is key to its biological function. Therefore knowing the conformation of tBid on the membrane will be the first step toward understanding its crucial role in triggering apoptosis. Here, we present NMR characterization of the structure and dynamics of human tBid in 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] micelles. Our data showed that tBid is monomeric with six well defined α-helices in the micelles. Compared with the full-length Bid structure, a longer flexible loop between tBid helix α4 and α5 was observed. Helices in tBid do not pack into a compact-fold but form an extended structure with a C-shape configuration in the micelles. All six tBid helices were shown to interact with LPPG micelles, with helix α6 and α7 being more embedded. Of note, the BH3-containing helix α3, which was previously believed to be exposed above the membrane surface, is also membrane associated, suggesting an “on the membrane” binding mode for tBid interaction with Bax. Our data provided structural details on the membrane-associated state of tBid and the functional implications of its membrane-associated BH3 domain.
Introduction
Apoptosis (programmed cell death) plays an important role in tissue development and maintenance (1). A group of apoptosis regulator proteins, the Bcl-2 (B-cell lymphoma 2) family proteins, were discovered to work coordinately to modulate mitochondrial outer membrane permeabilization (MOMP)2 (2–4). Bcl-2 family members share at least one of four BH (Bcl-2 homology) domains and many of them possess a transmembrane domain at the C terminus. Based on their apoptotic function, Bcl-2 family proteins are classified into two groups: pro-apoptotic and anti-apoptotic proteins. A subgroup of the pro-apoptotic proteins, containing only the BH3 domain (e.g. Bid, Bim, and PUMA), can directly interact with pro-apoptotic proteins Bax/Bak and promote their homo-oligomerization on the membrane and lead to MOMP (5–7). This represents the major process of the direct activation model for apoptosis initiation. Another competing model, the indirect activation (or displacement) model, suggests anti-apoptotic proteins (e.g. Bcl-2, Bcl-xL, Bcl-w, and Mcl-1) can heterodimerize with Bax/Bak to suppress their pro-apoptotic activity (8). Within this model, a sensitizing role was implied for the BH3-only proteins (e.g. Bad), which can bind anti-apoptotic proteins, displace Bax/Bak, and indirectly promote Bax/Bak oligomerization and MOMP (9–16). Cumulative evidences imply mitochondrial membrane insertion of many Bcl-2 family proteins is critical for their apoptotic functions and the translocation from cytosol to mitochondrial membrane could account for totally different inter-protein interactions among Bcl-2 family members (13, 17, 18). To accommodate such effects in the context of membrane association, the “embedded together” model was proposed with the recognition that final steps of Bax/Bak activation and MOMP take place on the membrane (13). Currently, little structural information is available on the membrane-associated state of Bcl-2 family proteins compared with their well studied cytosolic forms (19).
The BH3-only protein, Bid, was shown to bridge the cross-talk between extrinsic and intrinsic pathways of apoptosis through its cleavage by caspase-8 (20–22), a downstream mediator of Fas or the tumor necrosis factor (TNF) death receptor signaling pathway (23, 24). The proteolytic cleavage of cytosolic Bid by activated caspase-8 results in two fragments: p7 (7 kDa) and p15 (15 kDa) (21, 25). After cleavage, the C-terminal fragment (p15), also known as tBid (truncated Bid), translocates to the mitochondrial outer membrane (21). The membrane association of tBid was shown to be important for the efficient recruitment of cytosolic Bax to the mitochondrial outer membrane (18, 26). Through interaction with the tBid BH3 domain, Bax was suggested to undergo conformational changes, membrane insertion, and homo-oligomerization at the mitochondrial outer membrane, mediating MOMP and cytochrome c release (17, 18, 26–28). However, studies also suggested tBid could bypass Bax/Bak and directly promote the mitochondrial permeabilization by tBid alone with the induction of the negative membrane curvature (29–31).
In the cytosol, both human and mouse Bid contain eight α-helices with two hydrophobic helices (helix α6 and α7) forming the core of the protein (32, 33). After caspase-8 cleavage, the Bid N-terminal p7 fragment retains tight association with tBid through primarily hydrophobic interactions (32). Earlier studies suggested that tBid dissociated from the p7 fragment upon contact with the hydrophobic membrane and exposed its hydrophobic p7 binding pocket, promoting tBid to undergo conformational changes on the membrane (34–36). In vitro, isolated tBid was shown to bind to membranes quickly and is competent to activate Bax-mediated membrane permeabilization (18). A previous solid-state NMR study suggested that tBid associated with the membrane with an orientation parallel to the membrane surface and without membrane-traversing helix insertion (37). Additionally, membrane-associated tBid showed helical propensity at Bid helices α6–8 regions with helix α6 tilted into the membrane based on the EPR studies (34). However, these studies did not provide specific information on tBid helical boundaries, the tertiary-fold of tBid, and its overall interaction with the membrane. Currently, a proposed model indicated that tBid helices α6–8 are membrane associated, leaving the tBid BH3 domain fully exposed above the membrane surface for potential interactions with other proteins (35). This model is useful to explain some of the observed tBid functional data in solution but provided limited information regarding tBid function on the membrane (35). The exposed conformation of tBid BH3 domain above the membrane surface is energetically unfavorable unless stabilized by other proteins. It is inconsistent with the finding that the tBid BH3 domain peptide was membrane associated and fully competent to activate Bax (38). Thus, it is still not clear whether the tBid BH3 domain forms a helix on the membrane and whether its hydrophobic residues that were believed to be involved in the protein interactions in solution behave differently on the membrane by interacting with the membrane instead (20).
To address these questions with more structural details, we chose to determine the full-length tBid structure on the membrane along with the characterization of tBid regions that are interacting with the membrane. Here, we used solution NMR methods to characterize human tBid in membrane mimic 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)] (LPPG) micelles. Our data showed tBid preserved all six Bid helices (α3 to α8), including the BH3-containing helix α3. Compared to full-length Bid, the biggest difference was observed for tBid helix α5, which is a shorter helix with a longer flexible loop connecting it to helix α4. tBid possessed a C-shape conformation with a close but no direct contact between N and C termini of tBid in LPPG micelles as evidenced by paramagnetic relaxation enhancement (PRE) measurements. Furthermore, extensive interactions with LPPG micelles were observed for all six tBid helices. Specifically, the BH3-containing helix α3, which was previously believed to be exposed outside the membrane, is also membrane associated. The protein-micelle interaction and protein backbone dynamics data suggested helices α6 and α7 are likely to be more embedded into the micelles than the rest of the helices. Our findings provide detailed structural information of membrane-associated tBid and suggest the membrane-associated feature of tBid BH3 domain favors an “on the membrane” binding mode with Bax.
EXPERIMENTAL PROCEDURES
Cloning and Sample Preparation
The pET15b-BidFL vector carrying human Bid cDNA was used to produce full-length Bid protein. To produce tBid in a more efficient manner without caspase-8 cleavage, we replaced the caspase-8 cleavage site with a TEV protease cleavage site by mutating the pET15b-BidFL vector (53YDELQTDG to 53DENLYFQG). Thus the modified full-length Bid can be cleaved by TEV protease and the resulting cleavage product tBid will be the same as the native tBid. The modified plasmid (pET15b-BidTEV) was transformed into Escherichia coli BL21(DE3) cells to express recombinant protein. Isotopically (15N or 15N,13C) labeled proteins were produced in isotope-enriched M9 minimal media. The perdeuterated tBid sample was grown in 99% D2O minimal media. The E. coli cells were grown at 37 °C, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at A600 ∼0.8, and cell growth was continued overnight at 18 °C. The cells were harvested by centrifugation, resuspended in lysis buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 5 mm imidazole, 1 mm PMSF), and sonicated. The lysate was centrifuged at 34,000 × g for 30 min at 4 °C and the supernatant was loaded to Ni2+ affinity chromatography. The His6-tagged full-length BidTEV was eluted using elution buffer (20 mm Tris-HCl, pH 8.0, 500 mm NaCl, 250 mm imidazole) and dialyzed into buffer A (20 mm Tris-HCl, pH 8.0) before it was further purified by an ion-exchange chromatography on a HiTrap 1-ml Q column (GE Healthcare) with a linear salt gradient. The purified full-length BidTEV was pooled and mixed with 200 units of AcTEV protease (Invitrogen) for cleavage by dialyzing against 1 liter of TEV cleavage buffer (50 mm Tris-HCl, pH 8.0, 0.5 mm EDTA, 1.0 mm DTT) overnight at room temperature. The cleaved protein mixture was loaded onto the Ni2+ affinity chromatography and tBid was separated by washing with lysis buffer with 1.2% (w/v) n-octyl-β-d-glucoside. The fraction containing purified tBid was collected and dialyzed against buffer (20 mm Tris-HCl, 100 mm NaCl, pH 8.0) to remove detergent n-octyl-β-d-glucoside. After dialysis, tBid precipitated out of solution and was collected by centrifugation. The NMR sample of tBid at 0.5–1.0 mm was prepared by dissolving the protein pellet into NMR buffer (50 mm potassium phosphate, pH 6.6) supplemented with 8% (160 mm) LPPG, 10% D2O, and 0.02% sodium azide. The tBid samples in LPPG micelles were kept at 45 °C at all times for better sample stability.
