Abstract
In bacteria, transcriptional regulation is a key step in cellular gene expression. All bacteria contain a core RNA polymerase that is catalytically competent but requires an additional σ factor for specific promoter recognition and correct transcriptional initiation. The RNAP core is not able to selectively bind to a given σ factor. In contrast, different σ factors have different affinities for the RNAP core. As a consequence, the concentration of alternate σ factors requires strict regulation in order to properly control the delicate interplay among them, which favors the competence for the RNAP core. This control is archived by different σ/anti-σ controlling mechanisms that shape complex regulatory networks and cascades, and enable the response to sudden environmental cues, whose global understanding is a current challenge for systems biology. Although there have been a number of excellent studies on each of these σ/anti-σ post-transcriptional regulatory systems, no comprehensive comparison of these mechanisms in a single model organism has been conducted. Here, we survey all these systems in E. coli dissecting and analyzing their inner workings and highlightin their differences. Then, following an integral approach, we identify their commonalities and outline some of the principles exploited by the cell to effectively and globally reprogram the transcriptional machinery. These principles provide guidelines for developing biological synthetic circuits enabling an efficient and robust response to sudden stimuli.
Keywords: Anti-sigma factors, Escherichia coli, Homeostasis, Negative feedback, Sigma factors, Transcriptional regulation.
INTRODUCTION
In bacteria, the regulation of gene expression is the basis for adaptability, morphogenesis, and cellular differentiation. From all the different regulatory layers, regulation of transcription initiation is a very important step for controlling gene expression. All eubacteria contain a single ∼380-kDa multi-subunit α2ββ´ω RNA polymerase (RNAP) core enzyme, which is catalytically competent and able to recognize DNA non-specifically. However, it requires an additional dissociable σ factor for specific promoter recognition and correct transcription initiation. Most σ factors belong to the σ70family, whose members contain two domains with a high degree of sequence and structural conservation: 1) σ2, which binds the RNAP β′ subunit coiled-coil and the -10 promoter element, and 2) σ4, which binds the RNAP β subunit flap and -35 promoter elements. An additional σ family, σ54, does not share any significant sequence similarity with the σ70 family and is functionally distinct [1].
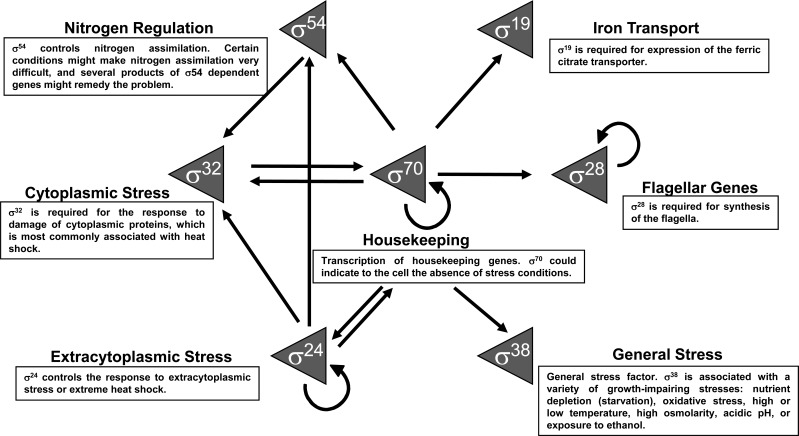
In Escherichia coli, the vast majority of transcriptional “housekeeping” is accomplished by the RNAP core in complex with σ70. However, six other σ factors provide selectivity for transcribing different sets of genes. These genes, required for adaptive responses, have related functions and are transcribed in response to specific stimuli (Fig. 1): σS (σ38) is the general stress factor associated with stationary phase and a variety of growth-impairing stresses; σH (σ32) controls heat shock responsive promoters; σF (σ28) is used for flagellum-related functions; σE (σ24) controls responses to extracytoplasmic or extreme heat shock stress; σFecI (σ19) is required in iron transport for transcription of the ferric citrate transporter; and σN (σ54) controls promoters for nitrogen assimilation. This repertoire of alternative σ factors reflects the lifestyle of E. coli as a gut commensal bacterium [2].
Fig. (1).
σ factors functions and their relationships at the transcriptional level. The arrows indicate the transcription interactions between σ factors.
