Abstract
Surface proteins of Gram-positive bacteria play crucial roles in bacterial adhesion to host tissues. Regarding commensal or probiotic bacteria, adhesion to intestinal mucosa may promote their persistence in the gastro-intestinal tract and their beneficial effects to the host. In this study, seven Lactococcus lactis strains exhibiting variable surface physico-chemical properties were compared for their adhesion to Caco-2 intestinal epithelial cells. In this test, only one vegetal isolate TIL448 expressed a high-adhesion phenotype. A nonadhesive derivative was obtained by plasmid curing from TIL448, indicating that the adhesion determinants were plasmid-encoded. Surface-exposed proteins in TIL448 were analyzed by a proteomic approach consisting in shaving of the bacterial surface with trypsin and analysis of the released peptides by LC-MS/MS. As the TIL448 complete genome sequence was not available, the tryptic peptides were identified by a mass matching approach against a database including all Lactococcus protein sequences and the sequences deduced from partial DNA sequences of the TIL448 plasmids. Two surface proteins, encoded by plasmids in TIL448, were identified as candidate adhesins, the first one displaying pilin characteristics and the second one containing two mucus-binding domains. Inactivation of the pilin gene abolished adhesion to Caco-2 cells whereas inactivation of the mucus-binding protein gene had no effect on adhesion. The pilin gene is located inside a cluster of four genes encoding two other pilin-like proteins and one class-C sortase. Synthesis of pili was confirmed by immunoblotting detection of high molecular weight forms of pilins associated to the cell wall as well as by electron and atomic force microscopy observations. As a conclusion, surface proteome analysis allowed us to detect pilins at the surface of L. lactis TIL448. Moreover we showed that pili appendages are formed and involved in adhesion to Caco-2 intestinal epithelial cells.
Proteins located at the surface of Gram-positive bacteria play crucial roles in bacterial interactions with biotic or abiotic environments. Surface proteins have been particularly studied in pathogen species and are commonly named adhesins because of their involvement in adhesion to host cells or tissues, which promotes subsequent colonization and invasion (1). Surface-located proteins have been the subject of proteomic studies with specific methodologies to target them taking advantage of the advanced mass spectrometry techniques (2–6). The surface proteome of nonpathogen Gram-positive species is far less explored. However, more recently, adhesion to intestinal mucosa was also proposed to contribute to mucosa colonization and persistence of commensal or probiotic bacteria in the gastro-intestinal tract of their host (7–9).
Lactic acid bacteria (LAB) are Gram-positive bacteria widely used in food fermentation and are also components of the human gut microbiota. Also, certain LAB strains have been selected as probiotics that confer beneficial health effect to their host after their consumption. Lactococcus lactis is a LAB species used as starter in dairy fermentations. In addition, because of its generally recognized as safe status, it has been proposed as a vehicle to deliver antigens or therapeutic molecules in vivo in the host gastro-intestinal tract (10). Besides, a few reports described L. lactis strains with properties typically attributed to probiotic strains such as tolerance to low pH and bile, adhesion to epithelial cells or to mucus and exclusion of pathogens (11, 12); also in vivo effects were reported such as anti-allergenic properties (13).
Genetic and phenotypic diversity among L. lactis strains has been highlighted in previous studies (14–16). Two subspecies are distinguished: the cremoris subspecies is isolated only from raw milk and dairy products whereas the lactis subspecies is found in various environments including animal sources, dairy products, and plant surfaces. In agreement with these different ecological niches, genome-based analysis of L. lactis subsp. lactis strains revealed a genome size variability of 20%, which suggests a large pan-genome for this subspecies (14). Plant isolates have been shown to possess additional capabilities compared with milk-derived strains such as fermentation of more diverse carbohydrates, flavor-forming abilities or stress resistance (17). Finally, clinical strains have been isolated from human immunocompromised patients (18).
In a previous study, the diversity of L. lactis strains at the level of their surface physicochemical properties was highlighted (19). Most of the 50 studied strains could be classified in three groups: strains with highly hydrophobic surface, strains with electronegative and hydrophilic surface and strains with low charge surface. In addition, the surface physicochemical properties were correlated with the adhesion properties to a solid polystyrene surface. The differences observed among L. lactis strains at the level of their cell surface charge or hydrophobicity suggest that different cell wall components are exposed at the outermost layer of their cell wall, which influence bacterial adhesion to both abiotic and biotic surfaces.
In this study, among a group of L. lactis strains with different cell surface physicochemical properties, we selected one L. lactis strain highly adhesive to Caco-2 intestinal epithelial cell line. In order to search for protein determinant(s) of the bacterial surface involved in adhesion to Caco-2 cells, we applied a proteomic approach based on “shaving” of the bacterial surface with trypsin and MS identification of the resulting peptides. This allowed us to identify two candidate adhesins, a pilin-like protein and a mucus-binding (MUB)1 protein, at the surface of the adhesive strain. In addition, we showed that pilins are polymerized in L. lactis TIL448 and exposed at the bacterial surface and that they are responsible for the adhesive phenotype. To our knowledge, this study is the first description of pili synthesized by a natural L. lactis isolate with a functional role in adhesion to human host cells.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Culture
Bacterial strains and plasmids used in this study are listed in Table I. Strains were provided by the INRA collection (Jouy-en-Josas, France). Escherichia coli strains were grown at 37 °C in Luria-Bertani (LB) medium with shaking (220 rpm). When required, erythromycin or chloramphenicol was added to the medium to a final concentration of 150 or 10 μg/ml, respectively. L. lactis strains were grown in M17 medium (Difco) supplemented with 0.5% (w/v) glucose (M17Glc) at 30 °C. When required, erythromycin or chloramphenicol was added to the medium to a final concentration of 5 μg/ml. Growth was monitored by measuring the optical density at 600 nm (OD600) with a spectrophotometer (Uvicon 931, Biotek Kontron).
Table I. Strains and plasmids used in this study.
| Strains and plasmids | Other name, relevant genotype or phenotype | Source/Reference |
|---|---|---|
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac–proAB) F'[traD36 proAB+ lacIq lacZΔM15] | |
| TG1repA+ | TG1 derivative with repA gene integrated into the chromosome, allowing replication of L. lactis plasmids | P. Renaulta |
| L. lactis | ||
| MG1363 | L. lactis subsp. cremoris, plasmid-free and prophage-cured derivative of NCDO712, hydrophilic and electronegative surface | (19, 49) |
| TIL256 | C2, L. lactis subsp. cremoris, dairy strain, hydrophobic and electronegative surface | (19) |
| TIL378 | NCDO2118, L. lactis subsp. lactis isolated from beans, hydrophilic and low surface charge | (19) |
| TIL445 | NCDO2108, L. lactis subsp.lactis isolated from peas, hydrophobic and electronegative surface | (19) |
| TIL664 | CNRZ739, L. lactis subsp.lactis, dairy strain, hydrophobic and electronegative surface | (19) |
| TIL1064 | KH, L. lactis subsp. cremoris, dairy strain, hydrophobic surface and low surface charge | (19) |
| TIL448 | NCDO2110, L. lactis subsp. lactis isolated from peas, hydrophobic and electronegative surface | (19) |
| TIL1229 | Derivative of TIL448 obtained by plasmid curing (one 25-kb plasmid lost) | This study |
| TIL1230 | Derivative of TIL448 obtained by plasmid curing (three plasmids of 25-, 32- and 48-kb lost) | This study |
| TIL1289 | TIL448 derivative obtained by disruption of yhgE2 gene | This study |
| TIL1290 | TIL448 derivative obtained by disruption of mub gene | This study |
| TIL1293 | MG1363 derivative overexpressing the pilin-gene cluster cloned in pILnew | This study |
| TIL1294 | MG1363 containing pILnew empty plasmid | This study |
| TIL1295 | TIL448 containing pGKV259 plasmid | This study |
| Plasmids | ||
| pES03 | Emr, replicative plasmid in L. lactis, containing gfpuv gene under the control of the constitutive promoter PldhL | A. Dufourb |
| pJIM2246 | Cmr, high copy number lactococcal cloning vector | (50) |
| pGFP | Cmr, pJIM2246 bearing PldhL-gfpuv cassette | This study |
| pGhost9 | Emr, lactococcal thermosensitive plasmid | (31) |
| pILnew | Emr, lactococcal high copy number cloning vector | (50) |
| pGKV259 | Emr, Cmr, lactococcal replicative vector | (51) |
a INRA, Jouy-en-Josas, France.
b Université de Bretagne Sud, Lorient, France.
General Molecular Biology Techniques
Molecular cloning techniques were performed according to standard procedures (20). Restriction enzymes (New England Biolabs, Ipswich, MA), TaqDNA polymerase (MP Biomedicals, Solon, OH) and Phusion® High-Fidelity DNA polymerase (Thermoscientific) were used as recommended by the manufacturers. Ligation was performed with Fast-Link™ DNA Ligation Kit (Epicenter). Oligonucleotides were purchased from Eurogentec. PCR was performed with a MasterCycler PCR system (Eppendorf). Except for large plasmids, DNA sequences were determined by GATC Biotech. Electrotransformation of L. lactis was performed as described elsewhere (21). Plasmids from L. lactis strains were extracted according to (22) except for sequencing.
Plasmid Curing
Plasmid elimination was performed by growing bacteria at different temperatures (30 °C or 37 °C) in presence of 5 μg/ml acridine orange (23). This procedure was repeated three times consecutively and serial dilutions of the final culture were plated on M17Glc agar plates. A total of 32 colonies were selected and analyzed for plasmid content. The whole procedure was repeated twice and led to the isolation of the two clones TIL1229 and TIL1230.
Plasmid Extraction and Sequencing
Plasmid DNA was prepared according the procedure described by Anderson and McKay (24). A final step of DNA cleaning was performed with a High Pure PCR Template Preparation Kit (Roche) according the manufacturer recommendations. Sequencing was performed on 50 μg of plasmid preparation extracted from the strain TIL1229 by Eurofins MWG Operon (Germany) using the GS FLX Titanium series chemistry technology. A total of 8296 reads were assembled in 309 contigs containing a total of 231,154 bp.
Detection of mRNA by RT-PCR
Total RNA was extracted from an exponential phase culture of L. lactis TIL448 (OD600 of 0.7) with TRIzol Reagent (Invitrogen) and cDNA was synthesized by reverse transcription as described previously (25). PCR was performed as described above on total DNA or cDNA with primers listed in supplemental Table S1. The L. lactis tuf gene, encoding the elongation factor TU, was used as a positive control in RT-PCR experiments as described previously (26). The absence of contamination of RNA samples by genomic DNA was checked by performing PCR with the different primer pairs on total RNA preparation.