NMR Spectroscopy
All NMR measurements were carried out on a Bruker Avance 600 MHz spectrometer with a room temperature probe, or a Bruker Avance 800 MHz or a Bruker Avance 900 MHz spectrometer with cryogenic probes. The following experiments were used for backbone resonances assignments: three-dimensional HNCA (39), CBCA(CO)NH (40), HNCACB (41), HBHA(CO)NH (42), and HNCO (43). Three-dimensional 15N-edited NOESY-HSQC (tmix = 70 ms) (42) and four-dimensional 13C/13C-edited NOESY (tmix = 70 ms) (44) experiments were acquired for inter-proton NOE distance restraints. The backbone 15N T1 measurement obtained using a 800 MHz spectrometer was recorded at various delays: 8, 120, 320, 600, 880, 1200, 1600, and 2240 ms (45). For 600 MHz, T1 delays used were 8.0, 128, 384, 560, 800, 1040, 1280, and 1600 ms. The 15N T2 measurement obtaining using a 800 MHz spectrometer was carried out using a Carr-Purcell-Meiboom-Gill pulse sequence at various total delays: 4.0, 12, 24, 36, 48, 68, 88, and 128 ms (45). For 600 MHz, T2 delays used were 8.0, 24, 40, 56, 72, 100, 132, and 192 ms. The PRE 1HN-Γ2 rates were determined from a two-time point (0.04 and 12 ms) interleaved measurement using a TROSY-based experiment (46). The protein backbone 15N relaxation dispersion experiments were carried out using two different τcp of 0.3 and 6 ms in the CPMG refocusing element with a constant delay time of 48 ms. All NMR data were processed using NMRPipe (47) and analyzed with NMRview (48) and PIPP (49). Experimental errors were estimated based on the spectral noise as described previously (50).
Characterization of Protein-Micelle Interaction
A 15N-labeled perdeuterated tBid sample was expressed in M9 minimal media supplemented with 99.9% D2O and 15N ammonium chloride. The three-dimensional 15N-edited NOESY-HSQC (tmix = 70 ms) experiment was acquired to obtain the NOE distance information between the protein backbone amide protons (HN) and the LPPG protons.
Site-directed Mutagenesis
tBid single cysteine mutants (S78C, Q136C, and Q180C) were prepared using a QuikChange site-directed mutagenesis kit (Stratagene). These point mutations were introduced using pET15b-BidTEV expressing plasmid as template. All mutants were confirmed by DNA sequencing.
Paramagnetic Relaxation Enhancement
Because there is no cysteine in tBid, non-native cysteines were introduced by site-directed mutagenesis at the respective sites mentioned above. The full-length Bid bearing the cysteine mutant was first expressed, purified, and cleaved as described above. After TEV cleavage, the protein sample was soaked with DTT at a final concentration of 20 mm to reduce any possible cysteine disulfide bonds. The protein was then exchanged into a reaction buffer (100 mm Tris-HCl, 1 mm diethylenetriaminepentaacetic acid, pH 8.5) using a PD-10 desalting column. Following that, parallel spin probe labeling reactions were carried out by mixing the protein sample with 20× molar excess of the paramagnetic probe MTSL (1-oxyl-2,2,5,5-tetramethyl-η3-pyrroline-3-methyl) methanethiosulfonate (Toronto Research Chemicals Inc.) or diamagnetic analog dMTSL (1-acetyl-2,2,5,5-tetramethyl-η3-pyrroline-3-methyl)methanethiosulfonate (Toronto Research Chemicals Inc.) at room temperature overnight. The stock of both MTSL and dMTSL reagents had been previously dissolved at 20 mg/ml in acetonitrile. After the reactions, excess spin label was removed by dialysis against a buffer (20 mm Tris-HCl, 100 mm NaCl, pH 8.0). Both the paramagnetic and diamagnetic protein samples were further purified in 1.2% n-octyl-β-d-glucoside as described above. Experimental errors for the PRE were estimated as previously described (51). Labeling of the tBid samples was confirmed by liquid chromatography/mass spectrometry. A two-dimensional 1H,15N-HSQC spectrum of each paramagnetic tBid sample was recorded and no obvious chemical shift changes observed, indicating spin labeling did not disrupt tBid structure. Solvent PRE experiments were performed by stepwise addition of the water-soluble spin label reagent gadodiamide (aqua[5,8-bid(carboxymethyl)-11-[2-(methylamino)-2-oxoethyl]-3-oxo-2,5,8,11-tetraazatridecan-13-oato(3-)-N5,N8,N11,O3,O5,O8, O11,O13]gadolinium hydrate; trade name Omniscan, GE Healthcare) into the NMR sample to arrive at final concentrations of 0.25, 0.5, 1.0, 2.0, and 5.0 mm. A reference spectrum was recorded prior to the addition of spin label reagent.
Structure Calculation
Cross-peak intensities from NOESY experiments were translated into a continuous distribution of proton-proton distances. Based on the assigned tBid backbone chemical shifts, the TALOS program (52) was used to predict φ and ψ dihedral angles. The statistically significant dihedral angles in regular α-helical secondary structure were used as structural restraints. Generic hydrogen bond distance restraints were imposed for residues located at well defined α-helical regions. Additionally, PRE relaxation rates were calibrated over residue pairs within one single helix and converted into distances for structure calculation. The backbone T1/T2 relaxation ratios were also implemented into the structure calculation as previously described (53). Planar energy restraints for tBid residues with strong micelle interactions were also applied by mimicking a close distance with the membrane interface (54). tBid structures were calculated using simulated annealing protocol by the program Xplor-NIH (55). The final structure calculations employed 749 inter-residual and 180 intra-residual interproton distance restraints, 80 hydrogen bond distance restraints, 137 PRE distance restraints, 72 φ and 72 ψ angle restraints, 59 T1/T2 ratio restraints, and 28 planar energy restraints (Table 1). Structure presentation was prepared using PyMOL (56).
TABLE 1.
Structural statistics of the human tBid in LPPG micelles
Statistics are given for the 20 lowest energy structures out of 200 calculated NMR structures.
| Distance restraints (Å) | |
| NOE (930)a | 0.021 ± 0.003 |
| Intraresidual NOE (180) | 0.007 ± 0.007 |
| Sequential NOE ( |i-j| = 1) (298) | 0.018 ± 0.003 |
| Medium range ( 2 ≤ |i-j| ≤ 5) (451) | 0.025 ± 0.006 |
| H-bonds (80) | 0.028 ± 0.004 |
| Paramagnetic relaxation enhancement (137) | 0.017 ± 0.006 |
| Dihedral angle restraints (°) | 0.207 ± 0.050 |
| φ (72) | 0.157 ± 0.054 |
| ψ (72) | 0.241 ± 0.070 |
| Other restraints | |
| Plane energy (Å) (28) | 0.75 ± 0.17 |
| T1/T2 ratio (59) | 25.48 ± 3.62 |
| Coordinate precision r.m.s. deviationb | |
| Backbone (Å) | 3.64 ± 1.37 |
| Heavy atoms (Å) | 4.24 ± 1.36 |
| Ramachandran plot | |
| Most favored regions | 79.0 ± 2.3% |
| Allowed regions | 14.1 ± 2.4% |
| Generously allowed regions | 4.0 ± 2.0% |
| Disallowed regions | 2.9 ± 1.5% |
a Number in parentheses corresponds to the total number of restraints.
b The r.m.s. deviation values were calculated by superimposing all six α-helices.