In growing E. coli cells, the number of RNAP holoenzymes involved in transcription is 1300 molecules per cell. There are also other 700 molecules per cell as free RNAP that has to be bound to any σ factor but is not involved in transcription [3]. Therefore, total RNAP is a limited resource. When a RNAP holoenzyme finishes the transcription of a given transcriptional unit, the σ factor dissociates from the RNAP core, and then other σ factors are able to compete for binding to RNAP core. The new RNAP holoenzyme incorporates to the pool of free RNAP [3]. Moreover, while RNAP core is not able to selectively bind to any σ factor, different σ factors have different affinities for RNAP core. As a consequence, the concentration of alternate σ factors requires strict regulation in order to properly control the delicate interplay favoring the competence for the scarce RNAP core. The concentration of σ factors is partially controlled by two types of protein-protein interaction regulatory elements: 1) anti-σ factors, which bind to and inhibit their cognate σ factor, and 2) adaptor proteins, which bind to their cognate σ factor and proteolyzed it. From a systems-level viewpoint, despite their mechanistic differences both are mechanisms of protein sequestration that have the same regulatory effect of removing the active σ factor from the cytoplasm, allowing the reprogramming of the basal transcriptional machinery towards a different set of target genes. Thus, for pragmatic reasons and taking an integrative approach, both regulatory elements will be called anti-σ factors in this review. Recently, it has been demonstrated that protein sequestration potentially can elicits an ultrasensitive response with a high apparent Hill-coefficient contrary to a traditional graded response [4]. In the context of the interaction between σ factors and their cognate anti-σ factors, this ultrasensitive response could enable an all-or-none response that drastically alters global gene expression. Moreover, the presence of feedback circuits enables the robust adaptability via a homeostatic response of E. coli to sudden environmental changes [1].
Since the great French scientist Claude Bernard (1813-1878), founder of modern experimental physiology, considered the stability of the miliéu interieur (internal environment), homeostasis mediated by feedback has been confirmed as one of the fundamental and ubiquitous mechanisms underlying life. One hundred years after Bernard’s seminal idea, cybernetics was developed by Norbert Wiener through formalizing the concept of feedback and mathematically demonstrating that negative feedback is a necessary condition to give rise to a teleological (goal-based) behavior in any system. In this control scheme, robust adaptation is created by feeding the output back into the input of the system, enabling it to respond to minimal deviations from the goal [5, 6]. This principle can be found not only in any man-made device that must maintain an internal steady state according to a design, from cisterns to electronic devices, but also in biological systems to allow robust internal stability under changing conditions [7]. A delicate balance exists between fragility and robustness in feedback control circuits [6]. The robustness of the final steady state (i.e., the return to the output value existing before perturbation) in response to a stimulus, such as heat shock, will arise at the price of a delicate or highly susceptible response in the dynamics of the circuit when a deviation from the steady state is sensed [7].
Molecular studies have determined specific roles for products of individual genes, but genomics has shown the decline of the traditional roles that have been assigned to proteins, such as σ factors and regulatory proteins. Despite their different regulatory mechanisms both proteins exerttranscriptional control in terms of their causal effects and both are important to the understanding of the regulatory hierarchy governing a bacterium [8]. From the perspective of systems biology, research has moved from individual genes to the regulation of systems. Although there have been a number of excellent studies on each of these σ/anti-σ post-transcriptional regulation systems [9-16], no comprehensive comparison of these mechanisms has been conducted in a single model organism. Certainly, regulatory proteins, σ factors, anti-σ factors, cofactor metabolites, and sRNAs, among other factors, shape a multi-layered complex regulatory network whose global understanding is a current challenge for systems biology. Both σ factors and anti-σ factors form one of these layers. Therefore, in this review we analyzed two issues from an integrative perspective: 1) the differences between the cellular components controlling the expression and function of σ factors and 2) the common principles governing σ/anti-σ post-transcriptional control in E. coli.