Labeling of L. lactis Strains with Green-Fluorescent Protein (GFP)
The PldhL-gfpuv cassette containing the gfpuv gene (27) placed under the control of the constitutive promotor PldhL from Lactobacillus sakei was excised from pES03 plasmid (kind gift of A. Dufour, Université de Bretagne Sud, Lorient, France), after EcoRI digestion. The purified cassette PldhL-gfpuv was cloned into the EcoRI-linearized pJIM2246 vector. The resulting plasmid (pGFP) was produced in the L. lactis plasmid-free strain MG1363 and introduced into the different strains of L. lactis selected in this study. Chloramphenicol-resistant clones were selected, and GFP production was confirmed by epifluorescence microscopy observation. Plasmids were extracted from each selected clone to check by agarose gel that the global plasmid content of the initial strains was not modified by the introduction of the pGFP plasmid.
Adhesion of L. lactis to Caco-2 Intestinal Epithelial Cells
Cell culture-Caco-2 cell line (passages 25 to 80) were routinely grown as previously described (28) in Dulbecco's modified Eagle medium (DMEM) supplemented with 20% heat-inactivated (56 °C, 30 min) fetal calf serum (FCS) and 1% nonessential amino acids (Invitrogen, Carlsbad, CA). Cells were grown at 37 °C in a 10% CO2 - 90% air atmosphere.
In Vitro Adhesion Assay
Monolayers of Caco-2 cells were prepared on round glass coverslips placed in 12-well tissue culture plates (Corning). Cells were seeded at a concentration of 2 × 104 cells per cm2 and grown at 37 °C in a 10% CO2-90% air atmosphere. The Caco-2 monolayers at late post-confluence, that is, after 17 days of culture, were washed twice with pre-warmed (37 °C) Dulbecco's phosphate-buffered saline DPBS (2.7 mm KCl, 1.5 mm KH2PO4, 8 mm Na2HPO4, 138 mm NaCl) (Invitrogen) containing 9 mm CaCl2 and 5 mm MgCl2. Mid-exponential phase cultures (OD600 of 0.7) of L. lactis strains were harvested and washed three times with PBS, and then resuspended in a pre-warmed (37 °C) mixture (50:50 v/v) of DPBS-CaCl2-MgCl2/DMEM to reach an OD600 of 1. Five hundred microliters of the bacterial suspension was added to each well of the plates containing the monolayer Caco-2 cells and the plates was incubated at 37 °C in a 10% CO2 - 90% air atmosphere. After 1 h of incubation, monolayers were washed five times with DPBS and bacterial adhesion was quantified either by fluorescent microscopy or by bacterial enumeration after plating.
Adhesion Quantification by Epifluorescence Microscopy
After the DPBS washes, the cell layers with adhered bacteria were fixed by incubation with 0.5 ml of 2% paraformaldehyde prepared in DPBS, washed three times with DPBS, and mounted onto a glass microscope slide with DABCO-PBS mounting medium (Fluka). Fluorescent bacteria were observed and images captured with an epifluorescence microscope (Olympus BX51, FITC filter). Ten random fields per coverslip were analyzed. Images were analyzed with the software UTHSCSA ImageTool (version 3.00, http://ddsdx.uthscsa.edu/dig/download.html) as previously described (29). Microbial adhesion was estimated as the percentage of surface covered by bacteria.
Adhesion Quantification by Counting Colony-Forming Units (cfus)
After the DPBS washes, Caco-2 layers were disrupted by addition of 1 ml of deionized sterile ice-cold water in each well and repeated pipetting. Serial dilutions of the suspension in PBS were plated on M17Glc agar with a spiral plater (Spiral System Inc.) and incubated for 48 h at 30 °C. Results were expressed in cfus of adhesive bacteria per well of culture plates.
All adhesion assays were done in duplicate and repeated with three independent bacteria and Caco-2 cultures.
Shaving of Bacterial Surface-Exposed Proteins
Surface-exposed proteins were digested on whole bacteria with trypsin according to the shaving method previously described (2, 3) with some modifications after optimization of the incubation time with trypsin to minimize L. lactis lysis. L. lactis cells grown in M17Glc medium at 30 °C were harvested during exponential growth phase (OD600 of 1.2) or after reaching stationary phase (OD600 of 2.5), by centrifugation at 5000 × g for 10 min at 4 °C. After two washes with cold PBS, bacteria (V x OD of 7.0) were resuspended in 0.5 ml of PBS containing 1.2 m sucrose and 1 mm CaCl2 and preheated at 37 °C. Six μg of trypsin (sequencing-grade modified, Promega) was added and the mixture was incubated 5 min at 37 °C under mild stirring (20 rpm, Eppendorf thermomixer). The enzymatic reaction was stopped by transferring on ice before centrifugation at 2000 × g for 10 min at 4 °C. Supernatant was collected and centrifuged again at 20,000 × g for 10 min at 4 °C. An aliquot of the second supernatant was analyzed by electrophoresis in a 0.7% agarose gel and stained with ethidium bromide to check the absence of detectable amounts of nucleic acids, taken as an indicator of bacterial lysis. Trypsin (1.6 μg/ml) was added again in the supernatant and further incubated for 2 h at 37 °C to obtain complete hydrolysis by trypsin of the peptides released from the whole bacteria. The obtained tryptic peptides were purified by HPLC using a C18 column (Aquapore reverse phase RP300, 2.1 × 30 mm, 7 μm, Applied Biosystem), and recovered by isocratic elution with a solution of 72/28/0.1% (H2O/ACN/formic acid). They were dried using speed-vacuum and then solubilized in 25 μl of loading buffer: 2% ACN, 0.08% TFA in H2O for LC coupled to tandem MS (LC-MS/MS) analysis. For comparative analysis, two independent biological replicates were prepared for each strain and each growth phase.
Protein Identification by LC-MS/MS and Databases Searching
Tryptic peptides were analyzed by LC-MS/MS with a nanoLC Ultra system (Ultimate 3000, Dionex) connected to a LTQ-Orbitrap Discovery mass spectrometer (Thermo Fisher Scientific) located on the proteomic platform PAPPSO (INRA, Micalis, France, http://pappso.inra.fr). The concentrated tryptic peptides were loaded onto a trapping column (C18 Pepmap100, 0.3 × 5 mm, 5 μm) at 20 μl/min by the loading solution (98/2/0.08% H2O/ACN/TFA). After 4 min, the pre-column was back-flush eluted onto the column. C18 Pepmap100 (0.075 × 150 mm, 3 μm; Dionex) at a flow rate of 300 nl/min by an ACN gradient from 0 to 36% of solution B (A: 98/2/0.1, B: 20/80/0.1% H2O/ACN/Formic acid). The spray was created by Silica Tips emitter (360 μm OD, 10 μm ID, New objective) with an applied voltage of 1.4 kV. Peptides were analyzed by a data-dependant method of acquisition controlled by Xcalibur 2.07 software version (Thermo Scientific). The four most intense precursor ions detected in the full MS analysis in the Orbitrap analyzer (m/z range: 300–1600) were selected and fragmented by CID onto the linear ion trap (LTQ XL) at a normalized collision energy of 35% with an isolation width of 2 amu. Only the doubly and triply charged ions were subjected to the CID fragmentation step. The m/z of the fragmented precursor was excluded to the analysis during 90 s with a tolerance of 10 ppm.
The raw files produced under Xcalibur were first converted in mzXML files with ReADW (http://sashimi.sourceforge.net) and in a second step, protein identification was performed with X!tandem software (X!tandem Cyclone 2011.12.01.1; http://www.thegpm.org) against a Lactococcus protein database (NCBI, 54381 proteins downloaded on June, 13th, 2012) and the six-frame translation ORFs deduced from the TIL1229 plasmid sequences obtained in this study (1854 translated protein sequences), associated to a classical proteomic contaminant database. The X!Tandem search parameters were trypsin specificity with two missed cleavages and variable oxidation states of Met. The semi-tryptic peptide detection was included by activating the option in the data refine mode. The mass tolerance was fixed to 10 ppm for precursor ions and 0.5 Da for fragment ions. The final search results were filtered using a multiple threshold filter applied at the protein level and consisting in the following criteria: protein log (E-value) = -3 identified with a minimum of two different peptides sequences, detected with a peptide E-value lower than 0.03. The False Discovery Rate was estimated for all samples by using a reverse database between the X!Tandem analysis to 0.04 and 0% at the peptide and protein levels respectively. Results from X!TandemPipeline 3.3.0 analysis (http://pappso.inra.fr/bioinfo/xtandempipeline/) of MS/MS data are available in (supplemental Table S4).
The mass-matching approach was completed by a de novo approach only for the unmatched spectra that were not used by X!tandem. The unmatched spectra were automatically translated into amino acid sequences by Pepnovo software (version 3.1). All sequences with a Pepnovo score higher than 80 were compared by fasts36 (version 36.2.6) against a Lactobacillales Uniref90 database (UNIREF, 266,612 proteins) in an iterative process with the matrix MDM20. The sequences corresponding to a validated protein with an E-value lower than 0.05 were eliminated before a next step of analysis of FASTS36. The de novo pipeline used was developed on the PAPPSO platform by Benoit Valot (http://pappso.inra.fr/bioinfo/denovopipeline/).
The detailed information on all the peptides and proteins identified in all liquid chromatography-tandem mass spectrometry (LC-MS/MS) runs were deposited online using PROTICdb database (30) at the following URL: http://moulon.inra.fr/protic/surftil448.
The putative localization of the experimentally detected proteins was predicted with the LocateP database corresponding to the four Lactococcus strains IL1403, MG1363, KF147 and SK11 (http://www.cmbi.ru.nl/locatep-db/cgi-bin/locatepdb.py).
Construction of L. lactis TIL448 Mutant Derivatives
An 813-bp internal fragment of yhgE2 gene was amplified by PCR from TIL448 total DNA with primers 5′-AAACTGCAGCAGGTAGTGCAAATGATGG-3′ (PstI site underlined) and 5′-ATATGCGGCCGCTTAAATAGCGATACCGAATACAGC-3′ (NotI site underlined). The resulting fragment was cloned into the PstI and NotI restriction sites of pGhost9 plasmid vector, a thermosensitive plasmid in L. lactis (31). The recombinant plasmid was produced in E. coli TG1repA+ and used to transform competent L. lactis TIL448 grown at 28 °C. Two recombinant clones resistant to erythromycin were selected. They were further cultured at 37.5 °C and integrants were selected after plating on erythromycin-containing medium. Plasmid integration was verified by PCR. One clone with disrupted yhgE2 was selected and named TIL1289. A similar strategy was used to inactivate muc gene encoding the putative MUB-protein. The oligonucleotides used to amplify a 732-bp internal fragment were 5′-AAACTGCAG CTGCTGTATTTGCTAGTACC-3′ (PstI site underlined) and 5′ATATGCGGCCGCTTATA GAGTCCCCATATGATCTTC-3′ (NotI site underlined). The resulting mutant was named TIL1290.