RESULTS
Secondary Structure of tBid in LPPG Micelles
tBid was produced by incubating purified modified full-length Bid with TEV protease instead of caspase-8 (25) (see “Experimental Procedures”). The choice of LPPG micelles to solubilize tBid for NMR characterization was made based on previous studies (37, 57) and a screen through various detergent micelles. The acquired two-dimensional 1H,15N-HSQC spectrum of tBid in NMR buffer with 8% LPPG at 45 °C showed varying peak intensities suggesting tBid residues might experience different effective rotational correlation times. This is likely due to their different states of association with the micelles. Analysis of the NMR datasets yielded near complete backbone and side chain resonance assignments of tBid. As many as 125 of 131 non-proline tBid backbone amides were assigned and only residues at the N terminus (Asn62 to His66) were not assigned due to the exchange with the solvent. The 1HN NMR resonances of tBid showed a small dispersion, indicating the presence of mostly helical and random coil conformation. To be noted, the tBid C-terminal region (Ser188–Asp195) branched with another set of distinct resonance peaks with slightly weaker intensities, suggesting the flexible C-terminal tail experienced at least two distinct conformations.
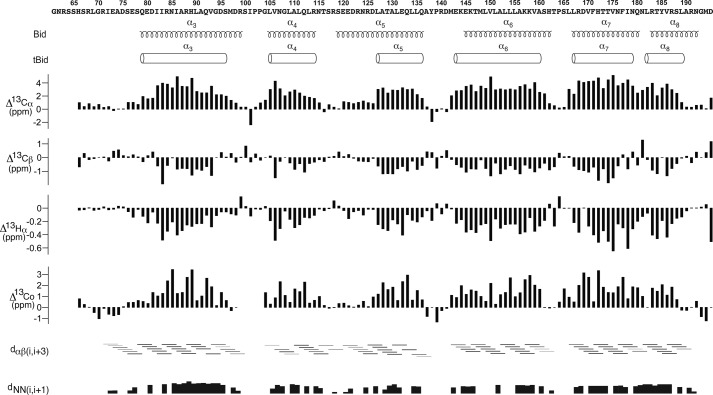
The assigned Cα, Cβ, Hα, and CO secondary chemical shifts indicated the presence of six α-helices (α3, 79–95; α4, 105–114; α5, 127–136; α6, 143–160; α7, 167–179; α8, 182–189) (Fig. 1), which are further corroborated by the inter-proton NOE connectivity data. The overall secondary structural composition is similar to the previously reported NMR structure of cytosolic full-length human Bid (Fig. 1), but there are some obvious differences in the helical boundaries between tBid and Bid. The most significant difference comes from helix α5. Compared with Bid, tBid helix α5 is a shorter helix and residues Ser119–Asp126 are in a loop conformation instead of α-helical conformation. Of note, weak helical inter-proton NOE distance (Hα(i) to Hβ(i+3)) were observed for these residues (Ser119–Asp126), suggesting a residual α-helical character (Fig. 1). This particular feature was also observed for tBid residues adjacent to the N-terminal end of helix α3.
FIGURE 1.
Secondary structures of human tBid in LPPG micelles. The secondary chemical shift index (13Cα, 13Cβ, 1Hα, and 13CO) defined the presence of six α-helices. The chemical shift differences were calculated by subtracting the average random coil chemical shift from the assigned tBid chemical shift values. The consecutive large positive bars in 13Cα,13CO and negative bars in 13Cβ,1Hα suggest the presence of the α-helical conformation. The defined α-helices were also indicated by the characteristic medium range inter-proton NOE connectivities of Hαi to Hβi+3 and strong NOE connectivities of HNi to HNi+1. The thickness of bars used to show NOE connectivities corresponds to the relative NOE intensity. The missing NOEs within the α-helical regions are due to overlapped peaks. The secondary structures of human Bid reported previously were also shown for comparison (32). Residue numbering is kept the same as the full-length Bid.
Backbone Dynamics of tBid in LPPG Micelles
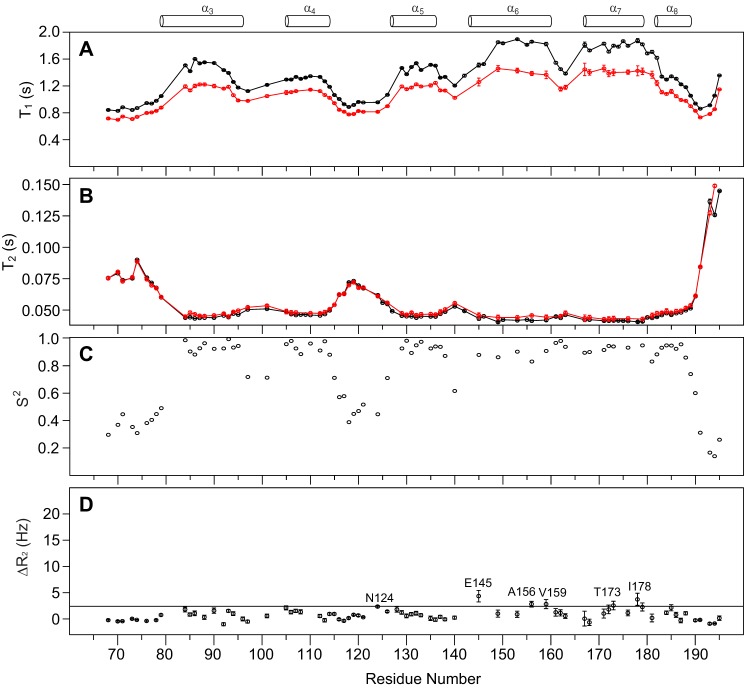
The backbone 15N T1 and T2 relaxations were measured for tBid at both 800 and 600 MHz proton resonance frequencies. The backbone 15N T2 relaxation data indicated that all tBid helical regions are rigid compared with the non-helical regions (Fig. 2B), and show similar diffusive behavior. This implies that these rigid helices tumble together with the micelle. In contrast, the loop between helices α4 and α5 (Asn115–Asp126) are highly flexible with high T2 and low T1 values (Fig. 2), similar to flexible N and C termini. The average T1 and T2 relaxation times for the rigid helical regions at 800 MHz frequency are 1.55 ± 0.21 s and 38.2 ± 4.1 ms, respectively. The calculated T1/T2 ratios (data not shown) gave an estimate of rotational correlation time (τc) of 16.0 ns, which is roughly equivalent to a globular protein with a molecular mass of 43 kDa in solution. The variations in T1/T2 ratios (data not shown) for the helices in tBid indicate that the molecule is quite anisotropic in LPPG micelles. It is also worth noting that tBid helices α6 and α7 have shorter 15N T2 values (33.3 ± 1.1 ms) and higher T1/T2 ratios (55.0 ± 2.9) at 800 MHz frequency than the rest of the helices observed (Fig. 2). The similar T1/T2 ratios also suggested helices α6 and α7 have comparable effective correlation times and orient similarly to each other with respect to the rotational diffusion axis. To identify residues that experience large amplitude fast motion, residue specific order parameters (S2) were calculated by fitting the relaxation data obtained at the two magnetic fields (Fig. 2C). As expected the exposed loops between helices α3-α4, α4-α5, and α5-α6 have low S2, in addition to the termini. All helices show no evidence of large amplitude fast motion. The initial probe for possible conformational exchange was done by evaluating the ratio of T2 values obtained at 600 versus 800 MHz (data not shown). Residues whose T2 ratio lies outside the 1.5 times S.D. from the average value are Asn85, Ile86, Leu149, Ala156, Gly193, Met194, and Asp195. Residues Asn85 and Ile86 are in helix α3, however, their T2 ratios are not significantly outside the cutoff. Although residues Leu149 and Ala156 are in α6, and Gly193, Met194, and Asp195 are at the C-terminal all show significant deviation. To identify additional residues that might be undergoing conformational exchange as well as to confirm those identified by the T2 ratio at the two magnetic fields, relaxation dispersion experiments were carried out. The relaxation dispersion data (Fig. 2D) suggested a few residues (Asn124, Glu145, Ala156, Val159, Thr173, and Ile178) undergo microsecond-millisecond time scale exchange. They are identified by choosing a cutoff of ΔR2 beyond the 1.5 times S.D. Residues Glu145, Ala156, and Val159 suggest tBid helix α6 is likely to experience some conformational exchange, which is consistent with the T2 ratio data. Of note, residue Asn124 at the loop between helices α4-α5 and Thr173 and Ile178 at helix α7 are also shown to undergo conformational exchange.
FIGURE 2.