ANTI-σ REGULATION
Adequately orchestrated gene expression is necessary for the successful adaptation and survival of cells. A strategy in bacteria to control gene expression is to displace the housekeeping σ subunit of the RNAP and replace it with an alternative σ factor. Then, with the new σ, the RNAP holoenzyme, can recognize and initiate transcription for a different type of promoter. Consequently, genes not being expressed can be transcribed drastically reprogramming global gene expression and enabling a proper response to an environmental cue [17]. To accomplish this, the activity of σ factors is mainly determined by both their intracellular levels and the activity of proteins called anti-σ factors. The latter binds to their cognate σ factor, blocking the formation of the transcription complex in the presence of a certain environmental cue [18]. This avoids the transcription of unnecessary genes in the new condition. Anti-σ factors regulate diverse processes including the stress response, flagellar biosynthesis, and enzymatic modification [19]. There are two classes of anti-σ factors: 1) the cytoplasmic anti-σ factors, and 2) the inner-membrane-bound anti-σ factors, also known as extra-cytoplasmic function (ECF) anti-σ factors [20]. The first class includes Rsd, HscC, DnaK, RssB, and FlgM and the second class contains RseA and FecR. Interestingly, all the anti-σ factors in E. coli are transcribed by the σ factor that they repress, creating a negative feedback circuit between the σ factor and its corresponding anti-σ.
REGULATION OF σ70 BY Rsd AND HscC
The transcription initiation of genes in E. coli required for growth under optimum conditions is ensured by the σ70 factor. σ70 has two anti-σ factors, Rsd and HscC, which disable σ70-dependent transcription at the onset of stationary phase and also possibly heat shock, respectively. The latter is suggested by in silico evidence [21], so further research is needed to validate this possibility.
The transcriptional control of rsd has been validated in vitro [22]. It is transcribed from two promoters, P1 and P2, which are dependent on σ38 and σ70, respectively. In the promoter region of hscC two promoters, σ70 and σ32, have been proposed by in silico methods [21]. For graphical schemes of the organization and genomic context of genes coding for σ factors and anti-σ factors here discussed the reader is referred to (Supplementary Fig. 1 (219KB, pdf) ).
In stationary phase, the putative anti-σ70 factor Rsd binds to the region 4 helix-turn-helix motif of σ70, which is responsible for -35 promoter recognition, blocking association with the RNAP core [17]. This also contributes to the preferential use of σ38, which is the σ factor used upon entry into stationary phase [17, 19, 20, 23-32]. Reminiscent of Rsd, other E. coli anti-σ factors (i.e., FlgM, RseA, FecR, DnaK and RssB) also bind to multiple regions of their cognate σ factors to prevent the holoenzyme formation [33].
In contrast, HscC forms a complex with σ70 and may function as a negative modulator of housekeeping gene expression. This anti-σ is highly homologous to DnaK and HscA [34], two members of the Hsp70 chaperone family proteins in E. coli. The overexpression of hscC causes severe inhibition of cell growth. Primer extension and beta-galactosidase assays have shown that overexpression of HscC reduces σ70-dependent promoter activity. An in vitro transcription assay revealed that HscC inhibits σ70-dependent transcription of the groESL promoter. In addition, co-purification analysis showed that σ70 co-eluted with HscC [34]. HscC possesses the typical structural features of an Hsp70 protein, such as an ATPase domain, a substrate-binding domain, and a C-terminal domain [34]. However, rather than the substrate-binding domain, the ATPase domain of HscC contributes to its functional specificity for σ70, as suggested by the fact that chimeric proteins carrying the ATPase domain of HscC combined with other domains of either HscC or DnaK form complexes with σ70 [34]. Promoter mutation analyses with wild-type and σ38 mutant backgrounds have suggested that P2 is the primary promoter for the transcription of rsd [35]. However, the contribution of σ38 to P1 expression appears to be limited to a short period during the transition from exponential growth to stationary phase, with a maximal relative Rsd intracellular level that is only 20% of the σ70 level [36, 37]. Another report proposes that the levels of both Rsd and σ70 molecules in E. coli during stationary phase are similar, and that their association is significantly higher [33]. The mutation or deletion of Rsd did not reduce growth or alter the transcript levels of E. coli during stationary phase [37, 38]. Nevertheless, Rsd overexpression or a σ70 highly affine Rsd mutant resulted in the up-regulation of several genes transcribed by σ38, indicating that Rsd enables the redistribution of RNAP to σ38-specific promoters by sequestering σ70 [38]. These observations suggest that the main function of Rsd is to promote σ38 transcription. In fact, the transcription mediated by other alternative σ factors, such as σ24, σ32 and σ54, was also favored by Rsd overexpression [37-41]. Future studies are thus likely to focus on how the Rsd-binding activity is controlled in the exponential and stationary phases [35]. Although rsd transcription is not initiated primarily by σ70 but also by σ38, the fact that σ38 is only transcribed by σ70 allows the formation of an indirect negative feedback circuit between σ70 and Rsd that could contribute to its regulation, which supports cellular homeostasis (Fig. 2).