Overexpression of the Pili-Encoding Gene Cluster in L. lactis MG1363
A 5910-bp fragment encompassing the identified promoter and the four genes of the pili-encoding cluster was amplified by PCR with PCR Extender System (5Prime) with oligonucleotides 5′-CGGCTCGAGACCTTAGAAAGCTAGTAACG-3′ (XhoI site underlined) and 5′-ATATGCGGCCGCTAGAACATTAGAAAACTTCGC-3′ (NotI site underlined). The resulting fragment was further ligated with pILnew high copy number plasmid, predigested with XhoI and NotI restriction enzymes. The ligation mixture was used to transform L. lactis MG1363 competent cells. One clone with the expected recombinant plasmid was selected and checked by sequencing.
Immunoblotting
Cell envelope preparation - Bacteria (5 ml) were grown in M17Glc medium until stationary phase and harvested by centrifugation. The cell pellet was washed once with 50 mm Tris-HCl, pH 7.0 and resuspended in 1 ml of 50 mm Tris-HCl pH 7.0 containing 2 mm AEBSF at 4 °C. Bacteria were disrupted with glass beads using a Fastprep system (Qbiogene). The suspension was recovered and centrifuged at low speed (1,000 g) for 2 min at 4 °C to remove glass beads and unbroken bacteria. The supernatant was further centrifuged at 16,000 × g for 15 min at 4 °C. The pellet containing bacterial cell envelopes was digested with lysozyme (1 mg/ml) and mutanolysin (200 u.ml−1) in 50 mm Tris-HCl pH 7.0 containing 5 mm MgCl2 for 3 h at 37 °C. The samples were then centrifuged 10 min at 10,000 × g and the supernatants containing cell wall proteins were analyzed by Western blotting.
Generation of Pilin-Specific Antibodies
The 16-amino acid peptide DSTNTPDTSSVREIT(C) derived from YhgE2 sequence (positions 35 to 49) and with a C-terminal Cys, was synthesized and coupled to keyhole limpet hemocyanin carrier protein. Antibodies were raised against the conjugate in rabbits according to a Speedy 28-day protocol (Eurogentec). Antibodies specific for the peptide were purified by affinity chromatography by Eurogentec.
SDS-PAGE and Western Blotting
Proteins of the cell-envelope extracts were separated by SDS-PAGE with a gradient 4–12% polyacrylamide gel (Criterion XT Bis-Tris, Bio-Rad). After separation, proteins were transferred onto nitrocellulose membrane (Amersham Biosciences Hybond ECL) with a semidry apparatus (Bio-Rad). Membrane was incubated successively with purified anti-peptide antibodies, goat anti-rabbit antibodies coupled to horseradish peroxidase and revealed by chemiluminescence with ECL-plus kit (GE Healthcare) according to the manufacturer's instructions. The resulting light was detected with a Chemidoc system (ScienceTec).
Negative Staining and Electron Microscopy
Bacteria were grown overnight in M17Glc medium, harvested by low speed centrifugation (3000 × g for 2 min, 4 °C), washed in PBS and resuspended in PBS. A drop (10 μl) was deposited on a 200-mesh cooper carbon grid. Staining was performed by incubation of the samples for 20 s in 1% phosphotungstic acid. Grids were examined with a Zeiss EM902 electron microscope operated at 80 kV (Carl Zeiss - France), and images were acquired with a charge-coupled device camera (Megaview III) and analyzed with ITEM Software (Eloïse, France) on the MIMA2 Platform, INRA, Jouy-en-Josas, France (www6.jouy.inra.fr/mima2).
Atomic Force Microscopy (AFM)
L. lactis cells were grown to exponential phase (OD600 of 0.6–0.8) in M17Glc medium at 37 °C. Bacterial cells were harvested by low speed centrifugation (3000 × g for 2 min, 4 °C) to avoid damage of the pili. Each pellet was washed twice in acetate buffer (150 mm sodium acetate, pH 4.75) and was finally resuspended in 2 ml acetate buffer.
AFM contact mode images were obtained in air at room temperature using a Nanoscope V Multimode AFM and MLCT cantilevers (Veeco Metrology Group, Santa Barbara, CA). Cells were immobilized by drying on a mica surface. Basically, 100 μl of a concentrated bacterial cell suspension was dropped on the mica surface and let to incubate for 2 h. The mica was then gently rinsed three times in deionized water and the samples were dried in an incubator at 37 °C for 2 h.
RESULTS
A Natural Isolate of L. lactis Adhering to Intestinal Epithelial Cells
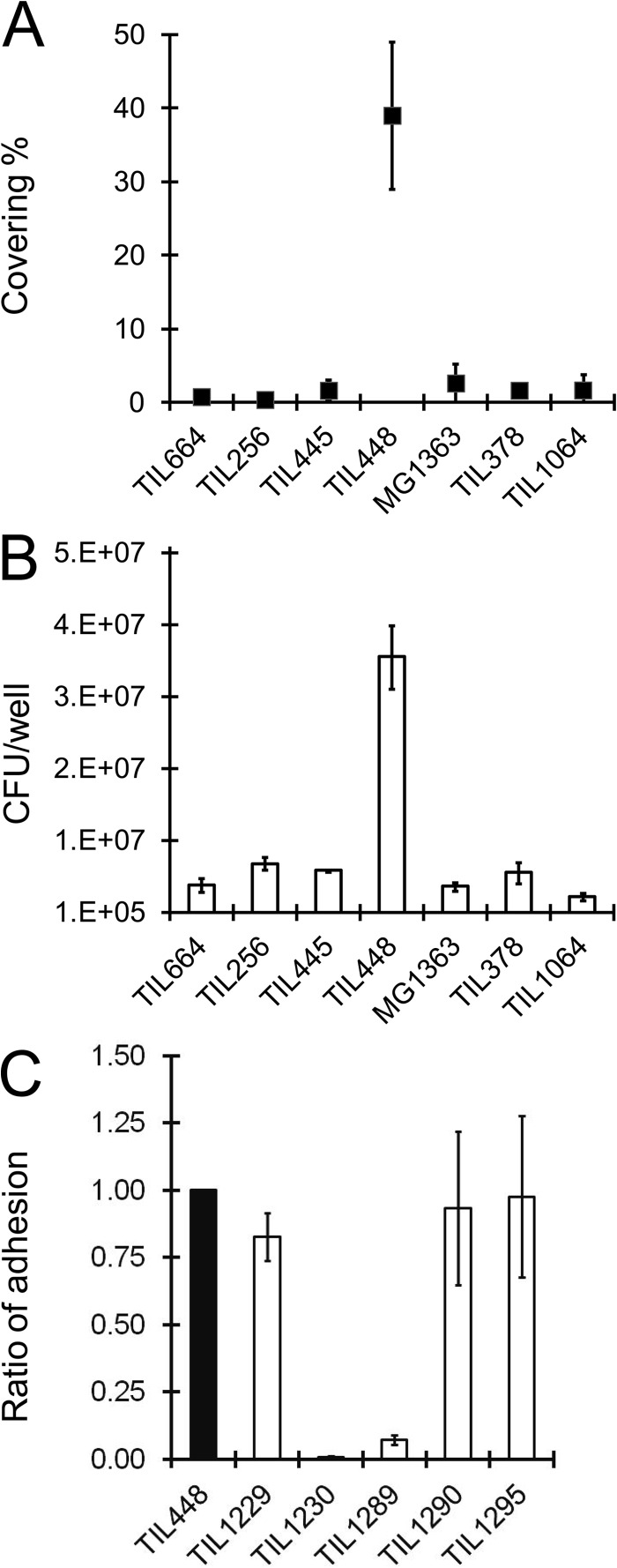
Seven L. lactis strains with different surface physico-chemical properties (19) (Table I) were compared for their ability to adhere to Caco-2 human intestinal epithelial cells. In order to quantify adhesion, bacteria were labeled with GFP and observed by epifluorescence microscopy; alternatively adhered bacteria were enumerated after plating. Among the seven tested strains, L. lactis subsp. lactis TIL448, a vegetal isolate characterized previously as a very hydrophobic strain, adhered strongly to Caco-2 cells as shown both by immunofluorescence microscopy observation and enumeration (Fig. 1A, 1B). In contrast, three other strains (MG1363, TIL378, and TIL1064) with hydrophilic surface did not exhibit the adhesion phenotype, as well as three other strains (TIL256, TIL445, and TIL664) with hydrophobic surface (19) (Fig. 1A, 1B). All these results suggested that the original adhesion properties of TIL448 were not resulting from its hydrophobic surface but more probably from a specific determinant present at the TIL448 cell surface and absent in the other tested strains.
Fig. 1.
Adhesion of the L. lactis strains to Caco-2 intestinal epithelial cells. A, B, adhesion of natural L. lactis isolates with different surface physico-chemical properties (see Table I). A, bacteria were labeled with GFP and adhesion was quantified as the percentage of surface covered by bacteria estimated from epifluorescence microscopy images. B, adhesion of non-labeled bacteria was quantified in parallel by bacterial enumeration and expressed as cfus/well. C, adhesion of TIL448 and its derivatives was quantified by enumeration and the results were normalized on the reference strain TIL448 to be presented as a ratio.
Natural L. lactis strains are known to contain plasmids which are partly responsible for the genomic intraspecies variability (14). Thus we hypothesized that the adhesive character of TIL448 could be plasmid-encoded. The plasmid content of TIL448 was previously analyzed and consists in five plasmids with sizes ranging from 2.5 to 90 kb ((14); P. Le Bourgeois, personal communication). By acridine orange treatment, we obtained two distinct plasmid-cured derivatives, TIL1229 and TIL1230 (Table I). Interestingly, adhesion to Caco-2 cells of TIL1230 lacking three plasmids decreased dramatically compared with the wild-type TIL448 whereas adhesion of TIL1229 lacking only one plasmid was similar to that of the wild type (Fig. 1C). These results indicated that the adhesion properties of L. lactis TIL448 were plasmid-encoded.