Backbone 15N relaxation dynamics of human tBid in LPPG micelles. tBid 15N T1 (A) and T2 (B) relaxation times at two magnetic fields of 600 MHz (red) and 800 MHz (black) and the fitted backbone order parameter S2 using measured T1 and T2 data at both magnetic fields (C) are plotted as a function of residue number, indicating all six (α3–8) helical regions were shown to be rigid. The shorter T2 values (B) for both helices α6 and α7 suggested they are likely more embedded in the micelle. Error bar for the 15N T1 and T2 relaxation data were estimated as previously described (50). D, the residues undergoing microsecond-millisecond conformational exchange identified using the backbone 15N R2-CPMG rate differences, ΔR2, the difference between measurements at two different τcp values (ΔR2 = R2 (6 ms) − R2 (0.3 ms)). The cutoff line corresponds to the ΔR2 threshold of 1.5 times the S.D. The missing values are due to resonance peak overlap. Secondary structure elements of tBid are indicated at the top for reference.
tBid Adopts a C-shape Conformation in LPPG Micelles
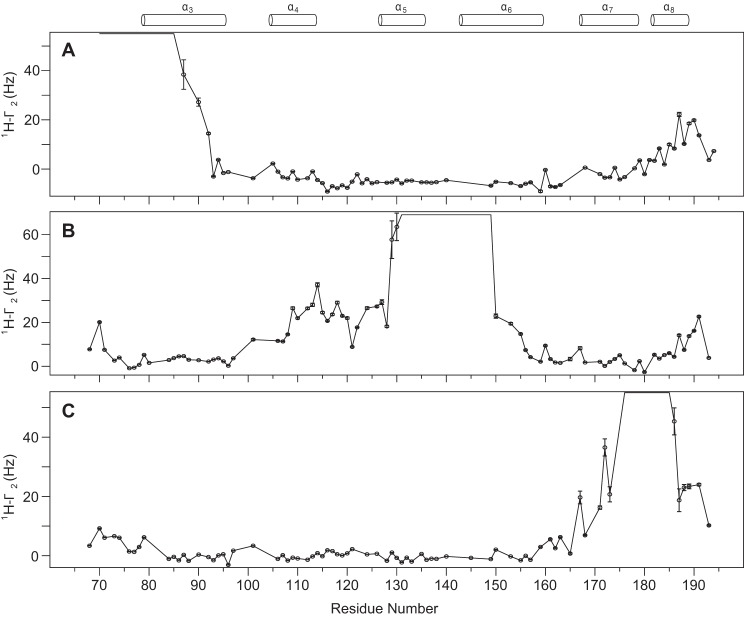
The near complete assignments of the inter-proton cross-peak NOE dataset yielded 931 NOEs. Analysis of these inter-proton NOEs did not produce any long-range contacts (greater than 5 residues apart). The absence of long-range NOEs renders the determination of tertiary fold of the six α-helices tBid structure impossible. Therefore, longer distance oriented PRE experiments were performed. Paramagnetic spin labels were incorporated at three tBid sites with cysteine mutations (S78C, Q136C, and Q180C). The backbone 1HN-Γ2 rate measurements were carried out and the results showed residues close to the paramagnetic center experienced obvious enhancement (Fig. 3). For the S78C sample, weak enhancements were shown for residues at the tBid C terminus, suggesting a close conformation but with no NOE contact (<5 Å) between the tBid N and C terminus (Fig. 3). Consistently, weaker PRE effects were shown for tBid N-terminal residues to the C-terminal spin label at residue 180 (Q180C sample). For Q136C, weak PRE effects were observed for residues in helices α4 and α5 as well as some residues in the N and C termini (Fig. 3). Another possible contribution to the observed PRE effects is due to intermolecular contacts. To address whether tBid homo-oligomerizes in LPPG micelles, PRE measurements by mixing 15N-labeled wild type tBid with spin-labeled 14N-tBid at equal molar ratios were carried out and no obvious enhancement was observed for both S78C and Q136C samples (data not shown). This implied no inter-molecular contact between tBid monomers and therefore tBid does not homo-oligomerize and is monomeric in LPPG micelles.
FIGURE 3.
Measured PRE effects for human tBid cysteine mutants in LPPG micelles at three different sites. The experimental 1HN-Γ2 values for S78C (A), Q136C (B), and Q180C (C) were plotted as the function of residue number. Residues experiencing significant PRE effects are mainly localized to residues that are close to the paramagnetic center. For the S78C sample (A), weak PRE effects for residues in the C terminus included helix α8. C, albeit even weaker, similar weak PRE effects were also observed between the tBid N terminus and C terminus. For the Q136C sample (B), weak PRE effects were observed for residues in helices α4 and α4 as well as some residues in flexible N and C termini. Error bars were estimated as previously described (51). Secondary structure elements of tBid are indicated at the top for reference.
All tBid Helices Are Involved in the Interaction with LPPG Micelles
To understand the molecular interaction between tBid and LPPG micelles, a perdeuterated 15N-labeled tBid was prepared in LPPG micelles. The three-dimensional 15N-edited NOESY-HSQC experiment was carried out to detect direct NOE interactions between the protein backbone amide protons and protons from LPPG micelles. The LPPG molecule possesses a long acyl chain with abundant methylene protons, which gives a dominant 1H NMR signal at a chemical shift of 1.26 ppm (Fig. 4A). Extensive analysis of the NOE cross-peaks at this chemical shift yielded a residue-specific protein-micelle interaction profile. Due to overlaps with the signals from some tBid Thr Hγ2 or Ile Hγ1 protons because of incomplete deuteration, we simply omitted those residues for further consideration of possible protein-micelle interactions. In the end, 28 tBid residues with obvious protein-micelle interactions were identified (Fig. 4B). These tBid residues are mostly hydrophobic residues and are scattered over all six tBid helices: α3 (Ile86, Ala87, Leu90, Val93, and Gly94), α4 (Val106, Leu109, Ala110, Leu113), α5 (Leu127, Leu131, Leu135), Tyr138, α6 (Leu149, Leu153, Ala156, Lys157, Val159, Ala160), Ser161, α7 (Leu167, Arg168, Phe171, His172, Val175, Asn179), and α8 (Leu182, Val186). This strongly suggested those rigid tBid helices are embedded in LPPG micelles through extensive hydrophobic interactions. Among those identified, residues in helix α6 and α7 showed noticeably higher relative NOE intensities, suggesting both helices are more embedded in LPPG micelles than other tBid helices (Fig. 4B). This is consistent with previous EPR studies that showed tBid helices α6–8 are membrane-associated (34). Of note, three positively charged residues (Lys157, Arg168, and His172) in these two helices also showed strong micelle interactions. Surprisingly, the BH3 domain containing helix α3, which was previously proposed to be exposed and outside the membrane surface, also show extensive micelle interactions (Fig. 4A). Taken together, tBid helices α3–5 and α8 showed a consistent micelle interaction pattern for hydrophobic residues located at one side of the helix, suggesting tBid helices are embedded parallel to the membrane surface. In contrast, residues (Ser76, Glu77, Ser78, Gln79, Arg118, Ser119, Glu120, Glu121, Asn124, and Gly193) in tBid flexible regions including the long loop (Asn115–Asp126) between helix α4 and α5 clearly did not show any obvious interaction with the micelles. This observation was further corroborated with the strong protein-water interaction for this flexible region (data not shown). In addition, the solvent PRE experiments were carried out and showed consistent limited water accessibility, which is a weak solvent PRE effect, for residues that are lipid embedded for all tBid helices and a strong PRE effect for water-exposed residues (Fig. 4C).
FIGURE 4.
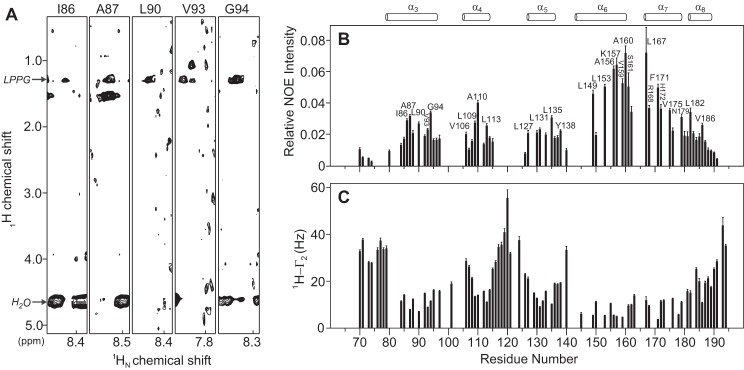
Interactions between human tBid and LPPG micelles. A, illustration of inter-proton NOEs between the tBid BH3 domain and LPPG micelles. A selection of the 15N-edited three-dimensional NOESY-HSQC strips of the tBid BH3 domain, containing helix α3 residues, showing the intermolecular NOEs to the LPPG acyl chain 1H at a chemical shift of 1.26 ppm. Among these residues, Ile86, Ala87, Leu90, Val93, and Gly94 showed obvious NOE cross-peaks to LPPG micelles. Conversely, these residues did not show an obvious cross-peaks signal to water (4.61 ppm). B, relative peak intensities are calculated by taking the ratios of measured NOE intensities between protein backbone amides (1HN) and the LPPG acyl chain methylene (CH2) groups at 1.26 ppm against the diagonal peak intensities. These relative peak intensity ratios are used to quantify the strength of interaction between various sites in tBid and micelles. Residues showing obvious protein-lipid interactions are from all six tBid helices (α3–8). C, the experimental solvent PRE measurement for tBid backbone amide protons. The measured 1HN-Γ2 PRE values in the presence of a water-soluble paramagnetic probe showed obvious relaxation enhancement for residues that are water exposed and weak enhancement for residues that are embedded in the lipid. Secondary structure elements of tBid are indicated at the top for reference.