Analysis of the Surface Proteome of the Adhesive L. lactis TIL448 and Comparison with a Nonadhesive Derivative
Surface proteins were previously reported to confer high hydrophobicity to the bacterial cell surface (29) and certain of them are known as adhesins. To identify surface proteins in L. lactis TIL448 that could be responsible for adhesion to Caco-2 cells, we applied a proteomic approach consisting in digesting surface-exposed proteins on whole bacteria with a proteolytic enzyme, followed by the identification of the released peptides by LC-MS/MS. This “shaving” method has revealed to be a powerful approach to identify surface-exposed proteins in several Gram-positive bacteria (2–6).
As a first step, we tested different incubation times of L. lactis cells with trypsin to minimize cell lysis. These tests were performed with L. lactis MG1363, which genome sequence is available. From these results (supplemental Fig. S1), we chose a short time of incubation with trypsin (5 min) allowing detection of extracellular proteins and minimizing cytoplasmic protein detection.
We then applied a comparative proteomic analysis to L. lactis TIL448 and its nonadhesive derivative TIL1230. Whole bacterial cells from both strains were harvested in exponential and stationary phase and were treated with trypsin in the conditions selected above. The tryptic peptides released from both strains in both growth stages were analyzed by LC-MS/MS. In parallel, because the complete genome sequence of TIL448 was not available, plasmids from TIL448 were partially sequenced. The tryptic peptides were then identified by a mass-matching approach against a database including all protein sequences derived from Lactococcus sequences and from the sequence data that we obtained from TIL448 plasmid partial sequencing. Also, de novo peptide sequencing followed by homology-based protein identification was performed using a home-developed de novo pipeline, to identify more distantly related proteins against a Lactobacillales protein database.
A total of 81 proteins could be identified in TIL448 by mass matching (Table II), including 32 proteins predicted to be located at the bacterial surface by LocateP, consisting of two LPXTG-proteins, 13 lipoproteins, 13 proteins with an N-terminal transmembrane anchor, and four secreted proteins. The other identified proteins include mainly ribosomal proteins (28 sequences) and other intracellular predicted proteins most of which known as moon-lighting proteins (32). The de novo analysis allowed (1) to increase the number of peptide sequences associated to the identification of several known surface proteins such as Usp45 (the major secreted protein by Lactococcus lactis) or d-Ala-d-Ala-carboxypeptidase DacA, and (2) to identify a few other proteins such as the hypothetical lipoprotein YufC and the peptidoglycan-deacetylase annotated XynD (supplemental Table S2). Most of the proteins identified by both approaches were detected in both strains, TIL448 and TIL1230 (Table II and supplemental Table S2). Interestingly, four proteins encoded by plasmids were detected in TIL448 but were absent in TIL1230 (Table II and supplemental Table S3). Blast searches identified similarities between the first protein (named YhgE2) and the chromosomally encoded L. lactis YhgE protein identified recently as a pilin in L. lactis IL1403 (33). The second protein exhibited high sequence similarity with putative MUB-proteins (34) of Leuconostoc citreum KM20 and Lactobacillus fermentum. It contains two MucBP domains (PF06458) (positions 716–817 and 971–1066) followed by a LPXTG motif for peptidoglycan anchoring at the C-terminal end, a N-terminal signal peptide followed by 14 Ser,Thr-rich repeated motifs of 11 amino-acids (with the following consensus sequence SSA(V,T)NA(D,E)(S,T)T(S,G)A); also two completely identical 85-amino acid repeated sequences preceding the MucBP domains (positions 599–683 and 880–964) are present (supplemental Fig. S2). The two last proteins were identified as a peptide transport system (OppA-like) and a nisin-resistance protein respectively, but were detected by a lower number of peptides (Table II). In other Gram-positive bacteria, both pili appendages and MUB-proteins have been shown to confer adhesive properties to bacterial cells (1, 9, 35). These two proteins were thus excellent candidates to be tested for their involvement in adhesion of L. lactis TIL448 to Caco-2 intestinal epithelial cells.
Table II. Surface proteins identified in adhesive L. lactis TIL448 and non-adhesive TIL1230 harvested in exponential phase or stationary phase. Surface-exposed proteins were shaved from whole bacteria with trypsin and peptides were identified by LC-MS/MS followed by Mass Matching data analysis with X!tandem against a database including all Lactococcus protein sequences and the sequences deduced from data obtained in this study from the partial sequencing of L. lactis TIL448 plasmids. Protein sequence % coverage is presented in supplemental data file and in supplemental Table S3 (for plasmid-encoded proteins).
| Locus_tag or contig n°a | Predicted function | Prediction locatePb | Score log(E-value)c | Number of peptidesd |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| TIL448 |
TIL1230 |
||||||||||
| Exp | Stat | Exp | Stat | ||||||||
| Contig 324 | Pilin | LPXTG | −124.8 | 7 | 6 | 13 | 15 | ||||
| Contig 608 | Mucus-binding protein (fragment) | LPXTG | −23.1 | 2 | 2 | 3 | 3 | ||||
| LLKF_0399 | Oligopeptide ABC transporter, substrate-binding protein (OptA) | Lipo | −383.6 | 4 | 2 | 29 | 32 | 49 | 43 | 15 | 20 |
| Contig 352 | Peptide transport system, OppA-like (fragment) | Lipo | −23.9 | 2 | 4 | ||||||
| LL0330 | Ferrichrome ABC transporter substrate binding protein (FhuD) | Lipo | −151.3 | 11 | 10 | 10 | 11 | 15 | 14 | 9 | |
| LL1124 | Fumarate reductase flavoprotein subunit (FrdC) | Lipo | −186.7 | 3 | 2 | 5 | 8 | 6 | 23 | 24 | |
| LL0563 | Glutamate or arginine ABC transporter substrate binding protein | Lipo | −60.8 | 1 | 2 | 6 | 7 | 5 | 4 | 1 | |
| LLKF_1544 | Basic membrane lipoprotein | Lipo | −56.3 | 2 | 3 | 2 | 4 | 5 | 4 | 2 | 3 |
| LLKF_1405 | Manganese ABC transporter, substrate-binding lipoprotein | Lipo | −34.1 | 1 | 1 | 3 | 1 | 5 | 6 | ||
| llmg_0739 | Maltose ABC transporter substrate binding protein | Lipo | −38.9 | 6 | 5 | 2 | 2 | ||||
| LL0937 | Amino acid ABC transporter substrate binding protein | Lipo | −26.4 | 3 | 2 | 2 | 2 | 3 | 3 | ||
| LL1112 | Thiamine biosynthesis lipoprotein (ApbE2) | Lipo | −30.6 | 3 | 4 | 5 | |||||
| LL1725 | Protein maturation protein | Lipo | −13.5 | 3 | 1 | 2 | 2 | ||||
| LLKF_1990 | Phage protein | Lipo | −32.3 | 1 | 1 | 1 | 4 | 1 | |||
| LL2051 | Amino acid ABC transporter substrate binding protein | Lipo | −13.5 | 1 | 3 | ||||||
| LL0320 | Outer membrane lipoprotein precursor | N-anchor | −69.3 | 5 | 8 | 4 | 5 | 5 | 4 | 4 | 8 |
| LL1831 | Unknown protein (YsjH) | N-anchor | −48.9 | 5 | 5 | 1 | 3 | 6 | 6 | ||
| LLKF_2122 | Hypothetical protein | N-anchor | −29.7 | 6 | 5 | 4 | 5 | ||||
| LL0944 | Unknown protein | N-anchor | −26.3 | 1 | 1 | 5 | 5 | ||||
| LLKF_0394 | Penicillin-binding protein, transpeptidase (Pbp2B) | N-anchor | −48.3 | 2 | 5 | 5 | |||||
| LL0016 | Hypothetical protein | N-anchor | −14.9 | 1 | 1 | 2 | 1 | 2 | 2 | ||
| LL1955 | Unknown protein | N-anchor | −10.9 | 2 | 1 | 1 | 2 | 2 | |||
| LL0321 | Outer membrane lipoprotein precursor | N-anchor | −12.4 | 2 | 2 | 1 | 1 | ||||
| LL0156 | Hypothetical protein | N-anchor | −15.5 | 2 | 2 | 2 | |||||
| LL1125 | Unknown protein | N-anchor | −20.3 | 1 | 4 | 2 | |||||
| LL1258 | d-alanine transfer protein | N-anchor | −10.9 | 3 | |||||||
| LL0105 | Putative xylanase/chitin deacetylase | N-anchor | −27.1 | 2 | 1 | 1 | 1 | 2 | 1 | 4 | |
| LL2231 | Cell shape determining protein | N-anchor | −40.7 | 2 | 5 | 3 | |||||
| Contigs 347/351 | Glucansucrase α amylase (fragments) | Sec | −188.4/−65.2 | 14 | 11 | 20 | 23 | 16 | 16 | 1 | 5 |
| LLKF_2530 | d-Ala-d-Ala-serine-type carboxypeptidase (DacA) | Sec | −76.7 | 6 | 3 | 6 | 5 | 6 | 8 | 2 | 2 |
| LLKF_0911 | Peptidoglycan hydrolase with CHAP domain (Usp45) | Sec | −23.7 | 1 | 2 | 5 | 2 | ||||
| lilo_0227 | N-acetylmuramidase | Sec | −16.6 | 2 | 2 | ||||||
| LACR_1287 | Spermidine/putrescine-binding periplasmic protein | Sec | −5.7 | 2 | |||||||
| Contig 597 | Nisin resistance protein, NisR | Intracellular | −10.6 | 2 | 2 | ||||||
| LL1739 | 30S ribosomal protein S20 | Intracellular | −29.1 | 4 | 4 | 3 | 4 | 2 | 2 | ||
| LACR_2382 | Ribosomal protein L15P | Intracellular | −32.4 | 6 | 5 | 1 | 1 | 2 | 2 | ||
| LACR_2399 | Ribosomal protein L2P | Intracellular | −35.1 | 4 | 2 | 1 | 2 | 2 | 2 | ||
| LACR_2597 | Ribosomal protein S12P | Intracellular | −32 | 6 | 4 | 2 | 1 | 1 | 1 | ||
| LL0284 | Ribosomal protein S4 | Intracellular | −19.4 | 5 | 3 | 1 | 2 | 1 | 1 | ||
| LACR_2455 | Ribosomal protein S2P | Intracellular | −21.9 | 4 | 4 | 2 | 2 | ||||
| LL0874 | Ribosomal protein L19 | Intracellular | −23.1 | 5 | 4 | 1 | 2 | 1 | |||
| LL2081 | Ribosomal protein S5 | Intracellular | −28.4 | 3 | 5 | 1 | 1 | 1 | 1 | ||
| LACR_2402 | Ribosomal protein S10P | Intracellular | −19.3 | 2 | 3 | 2 | 1 | 2 | 3 | ||
| LL2082 | Ribosomal protein L18 | Intracellular | −18.3 | 3 | 2 | 1 | 2 | 1 | 1 | ||
| LACR_2394 | Ribosomal protein L29P | Intracellular | −15.2 | 4 | 3 | 1 | 1 | 1 | |||
| LL2095 | Ribosomal protein S19 | Intracellular | −21.8 | 3 | 4 | 1 | 1 | 1 | |||
| LACR_2396 | Ribosomal protein S3P | Intracellular | −22.5 | 2 | 2 | 2 | 2 | ||||
| LLKF_1334 | Ribosomal protein L12P | Intracellular | −28.1 | 2 | 3 | 1 | 2 | 1 | |||
| LACR_2596 | Ribosomal protein S7P | Intracellular | −22.1 | 1 | 2 | 2 | 1 | 1 | |||
| LL2098 | Ribosomal protein L4 | Intracellular | −15.8 | 2 | 2 | 2 | 1 | ||||
| LACR_2386 | Ribosomal protein L6P | Intracellular | −20.1 | 3 | 3 | 1 | |||||
| LACR_2583 | Ribosomal protein S9P | Intracellular | −13.5 | 3 | 3 | 1 | |||||
| LACR_0076 | Ribosomal protein L33P | Intracellular | −7.1 | 1 | 2 | 2 | 2 | ||||
| LACR_1387 | Ribosomal protein L10P | Intracellular | −18.3 | 3 | 2 | 1 | |||||
| LACR_0246 | Ribosomal protein S21P | Intracellular | −6.7 | 2 | 1 | 1 | |||||
| LACR_2400 | Ribosomal protein L23P | Intracellular | −18.8 | 1 | 2 | 1 | |||||
| LACR_1182 | Ribosomal protein L27P | Intracellular | −12.8 | 2 | 2 | ||||||
| LACR_2377 | Ribosomal protein S13P | Intracellular | −5.0 | 2 | 1 | ||||||
| llmg_2383 | 50S ribosomal protein L3 | Intracellular | −8.2 | 2 | 2 | ||||||
| LACR_1180 | Ribosomal protein L21P | Intracellular | −12.6 | 1 | 2 | ||||||
| LACR_0077 | Ribosomal protein L32P | Intracellular | −21.5 | 2 | |||||||
| LL2191 | Ribosomal protein S6 | Intracellular | −14.6 | 2 | |||||||
| LACR_2054 | Translation elongation factor 1A | Intracellular | −48.7 | 7 | 7 | 2 | 2 | 3 | 4 | ||
| llmg_2556 | Elongation factor EF-G | Intracellular | −6.2 | 2 | 2 | 1 | 1 | ||||
| LACR_2454 | Translation elongation factor Ts | Intracellular | −15.6 | 3 | |||||||
| LLKF_0787 | Translation initiation factor 2 | Intracellular | −9.4 | 3 | |||||||
| LL0551 | Trigger factor | Intracellular | −13.4 | 1 | 2 | ||||||
| LL1332 | Pyruvate kinase | Intracellular | −51.8 | 4 | 9 | 3 | 2 | ||||
| llmg_2167 | Fructose-bisphosphate aldolase | Intracellular | −46.7 | 7 | 5 | 3 | 1 | ||||
| LACR_2555 | Glyceraldehyde-3-phosphate dehydrogenase | Intracellular | −25.4 | 1 | 4 | 2 | 3 | 1 | 1 | 1 | 1 |
| LACR_2470 | Glucose-6-phosphate isomerase | Intracellular | −9.2 | 1 | 2 | 1 | 1 | ||||
| llmg_0127 | Phosphoenolpyruvate-protein phosphotransferase | Intracellular | −13.7 | 3 | 1 | ||||||
| LACR_0668 | Enolase | Intracellular | −8.7 | 3 | |||||||
| LACR_0382 | Phosphoglycerate mutase | Intracellular | −8.3 | 1 | 2 | ||||||
| LACR_0525 | Bacterial nucleoid protein Hbs | Intracellular | −16.4 | 4 | 4 | 3 | 3 | 4 | |||
| LACR_0833 | Hypothetical protein | Intracellular | −32.0 | 4 | 6 | 3 | 3 | ||||
| LACR_0248 | Phosphoglycerate kinase | Intracellular | −22.9 | 5 | 1 | 1 | 1 | 1 | |||
| LL0168 | Cold shock protein E | Intracellular | −13.3 | 2 | 2 | 1 | 1 | 2 | |||
| LLKF_0629 | Aminodeoxychorismate lyase family protein | Intracellular | −21.0 | 2 | 3 | 2 | |||||
| LACR_1455 | l-lactate dehydrogenase | Intracellular | −19.9 | 2 | 2 | 1 | 2 | ||||
| LL2155 | Alcohol-acetaldehyde dehydrogenase | Intracellular | −14.8 | 1 | 2 | 1 | 1 | ||||
| LACR_0822 | Acyl carrier protein | Intracellular | −7.1 | 1 | 1 | 1 | 2 | ||||
| LACR_1027 | Molecular chaperone | Intracellular | −13.3 | 2 | 1 | ||||||
| LACR_1818 | Cell division initiation protein | Intracellular | −11.1 | 2 | 1 | ||||||
| LL2110 | Unknown Protein | Membrane | −5.6 | 2 | 1 | ||||||
| LL1451 | Glycine betaine transport system. busAB | Membrane | −5.4 | 2 | |||||||
a Locus-tag of the identified protein in the database made of available Lactococcus protein sequences or number of the contig of the new sequence data from TIL448 plasmids. Proteins in bold are plasmid-encoded.
b LPXTG, sortase signal; Lipo, lipoprotein; N-anchor, single transmembrane domain at the N-terminus; Sec, secreted with cleavable signal peptide.
c Protein score calculated by X!tandem Pipeline as the product of unique peptide E-values in all sample (http://pappso.inra.fr/downloads/xtandem_pipeline.pdf).
d Number of distinct peptides identified for each strain and growth phase in two independent experiments. Exp, exponential phase culture; Stat, stationary phase culture.
The yhgE2-Pilin Locus Plays a Major Role in Adhesion to Caco-2 Cells
To examine whether the two identified surface proteins, YhgE2 and the MUB-protein were involved in adhesion of L. lactis TIL448 to Caco-2 cells, the two corresponding genes were disrupted in TIL448 by single cross over. Adhesion of the two resulting mutants, named TIL1289 (yhgE2-minus) and TIL1290 (mub-minus) to Caco-2 was tested. Since these mutants should be cultured in the presence of erythromycin (Table I), we first checked that a control erythromycin-resistant derivative of L. lactis TIL448 (TIL1295) adhered at the same level as the wild type TIL448 (Fig. 1C). As shown also on Fig. 1C, the pilin mutant TIL1289 exhibited a much reduced level of adhesion to Caco-2 cells compared with the wild-type TIL448; in contrast the adhesion level of the MUB-protein mutant TIL1290 was similar to the wild-type one. These results clearly show the crucial role of the yhgE2-pilin locus in adhesion of L. lactis TIL448 to Caco-2 cells whereas the muc locus does not contribute or very weakly.
Description of the Pili Locus
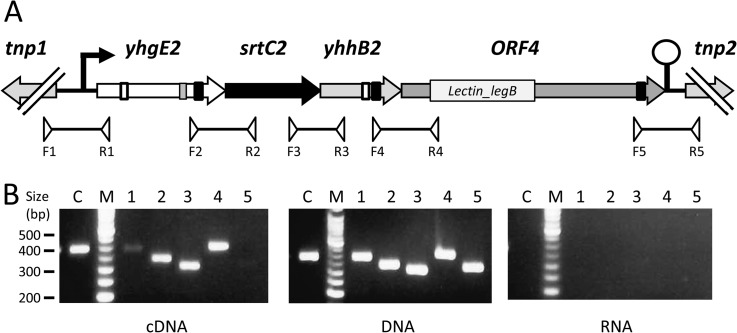
Sequence analysis of the 6.9-kb fragment obtained from plasmid sequencing and containing yhgE2 gene revealed a typical cluster of genes specifying sortase-dependent heterotrimeric pili (36) (Fig. 2A). YhgE2 which contains a typical WxxxVxVxVYPK pilin motif, an E-box motif YxLxETxAPxGY and a C-terminal LPXTG anchoring motif, is predicted to be the backbone pilin. The first gene (yhhA2 or srtC2) downstream of yhgE2 encodes a putative sortase C (SrtC) with a typical TLVTC motif containing the catalytic cysteine, involved in major pilin polymerization (37). The third gene (yhhB2) encodes a putative minor pilin with a pilin motif IYPK and an LPXTG motif. The last gene encodes a large LPXTG-containing protein (ORF4), with a lectin domain (PF00139 or Lectin_legB domain), which makes it as a putative candidate to be the tip pilin with carbohydrate-binding properties. These four genes are preceded by a putative promoter sequence and followed by a putative terminator and thus likely constitute an operon. RT-PCR experiments carried out on RNA extracted from TIL448 with the primer pairs shown on Fig. 2A, confirmed that the four genes: yhgE2, srtC2, yhhB2 and ORF4 are co-transcribed from a promoter located upstream of yhgE2 and their transcription ended at the putative predicted terminator (Fig. 2B). In addition, a fainter PCR product was detected with oligonucleotides F1 and R1 indicating transcription at a low level from another upstream-located promoter.
Fig. 2.
Schematic representation of the plasmid-encoded pilin gene cluster of L. lactis TIL448 and transcriptional analysis of the operon. A, the four genes are represented by arrows. The predicted promoter and terminator are indicated by a black arrow and a black stem-loop, respectively. Tnp, transposase. The presence of different motifs is indicated: LPXTG motif, black box; pilin motif, white box; E-box, gray box. Lectin_legB domain present at the N-terminus of ORF4 is indicated. B, RT-PCR analysis of the pilin gene cluster. cDNA obtained from RNA by reverse transcription, DNA and RNA were analyzed by PCR using the primer pairs F1-R1 (lanes 1), F2-R2 (lanes 2), F3-R3 (lanes 3), F4-R4 (lanes 4) and F5-R5 (lanes 5). Control experiments (C) were run with tuf primer pairs.
A chromosomal locus was recently characterized in L. lactis IL1403 which codes for pilus biogenesis; however pili synthesis could not be detected in this strain in standard laboratory growth conditions (33). The IL1403 chromosomal locus and the plasmidic one described in this study in TIL448 exhibit similarities but also marked differences. Sequence analysis revealed that YhgE2, YhhA2, and YhhB2 (Fig. 2A) exhibit identity but only at the level of 28, 53, and 38% respectively with their IL1403 chromosomal counterparts. In addition, the putative tip pilin encoded by the TIL448 gene cluster has no homolog in IL1403 chromosome or in other known L. lactis genome sequences. Lastly, the order of the genes in the operon is not conserved. Of note, the TIL448 plasmidic gene cluster is flanked by two transposase genes, indicating that the complete cluster could have been acquired by horizontal transfer by the L. lactis TIL448 strain.