Structure of tBid in LPPG Micelles
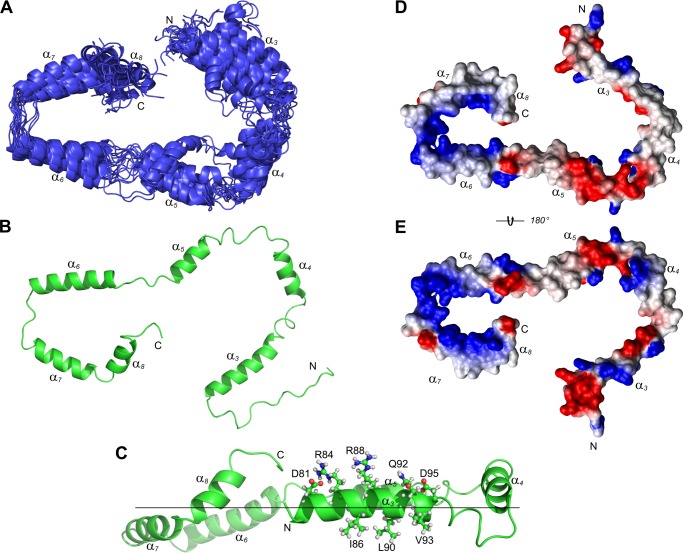
Structure calculation of tBid were carried out using the following NMR restraints: NOE-derived distances, TALOS-derived dihedral angles, generic hydrogen bonds, PRE-derived distances, and T1/T2 derived relaxation restraints (Table 1). With the assumption that tBid helices are embedded parallel to the membrane surface, tBid residues that showed a strong micelle interaction were also implemented into the structure calculation with residue-specific planar energy restraints. The calculated 20 lowest energy tBid structures show well defined individual helices (α3 to α8) with backbone r.m.s. deviations of 0.45 ± 0.16, 0.47 ± 0.13, 0.29 ± 0.09, 0.38 ± 0.15, 0.32 ± 0.10, and 0.41 ± 0.11 Å, respectively. As previously noted the absence of long-range NOE restraints left the tBid global conformation to be mainly defined by long-range PRE restraints. This resulted in calculated tBid structures with a backbone r.m.s. deviation of 3.64 ± 1.37 Å when superimposing all six tBid helices (Fig. 5A). The calculated tBid structures adopted an extended conformation with the N-terminal end being close but without direct atomic contact to the C terminus, thus forming a C-shape structure (Fig. 5, A and B). Relative to the membrane interface, tBid helices formed two faces that showed dramatic differences in electrostatic surface potentials (Fig. 5C). One side of the tBid structure that faces the membrane shows dominant hydrophobic patches, which correspond to hydrophobic tBid residues that showed strong interactions to the LPPG micelles (Fig. 5D). Distinct electrostatic surfaces were shown on the other side of the tBid structure with many charged surfaces including BH3 domain helix α3 (Fig. 5E). With micelle-interacting residues (Ile86, Ala87, Leu90, Val93, and Gly94) in helix α3 facing toward the membrane surface, charged or polar residues (Asp81, Arg84, Arg88, Gln92, and Asp95) are left facing away from the membrane surface for potential electrostatic interaction with other proteins (Fig. 5, B and C). Of note, strong positively charged surface areas were formed along tBid helices α6-α8 (Fig. 5E), which could potentially form electrostatic interactions with the negatively charged LPPG micelle head groups. This electrostatic interaction would prevent these helices to be completely buried inside the micelles. This is consistent with a previously suggested tBid conformation on the membrane by solid-state NMR studies (37). Overall, these structural features displayed by tBid in LPPG micelles are consistent with the fact that most tBid helices are amphipathic.
FIGURE 5.
Calculated human tBid structure shows a C-shape conformation. A, schematic representation of the 20 lowest energy structures of human tBid in LPPG micelles. The calculated tBid structures show six well defined helices α3–8 and the superposition to the mean structure on tBid helical regions results in a backbone r.m.s. deviation of 3.64 Å. Both N-terminal (Gly61–Ser78) and C-terminal (Ala190–Asp195) tails are unstructured and are omitted for clarity. Overall (B) tBid adopts a C-shape structure with both N and C termini in close conformation. C, tBid helices (α3–8) are parallel to the putative membrane surface with helices α6 and α7 more embedded. For the membrane-associated tBid helix α3, charged or polar residues (Asp81, Arg84, Arg88, Gln92, and Asp95) are facing away from the membrane surface and are exposed for potential interactions with other proteins. C, on the opposite side of helix α3 residues (Ile86, Ala87, Leu90, Val93, and Gly94) with strong micelle interactions are facing toward the membrane surface. The flexible N terminus (Gly61–Ser78) was omitted for clarity. D, electrostatic potential surface view of the side of the tBid structure facing the membrane shows dominant hydrophobic patches along the ordered tBid helices. E, the opposite side of the tBid surface shows strong charged areas along tBid helices with a continuous positive charged surface along the helices α6-α8. D and E, the flexible loop with a highly charged surface between helix α4 and helix α5 contains a number of charged residues.
DISCUSSION
The cleavage of full-length Bid by caspase-8 forms the activated tBid, which can be translocated to the mitochondrial outer membrane (21). The association of tBid to membrane is critical for the promotion of apoptosis (18). Upon interaction with the hydrophobic environment in vitro, tBid undergoes conformational changes for favorable lipid interactions (34). Similar to full-length Bid, tBid was shown to be highly helical in various membrane mimics (37, 58, 59). Previously, membrane-associated tBid was shown to be parallel to the membrane surface with no transmembrane helix and the region bound to the membrane was suggested to be located at helices α6-α8 (34, 37). Those studies did not provide details on tBid helical boundaries and the membrane-associated full-length tBid structure at the atomic level. Especially, no structural information on the functionally important BH3 domain is yet available. This renders understanding of the precise apoptotic function of tBid and its molecular interactions with Bax/Bak or Bcl-xL on the membrane impossible. To fill this gap, we determined the solution structure of full-length human tBid in a lipid membrane mimic, LPPG micelles by NMR.
Secondary structure characterization by NMR demonstrated that tBid has six well defined helices, which are mostly preserved from the full-length Bid (Fig. 1). However, some obvious differences were observed between full-length Bid and tBid in LPPG micelles. For instance, tBid residues (Ser119–Asp126) close to the N-terminal end of helix α5 did not form a rigid helix (Fig. 1). Compared to the full-length Bid structure, the loop between helices α4 and α5 is much shorter (32) than the loop in tBid. This extended tBid loop between helices α4 and α5 was shown to be highly flexible and did not show any interaction with the micelles, contrasting to the rigid helical conformation in Bid (Figs. 1 and 4). The highly flexible behavior of this loop is likely due to the presence of many charged residues (Arg118, Glu120, Glu121, Asp122, Arg123, Arg125, and Asp126), which could potentiate electrostatic interactions of tBid with other protein(s). Recent peptide screening proposed an important role for this flexible tBid region (60), through which tBid can directly interact with the protein MTCH2 (mitochondrial carrier homologue 2) (36, 61). The exposure of this flexible tBid loop enables a scenario where the interaction between tBid and MTCH2 is likely to occur at the mitochondria outer membrane.