Synthesis of Pili at the Surface of L. lactis TIL448
The synthesis of pili in TIL448 was investigated by Western blotting with specific anti-YhgE2 antibodies and by microscopic techniques. Also, we overexpressed the whole plasmidic pilin gene cluster in the plasmid-free strain MG1363 which is devoid of pili in laboratory growth conditions (see below), by placing this gene cluster on a high copy number plasmid.
Antibodies were raised against a 15-amino acid peptide derived from the N-terminal sequence of the mature putative backbone pilin YhgE2 after signal sequence cleavage. The synthesis of pili was first evaluated by Western blotting on cell wall extracts derived from wild-type TIL448, the two derived mutants TIL1289 and TIl1290 and the overexpressing strain TIL1293. As shown on Fig. 3 (lane 4), in the overexpresing L. lactis MG1363 strain (TIL1293), specific anti-peptide antibodies revealed a 50-kDa band corresponding most probably to the monomeric pilin, a likely degradation product at 45-kDa as well as high molecular weight (HMW) immunoreactive proteins, which likely correspond to polymeric forms of the YhgE2 pilin as previously described for other Gram-positive pili (9, 38). HMW immunoreactive proteins were also detected in the wild type L. lactis TIL448 as well as in the TIL1290 unrelated MUB-protein mutant (Fig. 3, lanes 1 and 3 respectively). In contrast, no immunoreactive band could be detected either in the yhgE2-negative mutant TIL1289 or in control MG1363 (Fig. 3, lanes 2 and 5 respectively) confirming the identity of the high molecular mass detected proteins.
Fig. 3.
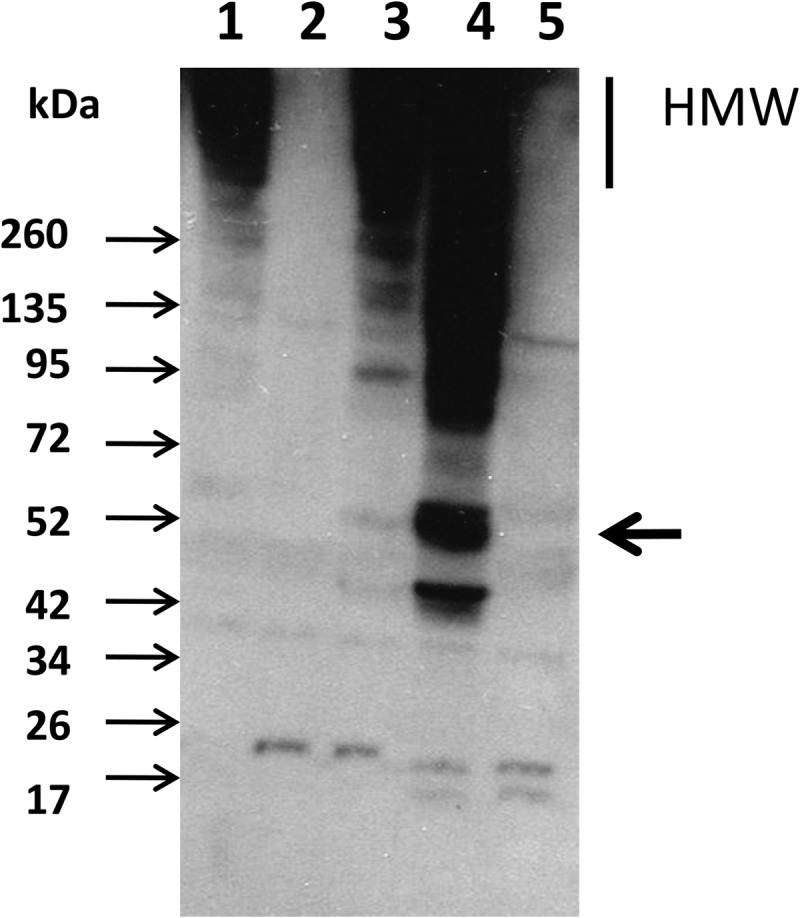
Detection of YhgE2 major pilin by immunoblotting in cell wall extracts. Cell-wall proteins were separated by SDS-PAGE, transferred to nitrocellulose membrane and probed with anti-YhgE2 specific antibodies. The expected migration of YhgE2 monomer is indicated by an arrow on the right side. High molecular weight (HMW) complexes correspond to polymerized YhgE2. Molecular masses are indicated in kDa on the left. Lane 1: wild-type TIL448; lane 2: yhgE2-negative mutant (TIL1289); lane 3: MUB-protein mutant (TIL1290); lane 4: MG1363 overexpressing the pilin gene cluster (TIL1293); lane 5: Control MG1363 with empty plasmid (TIL1294).
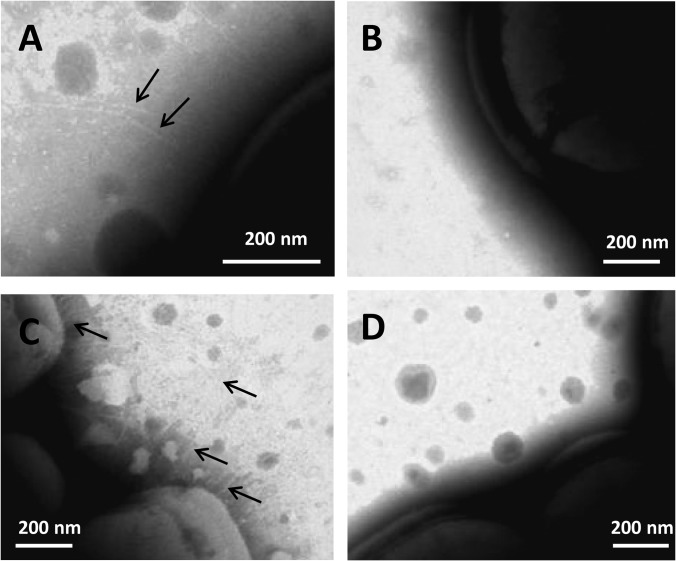
In order to confirm the presence of pili appendages at the surface of L. lactis TIL1293 and TIL448, bacteria were examined by electron microscopy after negative staining and by AFM. Negative staining revealed the presence of thin pili at the surface of TIL1293 overexpressing strain (Fig. 4C) and in a lower density at the surface of TIL448 (Fig. 4A). No similar structures could be detected when control MG1363 or TIL1289 negative mutant were observed confirming the nature of the observed structures (Fig. 4B and 4D respectively). The synthesis of pili was restored in TIL1289 mutant complemented with the four-gene cluster under the control of their native promoter (supplemental Fig. S3).
Fig. 4.
Electron micrographs of the L. lactis strains after negative staining. A, wild-type TIL448. B, yhgE2-negative mutant (TIL1289). C, MG1363 overexpressing the pilin gene cluster (TIL1293). D, Control MG1363 with empty plasmid (TIL1294). Pili are indicated by arrows.
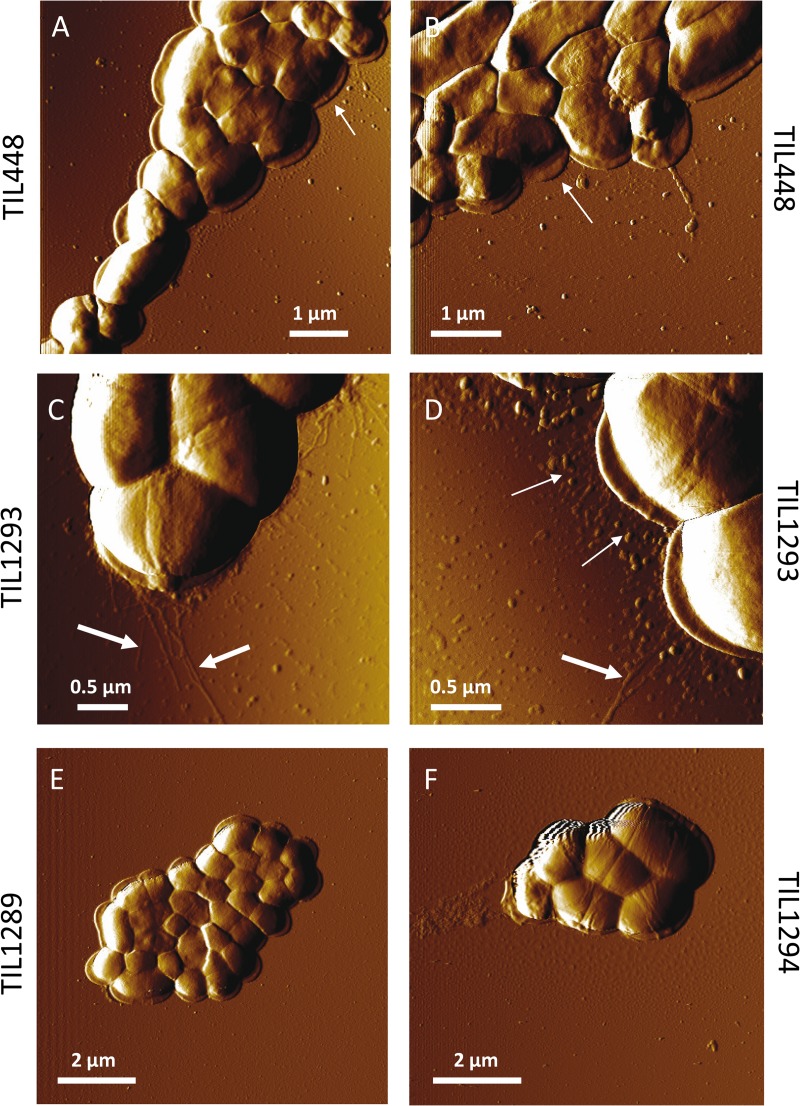
Furthermore, the strains were observed by AFM which was previously shown to be a potent technique to visualize pili at bacterial surfaces (39). AFM images revealed at the surface of the wild-type L. lactis TIL448 the presence of thin and short pili with an average length of 350 ± 90 nm (mean and standard deviation calculated on 40 measurements) (Fig. 5A, 5B) and a diameter that was too small to be measured. Thicker and longer pili with an average length of 1.83 ± 0.71 μm (mean and standard deviation calculated on 40 measurements) and an average diameter of 2.3 nm ± 0.1 nm (mean and standard deviation calculated on 40 measurements) were also imaged at the surface of the overexpressing strain TIL1293 in addition to these thin and short pili (Fig. 5C, 5D). No pili-like structures were detected on control strains TIL1289 and TIL1294 (Fig. 5E and 5F respectively).
Fig. 5.
AFM deflection images recorded in air for the L. lactis strains. A-B, wild-type TIL448. C-D, MG1363 overexpressing the pilin gene cluster (TIL1293). E, yhgE2-negative mutant (TIL1289). F, control MG1363 with empty plasmid (TIL1294). Thin and short pili are observed (thin arrows) for both TIL448 (A, B) and TIL1293 (C, D) cells whereas thick and long pili are only seen (thick arrows) for TIL1293 cells (C, D).
All these results indicate that the identified pilin-encoding gene cluster is functional and can lead to pili synthesis at the surface of L. lactis. To our knowledge, this study is the first description of pili synthesis by a natural isolate of L. lactis which confers it specific adhesion properties to human intestinal epithelial cells.
DISCUSSION
In this study, we have characterized a natural isolate of L. lactis with unexpectedly high adhesion properties to intestinal epithelial Caco-2 cells, as compared with other L. lactis strains. Thanks to a proteomic analysis of the surface-exposed proteins, we identified pilins at the surface of the adhesive L. lactis TIL448 strain. We have shown here that a functional plasmidic gene cluster specifying pili synthesis is expressed and leads to the formation of pili appendages that are responsible for the original adhesion properties of L. lactis TIL448 to human intestinal epithelial cells.
Proteomic analysis of the surface-exposed proteins combined with partial sequencing of L. lactis TIL448 plasmids revealed to be a straightforward and efficient approach to identify the adhesion determinants of TIL448. In addition, the comparative proteomic analysis that we performed on the adhesive L. lactis strain TIL448 and the non-adhesive derivative TIL1230 allowed us targeting directly proteins that are synthesized, surface-located and putative adhesins. This proteomic-based approach allowed bypassing a more fastidious and time-consuming one that would have consisted in complete sequencing of the large and not abundant plasmids of L. lactis TIL448 followed by systematic inactivation of all the genes predicted to encode surface proteins and characterization of the adhesive properties of the constructed mutants.
The “shaving” approach combined with MS/MS analysis proved to be very efficient in identifying surface-exposed proteins in the adhesive L. lactis TIL448 strain. Since the whole TIL448 genomic sequence was not available to identify proteins from MS/MS data, we used a mass matching approach against a database including all protein sequences derived from Lactococcus nucleotide sequences and from the sequence data that we obtained from plasmid sequencing in this study. This allowed us identifying 81 proteins including 32 surface-predicted proteins, among which only four plasmid-encoded proteins. A previous characterization of cell-surface proteins using shaving and LC-MS/MS was reported in L. lactis NZ9000 (40), a very close derivative of MG1363 with an available genome sequence. Remarkably, a similar total number of proteins could be identified in the NZ9000 study. This comparison validates our approach with a strain with a non-sequenced genome. On the basis of MS/MS data, YhgE2 pilin appears expressed at rather high level in TIL448. In contrast, the chromosomally encoded pilin YhgE was not detected in previous studies using a shaving approach in the plasmid-free L. lactis NZ9000 (40). These results are coherent with the absence of detection of pili-like structures by electron microcopy in the present study for MG1363 (Fig. 4D and Fig. 5F). In parallel, we tested a de novo approach that can be used as a complementary one to the mass matching approach for bacterial species with nonsequenced genomes. In this way, we could increase the number of peptides detected for certain proteins, such as for Usp45, a known abundant extracellular protein (41). Also, a few other extracellular proteins not identified by mass matching (such as YufC and XynD) were detected. Nevertheless, the plasmid-encoded proteins, YhgE2 pilin and MUB-protein, were too divergent at the amino acid sequence level, from the proteins of the same families encoded in the complete genome sequences of other L. lactis strains (only 28% sequence similarity between YhgE2 and YhgE), to be identified either by mass matching on Lactococcus sequence databases or by the de novo approach. They were detected by using the newly generated plasmid sequence data.
Most studies regarding pili in Gram-positive bacteria were conducted on pathogenic species, including Streptococci, Enterococci, Corynebacteria and Bacilli (36, 42, 43). They were shown to be involved in (1) adhesion to host epithelial cells and to proteins of the extracellular matrix such as fibronectin or collagen, (2) biofilm formation, and (3) stimulation of the host innate immune system; therefore they are considered as virulence factors (42). However, in commensal or probiotic bacteria such as lactobacilli or bifidobacteria, pili will have a rather positive effect by contributing to gut colonization and persistence in the gastro-intestinal tract (8, 9, 44). Regarding LAB, SpaCBA-type pili were discovered in Lactobacillus rhamnosus GG probiotic strain and were shown to be involved in adhesion to mucins and to intestinal epithelial Caco-2 cells as well as in biofilm formation and immune modulation (45, 46). More recently, pili were visualized by electron microscopy in L. lactis isolates but without identification of the genes involved in their synthesis (33). In contrast, expression of the chromosomal locus identified in the genome sequence of the plasmid-free strains IL1403 could not be demonstrated (33). To our knowledge, the present study is the first one to identify a plasmidic pilin gene cluster in a L. lactis strain, expressed in classical laboratory conditions and responsible for the synthesis of pili at the bacterial surface, which are involved in bacterial adhesion to intestinal epithelial cells.
Interestingly, the pilin YhgE2 protein was detected by a higher number of peptides in stationary phase cultures of TIL448 compared with exponential phase cultures whereas most other proteins were detected by similar number of peptides (Table II). These data suggest a higher expression level or accessibility of the YhgE2 pilin at the TIL448 surface in stationary phase compared with exponential phase.
In addition to the YhgE2 pilin, four other plasmid-encoded proteins were detected at the surface of L. lactis TIL448. One of them is the MUB-protein described above (supplemental Fig. S2), that exhibits high sequence similarity with MUB-proteins from L. fermentum and L. citreum. Also, a glucansucrase with high sequence similarity with lactobacilli glucansucrases (around 80% sequence identity with a Lactobacillus reuteri homolog) was detected. Of note no protein of these two families was detected by shaving of NZ9000 (40), although a MUB-protein is encoded by the chromosome of NZ9000 (LLNZ_12740) whereas no glucansucrase gene is present in the available L. lactis genome sequences. These results illustrate the high genetic and phenotypic diversity previously reported for the L. lactis species (14–16) with new functionalities revealed in our proteomic study such as mucosa adhesion or homopolysaccharide synthesis, that are encoded in its large pangenome.
L. lactis plant isolates are known to have a larger genetic and metabolic repertoire compared with dairy isolates, allowing their adaptation to the plant environment (15). In particular, they possess the enzymatic equipment required for degradation of complex polysaccharides which composed plant cell walls (17). The additional presence of surface structures such as pili can thus be proposed as another adaptation mechanism to the plant environment. Indeed, the tip pilin (ORF4) with a lectin-like domain could be involved in adhesion of the L. lactis vegetal isolate to plant cell walls through specific recognition of their constituent polysaccharides. Therefore we hypothesize that the ability of L. lactis TIL448 vegetal isolate to bind human gut epithelial cells results from the presence of similar sugar motifs at the surface of epithelial cells and plant cells.
MUB-proteins were previously identified as able to bind mucins which are highly glycosylated proteins (47, 48). Furthermore the tip pilin (ORF4) discovered in this study is endowed with a carbohydrate-binding domain. Further work will evaluate the ability of L. lactis TIL448 to bind mucins and the respective contribution of these two surface proteins.
Supplementary Material
Acknowledgments
We thank Véronique Monnet and Mireille Yvon (INRA, Micalis, France) for their constant support and helpful discussions. We thank Jean-Christophe Piard and Virginie Oxaran (INRA, Micalis, France) for helpful discussions, Stéphane Chaillou (INRA, Micalis, France) for precious advice for plasmid extraction and sequencing and Francis Repoila (INRA, Micalis, France) for his expert advice for RT-PCR experiments. We are indebted to Catherine Sapin (Saint-Antoine Hospital, France) for help with epithelial cell cultures. We thank Alain Dufour (Université de Quimper, France) for the gift of plasmid pES03. We warmly thank O. Langella (PAPPSO, INRA, France) for making proteomic data accessible in the Protic database.
Footnotes
* This work was supported by Institut National de la Recherche Agronomique (INRA) and Région Ile-de-France. J.A. received a fellowship from Région Ile-de-France (DIM Malinf). Work at the Université catholique de Louvain was supported by the National Foundation for Scientific Research (FNRS) and the Research Department of the Communauté française de Belgique (Concerted Research Action). Y.F.D. and P.H. are Senior Research Associate and Research Associate of the FNRS.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
The nucleotide sequences of the pilin-gene cluster and the mucus-binding protein have been deposited in the GenBank database under GenBank Accession Numbers KF234018 and KF234019 respectively.
1 The abbreviations used are:
- MUB
- mucus-binding
- LAB
- lactic acid bacteria
- MS/MS
- tandem mass spectrometry
- DMEM
- Dulbecco's modified Eagle medium
- DPBS
- Dulbecco's phosphate buffered saline
- cfu
- colony-forming unit
- AFM
- atomic force microscopy.