The analysis of tBid NOE data did not produce any long-range NOEs, indicating that six well defined tBid helices do not form a compact structure (Fig. 5). Long distance-oriented PRE measurements, however, showed some distances between the tBid N and C termini within 20–25 Å (Fig. 3). These distances impose a C-shape conformation for tBid on LPPG micelles surface. A rather similar result was also seen previously for the micelle-bound α-synuclein (62) with a contrast that tBid is more complex with 6 helices. The size of the LPPG micelle was estimated to have a diameter of ∼68 Å (63). At the longest circumference, it is considerably too large for tBid to wrap around it and form a C-shape configuration as described (Fig. 5). Based on our experimental data, however, we believe the C-shape conformation of tBid is likely due to its characteristic interaction with the curved LPPG micelle membrane within one constrained area on the micelle hemisphere. This is consistent with previous findings that tBid could colocalize with cardiolipin, a mitochondrial lipid that was previously suggested to induce mitochondrial membrane curvatures (5), at the mitochondrial outer membrane contact sites with “curvature stress” (64). Therefore, tBid could be attracted to the contact sites with membrane curvatures on the mitochondria outer membrane, hence sensitizing the mitochondrial membrane for Bax-induced membrane permeabilization.
The tBid α6 helix contains a stretch of hydrophobic residues (148MLVLALLLA156) and were predicted to be membrane-associated. Our data indeed showed strong micelle interactions for hydrophobic residues in tBid helix α6 and α7 as well (Fig. 4). The actual tBid membrane interaction site was suggested to be at the mitochondrial contact site with clustered negatively charged lipid cardiolipin (64–72). The negatively charged LPPG was chosen not only to provide the best working sample, but as a relevant mimic to the cardiolipin-rich mitochondrial membrane as well. Positively charged lysine residues (Lys157 and Lys158) in tBid helix α6 were suggested to be critical for a specific electrostatic interaction with the mitochondrial outer membrane (71, 72). Our data confirmed this hypothesis by showing a strong micelle interaction for tBid residue lysine 157 as well as charged residues Arg168 and His172 in helix α7 (Fig. 4). Consistently, the electrostatic surface map of the tBid structure further showed continuous positively charged surfaces along helices α6–8 (Fig. 5), which might form electrostatic interactions with negatively charged lipid cardiolipin headgroups on the membrane. Furthermore, this additional electrostatic interaction may account for our findings that both helix α6 and α7 are more membrane embedded than the rest of tBid helices (Figs. 2 and 4) and implicate a membrane recognition and docking role for these two helices. The recently reported fluorescence data showed tBid regions corresponding to Bid helix α4–7 are membrane embedded and also suggested a conformational equilibrium between two major states of tBid (36). Our structure, which show all tBid helices are membrane associated, is consistent with their findings. Furthermore, to be in equilibrium of multiple conformations, the tBid structure must have a considerable degree of flexibility. Our structure and dynamic data support this possibility. We showed that tBid adopted an extended membrane-associated structure that contains flexible loops connecting helices α3-α4, α4-α5, and α5-α6. In addition, we also showed that some residues in helix α6 (Leu149, Ala156, and Val159) and at its N-terminal (Glu145), the site of membrane targeting, undergo conformational exchange. This result seems to be supporting the conformational equilibrium observation by the fluorescence measurement.
A previous model proposed that the tBid BH3 domain was fully exposed and located above the membrane surface, poised for protein interactions with Bax/Bak or Bcl-xL (35). However, our studies revealed that the important BH3 domain containing helix α3 was membrane associated with hydrophobic residues facing toward the membrane surface (Figs. 4 and 5E). In contrast to the latest suggested solvent-exposed BH3 domain (36), our data presented an energetically more favorable conformation for the tBid BH3 domain that is membrane-associated due to its residue composition. Therefore, our structure supports a scenario that the interaction between the membrane-associated helical BH3 domain and Bax happens on the membrane surface. The on the membrane interaction is consistent with previous observations that membrane-targeted tBid BH3 peptides can still effectively interact with and activate Bax to permeabilize mitochondrial membranes (38, 73). Previous studies also suggested that Bax helix α1 interacts with the tBid BH3 domain as the first step in its tBid-induced activation (28, 74). Upon contact with the membrane surface, Bax was suggested to experience a reversible conformational change and its N terminus can get exposed and recognized by the conformation-specific antibody 6A7 (75). The subsequent membrane insertion and activation of Bax was shown to involve the interaction with membrane-associated tBid (18). It is possible that the tBid BH3 domain interacts with the transiently exposed Bax helix α1 on the membrane surface and this specific interaction can shift the reversible conformational change of membrane-encountered Bax into a membrane-associated state. In fact, a specific electrostatic interaction between tBid BH3 domain residue Arg84 and Bax helix α1 residue Asp33 was reported to be important for tBid-induced Bax activation in a previous study (74). Therefore, our results of the tBid structure in LPPG micelles in combination with the above reported findings lead to the possibility that charged residues in the tBid BH3 domain (Asp81, Arg84, Arg88, and Asp95) that were shown to face away from the membrane could potentially form electrostatic interactions with charged residues (Asp33 and Arg34) in Bax helix α1 on the membrane surface. If the conventional BH3 binding mode to Bax is required (19, 76–78), then the interaction has to occur in such a way to allow the hydrophobic tBid BH3 residues to be available to bind Bax. One possible scenario would be that the electrostatic interactions between the tBid BH3 domain and Bax helix α1 could promote displacement of the tBid BH3 residues from the lipid membrane, exposing them for binding to the Bax BH3 binding pocket.
For Bcl-xL, it is difficult to discuss its interaction with tBid on the membrane with very limited information on membrane-associated Bcl-xL. However, it is likely that a similar interaction between tBid and Bcl-xL on the mitochondrial membrane was adopted as in the tBid-Bax interaction given the structural similarity between Bax and Bcl-xL.
Overall, Our data provided novel molecular information regarding the structure and dynamics of tBid in LPPG micelles, which leads to a first step in understanding the tBid apoptotic function on the mitochondrial membrane. Although our findings did not directly address the favorability of a direct or indirect activation model, the membrane-embedded tBid BH3 domain suggested an on the membrane binding mode of tBid with Bax, thus leaning toward the previously proposed embedded together model (13). The recently solved x-ray structure of the Bax dimer promoted by the tBid BH3 peptide and detergent provided valuable information on the tBid-induced Bax activation (79). The reported binding mode with the tBid BH3 peptide sitting inside the hydrophobic BH3-binding cleft of Bax is essentially similar to inter-protein interactions among Bcl-2 family members in solution (19, 76, 77). Nevertheless, how the tBid BH3 domain, shown in our study to be associated with the micelles, can go into the BH3 binding pocket of Bax still remains unclear. Further elucidation of the tBid-Bax complex structure on the membrane will provide more molecular details on the mechanism of tBid-mediated Bax activation.
Acknowledgments
We are very grateful to Dr. Motoshi Suzuki for technical help and valuable discussion on the project and Dr. Charles D. Schwieters for help on restraint implementation in Xplor-NIH. We also thank Dr. Duck-Yeon Lee (Biochemistry Core Facility, NHLBI, National Institutes of Health) for expertise and advice regarding the mass spectrometry-related experiments performed in this paper. We thank Dr. Francesca Marassi for providing the Bid cDNA.
This work was supported, in whole or in part, by a National Institutes of Health grant from the Intramural Research Program of the NHLBI (to N. T.).
The atomic coordinates and structure factors (code 2M5I) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The chemical shifts can be accessed through the Biological Magnetic Resonance Bank (BMRB) under BMRB accession number 19054.
- MOMP
- mitochondrial outer membrane permeabilization
- tBid
- truncated Bid
- LPPG
- 1-palmitoyl-2-hydroxy-sn-glycero-3-[phospho-RAC-(1-glycerol)]
- PRE
- paramagnetic relaxation enhancement
- TEV
- tobacco etch virus
- HSQC
- heteronuclear single quantum coherence
- r.m.s.
- root mean square.
REFERENCES
- 1. Danial N. N., Korsmeyer S. J. (2004) Cell death. Critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 2. Wang X. (2001) The expanding role of mitochondria in apoptosis. Genes Dev. 15, 2922–2933 [PubMed] [Google Scholar]
- 3. Youle R. J., Strasser A. (2008) The BCL-2 protein family. Opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 4. Adams J. M. (2003) Ways of dying. Multiple pathways to apoptosis. Genes Dev. 17, 2481–2495 [DOI] [PubMed] [Google Scholar]
- 5. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 6. Wei M. C., Zong W. X., Cheng E. H., Lindsten T., Panoutsakopoulou V., Ross A. J., Roth K. A., MacGregor G. R., Thompson C. B., Korsmeyer S. J. (2001) Proapoptotic BAX and BAK. A requisite gateway to mitochondrial dysfunction and death. Science 292, 727–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marani M., Tenev T., Hancock D., Downward J., Lemoine N. R. (2002) Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol. Cell Biol. 22, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Structure of Bcl-xL-Bak peptide complex. Recognition between regulators of apoptosis. Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 9. García-Sáez A. J., Ries J., Orzáez M., Pérez-Payà E., Schwille P. (2009) Membrane promotes tBID interaction with BCL-XL. Nat. Struct. Mol. Biol. 16, 1178–1185 [DOI] [PubMed] [Google Scholar]
- 10. Kuwana T., Bouchier-Hayes L., Chipuk J. E., Bonzon C., Sullivan B. A., Green D. R., Newmeyer D. D. (2005) BH3 domains of BH3-only proteins differentially regulate Bax-mediated mitochondrial membrane permeabilization both directly and indirectly. Mol. Cell 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 11. Willis S. N., Fletcher J. I., Kaufmann T., van Delft M. F., Chen L., Czabotar P. E., Ierino H., Lee E. F., Fairlie W. D., Bouillet P., Strasser A., Kluck R. M., Adams J. M., Huang D. C. (2007) Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science 315, 856–859 [DOI] [PubMed] [Google Scholar]
- 12. Llambi F., Moldoveanu T., Tait S. W., Bouchier-Hayes L., Temirov J., McCormick L. L., Dillon C. P., Green D. R. (2011) A unified model of mammalian BCL-2 protein family interactions at the mitochondria. Mol. Cell 44, 517–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leber B., Lin J., Andrews D. W. (2007) Embedded together. The life and death consequences of interaction of the Bcl-2 family with membranes. Apoptosis 12, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Letai A., Bassik M. C., Walensky L. D., Sorcinelli M. D., Weiler S., Korsmeyer S. J. (2002) Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2, 183–192 [DOI] [PubMed] [Google Scholar]
- 15. Kim H., Rafiuddin-Shah M., Tu H. C., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2006) Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat. Cell Biol. 8, 1348–1358 [DOI] [PubMed] [Google Scholar]
- 16. Hsu Y. T., Wolter K. G., Youle R. J. (1997) Cytosol-to-membrane redistribution of Bax and Bcl-XL during apoptosis. Proc. Natl. Acad. Sci. U.S.A. 94, 3668–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bogner C., Leber B., Andrews D. W. (2010) Apoptosis. Embedded in membranes. Curr. Opin. Cell Biol. 22, 845–851 [DOI] [PubMed] [Google Scholar]
- 18. Lovell J. F., Billen L. P., Bindner S., Shamas-Din A., Fradin C., Leber B., Andrews D. W. (2008) Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell 135, 1074–1084 [DOI] [PubMed] [Google Scholar]
- 19. Petros A. M., Olejniczak E. T., Fesik S. W. (2004) Structural biology of the Bcl-2 family of proteins. Biochim. Biophys. Acta 1644, 83–94 [DOI] [PubMed] [Google Scholar]
- 20. Wang K., Yin X. M., Chao D. T., Milliman C. L., Korsmeyer S. J. (1996) BID. A novel BH3 domain-only death agonist. Genes Dev. 10, 2859–2869 [DOI] [PubMed] [Google Scholar]
- 21. Luo X., Budihardjo I., Zou H., Slaughter C., Wang X. (1998) Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 94, 481–490 [DOI] [PubMed] [Google Scholar]
- 22. Li H., Zhu H., Xu C. J., Yuan J. (1998) Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 94, 491–501 [DOI] [PubMed] [Google Scholar]
- 23. Boldin M. P., Goncharov T. M., Goltsev Y. V., Wallach D. (1996) Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 85, 803–815 [DOI] [PubMed] [Google Scholar]
- 24. Muzio M., Chinnaiyan A. M., Kischkel F. C., O'Rourke K., Shevchenko A., Ni J., Scaffidi C., Bretz J. D., Zhang M., Gentz R., Mann M., Krammer P. H., Peter M. E., Dixit V. M. (1996) FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death-inducing signaling complex. Cell 85, 817–827 [DOI] [PubMed] [Google Scholar]
- 25. Zha J., Weiler S., Oh K. J., Wei M. C., Korsmeyer S. J. (2000) Posttranslational N-myristoylation of BID as a molecular switch for targeting mitochondria and apoptosis. Science 290, 1761–1765 [DOI] [PubMed] [Google Scholar]
- 26. Ott M., Norberg E., Zhivotovsky B., Orrenius S. (2009) Mitochondrial targeting of tBid/Bax. A role for the TOM complex? Cell Death Differ. 16, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 27. Eskes R., Desagher S., Antonsson B., Martinou J. C. (2000) Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell Biol. 20, 929–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Epand R. F., Martinou J. C., Fornallaz-Mulhauser M., Hughes D. W., Epand R. M. (2002) The apoptotic protein tBid promotes leakage by altering membrane curvature. J. Biol. Chem. 277, 32632–32639 [DOI] [PubMed] [Google Scholar]
- 30. Schendel S. L., Azimov R., Pawlowski K., Godzik A., Kagan B. L., Reed J. C. (1999) Ion channel activity of the BH3 only Bcl-2 family member, BID. J. Biol. Chem. 274, 21932–21936 [DOI] [PubMed] [Google Scholar]
- 31. Grinberg M., Sarig R., Zaltsman Y., Frumkin D., Grammatikakis N., Reuveny E., Gross A. (2002) tBID homooligomerizes in the mitochondrial membrane to induce apoptosis. J. Biol. Chem. 277, 12237–12245 [DOI] [PubMed] [Google Scholar]
- 32. Chou J. J., Li H., Salvesen G. S., Yuan J., Wagner G. (1999) Solution structure of BID, an intracellular amplifier of apoptotic signaling. Cell 96, 615–624 [DOI] [PubMed] [Google Scholar]
- 33. McDonnell J. M., Fushman D., Milliman C. L., Korsmeyer S. J., Cowburn D. (1999) Solution structure of the proapoptotic molecule BID. A structural basis for apoptotic agonists and antagonists. Cell 96, 625–634 [DOI] [PubMed] [Google Scholar]
- 34. Oh K. J., Barbuto S., Meyer N., Kim R. S., Collier R. J., Korsmeyer S. J. (2005) Conformational changes in BID, a pro-apoptotic BCL-2 family member, upon membrane binding. A site-directed spin labeling study. J. Biol. Chem. 280, 753–767 [DOI] [PubMed] [Google Scholar]
- 35. Billen L. P., Shamas-Din A., Andrews D. W. (2008) Bid. A Bax-like BH3 protein. Oncogene 27, S93–104 [DOI] [PubMed] [Google Scholar]
- 36. Shamas-Din A., Bindner S., Zhu W., Zaltsman Y., Campbell C., Gross A., Leber B., Andrews D. W., Fradin C. (2013) tBid undergoes multiple conformational changes at the membrane required for Bax activation. J. Biol. Chem. 288, 22111–22127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong X. M., Choi J., Franzin C. M., Zhai D., Reed J. C., Marassi F. M. (2004) Conformation of membrane-associated proapoptotic tBid. J. Biol. Chem. 279, 28954–28960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oh K. J., Barbuto S., Pitter K., Morash J., Walensky L. D., Korsmeyer S. J. (2006) A membrane-targeted BID BCL-2 homology 3 peptide is sufficient for high potency activation of BAX in vitro. J. Biol. Chem. 281, 36999–37008 [DOI] [PubMed] [Google Scholar]
- 39. Grzesiek S., Bax A. (1992) Improved 3D triple-resonance NMR techniques applied to a 31-kDa protein. J. Magn. Reson. 96, 432–440 [Google Scholar]
- 40. Grzesiek S., Bax A. (1992) Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J. Am. Chem. Soc. 114, 6291–6293 [Google Scholar]
- 41. Wittekind M., Mueller L. (1993) HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the α- and β-carbon resonances in proteins. J. Magn. Reson. Ser. B 101, 201–205 [Google Scholar]
- 42. Bax A., Grzesiek S. (1993) Methodological advances in protein NMR. Acc. Chem. Res. 26, 131–138 [Google Scholar]
- 43. Kay L. E., Xu G. Y., Yamazaki T. (1994) Enhanced-sensitivity triple-resonance spectroscopy with minimal H2O saturation. J. Magn. Reson. Ser. A 109, 129–133 [Google Scholar]