REFERENCES
- 1. Kline K. A., Fälker S., Dahlberg S., Normark S., Henriques-Normark B. (2009) Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5, 580–592 [DOI] [PubMed] [Google Scholar]
- 2. Rodriguez-Ortega M. J., Norais N., Bensi G., Liberatori S., Capo S., Mora M., Scarselli M., Doro F., Ferrari G., Garaguso I., Maggi T., Neumann A., Covre A., Telford J. L., Grandi G. (2006) Characterization and identification of vaccine candidate proteins through analysis of the group A Streptococcus surface proteome. Nat. Biotechnol. 24, 191–197 [DOI] [PubMed] [Google Scholar]
- 3. Severin A., Nickbarg E., Wooters J., Quazi S. A., Matsuka Y. V., Murphy E., Moutsatsos I. K., Zagursky R. J., Olmsted S. B. (2007) Proteomic analysis and identification of Streptococcus pyogenes surface-associated proteins. J. Bacteriol. 189, 1514–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doro F., Liberatori S., Rodriguez-Ortega M. J., Rinaudo C. D., Rosini R., Mora M., Scarselli M., Altindis E., D'Aurizio R., Stella M., Margarit I., Maione D., Telford J. L., Norais N., Grandi G. (2009) Surfome analysis as a fast track to vaccine discovery: identification of a novel protective antigen for Group B Streptococcus hypervirulent strain COH1. Mol. Cell. Proteomics 8, 1728–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dreisbach A., van Dijl J. M., Buist G. (2011) The cell surface proteome of. Staphylococcus aureus. Proteomics 11, 3154–3168 [DOI] [PubMed] [Google Scholar]
- 6. Dreisbach A., Hempel K., Buist G., Hecker M., Becher D., van Dijl J. M. (2010) Profiling the surfacome of Staphylococcus aureus. Proteomics 10, 3082–3096 [DOI] [PubMed] [Google Scholar]
- 7. Ventura M., Turroni F., Motherway M. O., MacSharry J., van Sinderen D. (2012) Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20, 467–476 [DOI] [PubMed] [Google Scholar]
- 8. Hill C. (2012) Virulence or niche factors: what's in a name? J. Bacteriol. 194, 5725–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kankainen M., Paulin L., Tynkkynen S., von Ossowski I., Reunanen J., Partanen P., Satokari R., Vesterlund S., Hendrickx A. P., Lebeer S., De Keersmaecker S. C., Vanderleyden J., Hämäläinen T., Laukkanen S., Salovuori N., Ritari J., Alatalo E., Korpela R., Mattila-Sandholm T., Lassig A., Hatakka K., Kinnunen K. T., Karjalainen H., Saxelin M., Laakso K., Surakka A., Palva A., Salusjarvi T., Auvinen P., de Vos W. M. (2009) Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human- mucus binding protein. Proc. Natl. Acad. Sci. U.S.A. 106, 17193–17198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bermudez-Humaran L. G., Kharrat P., Chatel J. M., Langella P. (2011) Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microb. Cell Fact. 10, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vesterlund S., Karp M., Salminen S., Ouwehand A. C. (2006) Staphylococcus aureus adheres to human intestinal mucus but can be displaced by certain lactic acid bacteria. Microbiology 152, 1819–1826 [DOI] [PubMed] [Google Scholar]
- 12. Kimoto H., Kurisaki J., Tsuji N. M., Ohmomo S., Okamoto T. (1999) Lactococci as probiotic strains: adhesion to human enterocyte-like Caco-2 cells and tolerance to low pH and bile. Lett. Appl. Microbiol. 29, 313–316 [DOI] [PubMed] [Google Scholar]
- 13. Zuercher A. W., Weiss M., Holvoet S., Moser M., Moussu H., van Overtvelt L., Horiot S., Moingeon P., Nutten S., Prioult G., Singh A., Mercenier A. (2012) Lactococcus lactis NCC 2287 alleviates food allergic manifestations in sensitized mice by reducing IL-13 expression specifically in the ileum. Clin. Dev. Immunol. 2012, 485750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Passerini D., Beltramo C., Coddeville M., Quentin Y., Ritzenthaler P., Daveran-Mingot M. L., Le Bourgeois P. (2010) Genes but not genomes reveal bacterial domestication of Lactococcus lactis. PLoS One 5, e15306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Siezen R. J., Bayjanov J. R., Felis G. E., van der Sijde M. R., Starrenburg M., Molenaar D., Wels M., van Hijum S. A., van Hylckama Vlieg J. E. (2011) Genome-scale diversity and niche adaptation analysis of Lactococcus lactis by comparative genome hybridization using multi-strain arrays. Microb. Biotechnol. 4, 383–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan-a-Ram P., Cardoso T., Daveran-Mingot M. L., Kanchanatawee S., Loubiere P., Girbal L., Cocaign-Bousquet M. (2011) Assessment of the diversity of dairy Lactococcus lactis subsp. lactis isolates by an integrated approach combining phenotypic, genomic, and transcriptomic analyses. Appl. Environ. Microbiol. 77, 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Siezen R. J., Starrenburg M. J., Boekhorst J., Renckens B., Molenaar D., van Hylckama Vlieg J. E. (2008) Genome-scale genotype-phenotype matching of two Lactococcus lactis isolates from plants identifies mechanisms of adaptation to the plant niche. Appl. Environ. Microbiol. 74, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elliott J. A., Collins M. D., Pigott N. E., Facklam R. R. (1991) Differentiation of Lactococcus lactis and Lactococcus garvieae from humans by comparison of whole-cell protein patterns. J. Clin. Microbiol. 29, 2731–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Giaouris E., Chapot-Chartier M. P., Briandet R. (2009) Surface physicochemical analysis of natural Lactococcus lactis strains reveals the existence of hydrophobic and low charged strains with altered adhesive properties. Int. J. Food Microbiol. 131, 2–9 [DOI] [PubMed] [Google Scholar]
- 20. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning : a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press [Google Scholar]
- 21. Holo H., Nes I. F. (1989) High-Frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55, 3119–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Sullivan D J., Klaenhammer T. R. (1993) Rapid Mini-Prep Isolation of High-Quality Plasmid DNA from Lactococcus and Lactobacillus spp. Appl. Environ. Microbiol. 59, 2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mesas J. M., Rodriguez M. C., Alegre M. T. (2004) Plasmid curing of Oenococcus oeni. Plasmid 51, 37–40 [DOI] [PubMed] [Google Scholar]
- 24. Anderson D. G., McKay L. L. (1983) Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46, 549–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fouquier d'Herouel A., Wessner F., Halpern D., Ly-Vu J., Kennedy S. P., Serror P., Aurell E., Repoila F. (2011) A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 39, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sperandio B., Polard P., Ehrlich D. S., Renault P., Guédon E. (2005) Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187, 3762–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gory L., Montel M. C., Zagorec M. (2001) Use of green fluorescent protein to monitor Lactobacillus sakei in fermented meat products. FEMS Microbiol. Lett. 194, 127–133 [DOI] [PubMed] [Google Scholar]
- 28. Sapin C., Colard O., Delmas O., Tessier C., Breton M., Enouf V., Chwetzoff S., Ouanich J., Cohen J., Wolf C. (2002) Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J. Virol. 76, 4591–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Habimana O., Le Goff C., Juillard V., Bellon-Fontaine M. N., Buist G., Kulakauskas S., Briandet R. (2007) Positive role of cell wall anchored proteinase PrtP in adhesion of lactococci. BMC Microbiol. 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langella O., Valot B., Jacob D., Balliau T., Flores R., Hoogland C., Joets J., Zivy M. (2013) Management and dissemination of MS proteomic data with PROTICdb: example of a quantitative comparison between methods of protein extraction. Proteomics 13, 1457–1466 [DOI] [PubMed] [Google Scholar]
- 31. Biswas I., Gruss A., Ehrlich S. D., Maguin E. (1993) High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175, 3628–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bendtsen J. D., Kiemer L., Fausbøll A., Brunak S. (2005) Non-classical protein secretion in bacteria. BMC Microbiol. 5, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oxaran V., Ledue-Clier F., Dieye Y., Herry J. M., Pechoux C., Meylheuc T., Briandet R., Juillard V., Piard J. C. (2012) Pilus biogenesis in Lactococcus lactis: molecular characterization and role in aggregation and biofilm formation. PLoS One 7, e50989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Boekhorst J., Helmer Q., Kleerebezem M., Siezen R. J. (2006) Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152, 273–280 [DOI] [PubMed] [Google Scholar]
- 35. Lebeer S., Vanderleyden J., De Keersmaecker S. C. (2008) Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mandlik A., Swierczynski A., Das A., Ton-That H. (2008) Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 16, 33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dramsi S., Trieu-Cuot P., Bierne H. (2005) Sorting sortases: a nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 156, 289–297 [DOI] [PubMed] [Google Scholar]
- 38. Dramsi S., Caliot E., Bonne I., Guadagnini S., Prévost M. C., Kojadinovic M., Lalioui L., Poyart C., Trieu-Cuot P. (2006) Assembly and role of pili in group B streptococci. Mol. Microbiol. 60, 1401–1413 [DOI] [PubMed] [Google Scholar]
- 39. Tripathi P., Beaussart A., Andre G., Rolain T., Lebeer S., Vanderleyden J., Hols P., Dufrêne Y. F. (2012) Towards a nanoscale view of lactic acid bacteria. Micron 43, 1323–1330 [DOI] [PubMed] [Google Scholar]
- 40. Berlec A., Zadravec P., Jevnikar Z., Štrukelj B. (2011) Identification of candidate carrier proteins for surface display on Lactococcus lactis by theoretical and experimental analyses of the surface proteome. Appl. Environ. Microbiol. 77, 1292–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Asseldonk M., Rutten G., Oteman M., Siezen R. J., de Vos W. M., Simons G. (1990) Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95, 155–160 [DOI] [PubMed] [Google Scholar]
- 42. Danne C., Dramsi S. (2012) Pili of gram-positive bacteria: roles in host colonization. Res. Microbiol. 163, 645–658 [DOI] [PubMed] [Google Scholar]
- 43. Hendrickx A. P., Budzik J. M., Oh S. Y., Schneewind O. (2011) Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat. Rev. Microbiol. 9, 166–176 [DOI] [PubMed] [Google Scholar]
- 44. O'Connell Motherway M., Zomer A., Leahy S. C., Reunanen J., Bottacini F., Claesson M. J., O'Brien F., Flynn K., Casey P. G., Munoz J. A., Kearney B., Houston A. M., O'Mahony C., Higgins D. G., Shanahan F., Palva A., de Vos W. M., Fitzgerald G. F., Ventura M., O'Toole P. W., van Sinderen D. (2011) Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U.S.A. 108, 11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. von Ossowski I., Reunanen J., Satokari R., Vesterlund S., Kankainen M., Huhtinen H., Tynkkynen S., Salminen S., de Vos W. M., Palva A. (2010) Mucosal adhesion properties of the probiotic Lactobacillus rhamnosus GG SpaCBA and SpaFED pilin subunits. Appl. Environ. Microbiol. 76, 2049–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lebeer S., Claes I., Tytgat H. L., Verhoeven T. L., Marien E., von Ossowski I., Reunanen J., Palva A., Vos W. M., Keersmaecker S. C., Vanderleyden J. (2012) Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl. Environ. Microbiol. 78, 185–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Roos S., Jonsson H. (2002) A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148, 433–442 [DOI] [PubMed] [Google Scholar]
- 48. Van Tassell M. L., Miller M. J. (2011) Lactobacillus adhesion to mucus. Nutrients 3, 613–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gasson M. J. (1983) Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Renault P., Corthier G., Goupil N., Delorme C., Ehrlich S. D. (1996) Plasmid vectors for gram-positive bacteria switching from high to low copy number. Gene 183, 175–182 [DOI] [PubMed] [Google Scholar]
- 51. van der Vossen J. M., van der Lelie D., Venema G. (1987) Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53, 2452–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.