- 44. Clore G. M., Kay L. E., Bax A., Gronenborn A. M. (1991) Four-dimensional 13C/13C-edited nuclear Overhauser enhancement spectroscopy of a protein in solution. Application to interleukin 1β. Biochemistry 30, 12–18 [DOI] [PubMed] [Google Scholar]
- 45. Barbato G., Ikura M., Kay L. E., Pastor R. W., Bax A. (1992) Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy. The central helix is flexible. Biochemistry 31, 5269–5278 [DOI] [PubMed] [Google Scholar]
- 46. Iwahara J., Tang C., Marius Clore G. (2007) Practical aspects of 1H transverse paramagnetic relaxation enhancement measurements on macromolecules. J. Magn. Reson. 184, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe. A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 48. Johnson B. A., Blevins R. A. (1994) NMR view. A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603–614 [DOI] [PubMed] [Google Scholar]
- 49. Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) A common sense approach to peak picking two-, three- and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 50. Farrow N. A., Muhandiram R., Singer A. U., Pascal S. M., Kay C. M., Gish G., Shoelson S. E., Pawson T., Forman-Kay J. D., Kay L. E. (1994) Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry 33, 5984–6003 [DOI] [PubMed] [Google Scholar]
- 51. Iwahara J., Schwieters C. D., Clore G. M. (2004) Ensemble approach for NMR structure refinement against 1H paramagnetic relaxation enhancement data arising from a flexible paramagnetic group attached to a macromolecule. J. Am. Chem. Soc. 126, 5879–5896 [DOI] [PubMed] [Google Scholar]
- 52. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+. A hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ryabov Y., Schwieters C. D., Clore G. M. (2011) Impact of 15N R2/R1 relaxation restraints on molecular size, shape, and bond vector orientation for NMR protein structure determination with sparse distance restraints. J. Am. Chem. Soc. 133, 6154–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xu C., Gagnon E., Call M. E., Schnell J. R., Schwieters C. D., Carman C. V., Chou J. J., Wucherpfennig K. W. (2008) Regulation of T cell receptor activation by dynamic membrane binding of the CD3ϵ cytoplasmic tyrosine-based motif. Cell 135, 702–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schwieters C. D., Kuszewski J. J., Tjandra N., Clore G. M. (2003) The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson 160, 65–73 [DOI] [PubMed] [Google Scholar]
- 56. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 57. Krueger-Koplin R. D., Sorgen P. L., Krueger-Koplin S. T., Rivera-Torres I. O., Cahill S. M., Hicks D. B., Grinius L., Krulwich T. A., Girvin M. E. (2004) An evaluation of detergents for NMR structural studies of membrane proteins. J. Biomol. NMR 28, 43–57 [DOI] [PubMed] [Google Scholar]
- 58. Bleicken S., García-Sáez A. J., Conte E., Bordignon E. (2012) Dynamic interaction of cBid with detergents, liposomes and mitochondria. PLoS One 7, e35910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cho E. Y., Yun C. H., Ahn T. (2012) Effects of phospholipids on the functional regulation of tBID in membranes. Mol. Cell Biochem. 363, 395–408 [DOI] [PubMed] [Google Scholar]
- 60. Katz C., Zaltsman-Amir Y., Mostizky Y., Kollet N., Gross A., Friedler A. (2012) Molecular basis of the interaction between proapoptotic truncated BID (tBID) protein and mitochondrial carrier homologue 2 (MTCH2) protein. Key players in mitochondrial death pathway. J. Biol. Chem. 287, 15016–15023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zaltsman Y., Shachnai L., Yivgi-Ohana N., Schwarz M., Maryanovich M., Houtkooper R. H., Vaz F. M., De Leonardis F., Fiermonte G., Palmieri F., Gillissen B., Daniel P. T., Jimenez E., Walsh S., Koehler C. M., Roy S. S., Walter L., Hajnóczky G., Gross A. (2010) MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat. Cell Biol. 12, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ulmer T. S., Bax A., Cole N. B., Nussbaum R. L. (2005) Structure and dynamics of micelle-bound human α-synuclein. J. Biol. Chem. 280, 9595–9603 [DOI] [PubMed] [Google Scholar]
- 63. Göbl C., Dulle M., Hohlweg W., Grossauer J., Falsone S. F., Glatter O., Zangger K. (2010) Influence of phosphocholine alkyl chain length on peptide-micelle interactions and micellar size and shape. J. Phys. Chem. B 114, 4717–4724 [DOI] [PubMed] [Google Scholar]
- 64. Lutter M., Perkins G. A., Wang X. (2001) The pro-apoptotic Bcl-2 family member tBid localizes to mitochondrial contact sites. BMC Cell Biol. 2, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000) Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2, 754–761 [DOI] [PubMed] [Google Scholar]
- 66. Kim T. H., Zhao Y., Ding W. X., Shin J. N., He X., Seo Y. W., Chen J., Rabinowich H., Amoscato A. A., Yin X. M. (2004) Bid-cardiolipin interaction at mitochondrial contact site contributes to mitochondrial cristae reorganization and cytochrome c release. Mol. Biol. Cell 15, 3061–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu J., Weiss A., Durrant D., Chi N. W., Lee R. M. (2004) The cardiolipin-binding domain of Bid affects mitochondrial respiration and enhances cytochrome c release. Apoptosis 9, 533–541 [DOI] [PubMed] [Google Scholar]
- 68. Gonzalvez F., Bessoule J. J., Rocchiccioli F., Manon S., Petit P. X. (2005) Role of cardiolipin on tBid and tBid/Bax synergistic effects on yeast mitochondria. Cell Death Differ 12, 659–667 [DOI] [PubMed] [Google Scholar]
- 69. Tyurin V. A., Tyurina Y. Y., Osipov A. N., Belikova N. A., Basova L. V., Kapralov A. A., Bayir H., Kagan V. E. (2007) Interactions of cardiolipin and lyso-cardiolipins with cytochrome c and tBid. Conflict or assistance in apoptosis. Cell Death Differ. 14, 872–875 [DOI] [PubMed] [Google Scholar]
- 70. Manara A., Lindsay J., Marchioretto M., Astegno A., Gilmore A. P., Esposti M. D., Crimi M. (2009) Bid binding to negatively charged phospholipids may not be required for its pro-apoptotic activity in vivo. Biochim. Biophys. Acta 1791, 997–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Gonzalvez F., Pariselli F., Jalmar O., Dupaigne P., Sureau F., Dellinger M., Hendrickson E. A., Bernard S., Petit P. X. (2010) Mechanistic issues of the interaction of the hairpin-forming domain of tBid with mitochondrial cardiolipin. PLoS One 5, e9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Petit P. X., Dupaigne P., Pariselli F., Gonzalvez F., Etienne F., Rameau C., Bernard S. (2009) Interaction of the α-helical H6 peptide from the pro-apoptotic protein tBid with cardiolipin. FEBS J. 276, 6338–6354 [DOI] [PubMed] [Google Scholar]
- 73. Walensky L. D., Pitter K., Morash J., Oh K. J., Barbuto S., Fisher J., Smith E., Verdine G. L., Korsmeyer S. J. (2006) A stapled BID BH3 helix directly binds and activates BAX. Mol. Cell 24, 199–210 [DOI] [PubMed] [Google Scholar]
- 74. Cartron P. F., Gallenne T., Bougras G., Gautier F., Manero F., Vusio P., Meflah K., Vallette F. M., Juin P. (2004) The first α helix of Bax plays a necessary role in its ligand-induced activation by the BH3-only proteins Bid and PUMA. Mol. Cell 16, 807–818 [DOI] [PubMed] [Google Scholar]
- 75. Yethon J. A., Epand R. F., Leber B., Epand R. M., Andrews D. W. (2003) Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J. Biol. Chem. 278, 48935–48941 [DOI] [PubMed] [Google Scholar]
- 76. Liu X., Dai S., Zhu Y., Marrack P., Kappler J. W. (2003) The structure of a Bcl-xL/Bim fragment complex. Implications for Bim function. Immunity 19, 341–352 [DOI] [PubMed] [Google Scholar]
- 77. Yao Y., Bobkov A. A., Plesniak L. A., Marassi F. M. (2009) Mapping the interaction of pro-apoptotic tBID with pro-survival BCL-XL. Biochemistry 48, 8704–8711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gavathiotis E., Suzuki M., Davis M. L., Pitter K., Bird G. H., Katz S. G., Tu H. C., Kim H., Cheng E. H., Tjandra N., Walensky L. D. (2008) BAX activation is initiated at a novel interaction site. Nature 455, 1076–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Czabotar P. E., Westphal D., Dewson G., Ma S., Hockings C., Fairlie W. D., Lee E. F., Yao S., Robin A. Y., Smith B. J., Huang D. C., Kluck R. M., Adams J. M., Colman P. M. (2013) Bax crystal structures reveal how BH3 domains activate Bax and nucleate its oligomerization to induce apoptosis. Cell 152, 519–531 [DOI] [PubMed] [Google Scholar]