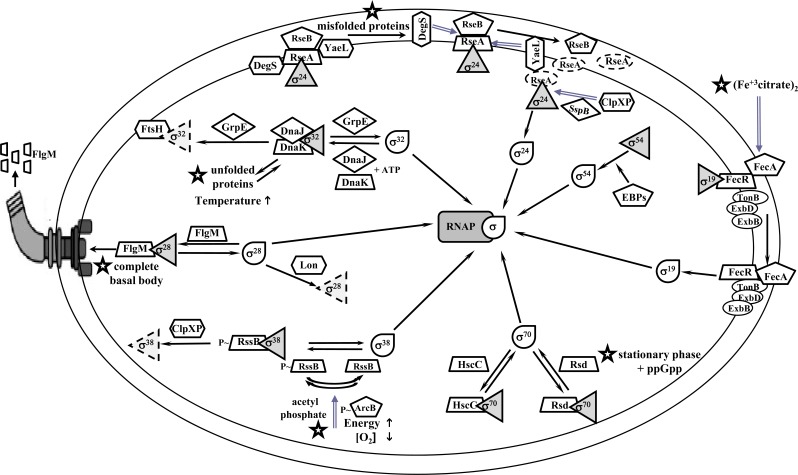
Fig. (2).
Regulation of σ factors by anti-σ factors in E. coli. The forms in figure represent: tear (σ factor active form), solid triangle (σ factor inactive form), dashed triangle (σ factor degraded form), star (environmental signal that releases the σ factor), trapezoid (anti-σ factor), pentagon (anti-σ factor sensor or modulator), solid ellipse (transducer signal complex), hexagon (protease), rhombus (chaperone), dashed ellipse (any protein in degraded form), rounded rectangle (RNAP core).
REGULATION OF σ38 BY RssB
The σ38 factor is the master regulator of the general stress response, transcribing more than 140 genes that confer resistance against adverse conditions, such as stationary phase, oxidative stress, UV radiation, heat shock, hyper-osmolality, acidic pH, and ethanol [23].
In evolutionary terms, this σ factor is closely related to σ70 and therefore does not have a full unique consensus sequence. Specific promoter selectivity is achieved by a broader tolerance for deviations from 1) the optimal promoter sequence, or 2) the exact geometric alignment of the -35 and -10 regions requiring an optimal spacer length of 17 bp [42]. Additionally, trans-acting regulatory factors, such as regulatory proteins or regulatory RNAs [24], that bind to a promoter region contribute to RNAP holoenzyme selectivity. However, the cellular levels of σ38 are believed to reach a maximum of one-third of those of σ70 [22, 25]. Given all these in addition to the lower affinity of σ38 for the RNAP core [3, 18, 28-31, 43], mechanisms ensuring that σ38 can recruit enough RNAP cores to carry out its transcriptional program are needed.
RssB (also named SprE) is a response regulator that functions as an anti-σ factor adaptor and plays a critical role in the control of cellular σ38 levels in E. coli. An rssB mutant has near constitutively high levels of σ38 but is impaired in the post-transcriptional regulation of σ38 [44]. Two promoters transcribe rssB: the first, P1, is a distal promoter transcribed by σ38, which also co-transcribes rssA, and the second, P2, is situated in the rssA-rssB intergenic region from which rssB is transcribed in a σ38-dependent fashion [45].
The proteolysis of σ38 depends on three factors: the ClpXP protease, RssB, and a “turnover element”. The interface with RssB is the lysine-173 of the σ38 amino acid sequence. Binding of RssB to σ38 is stimulated by phosphorylation of the RssB receiver domain, suggesting that environmental stress affects σ38 proteolysis by modulating RssB affinity for σ38 [46].
Therefore, the cellular levels of σ38 are regulated at different points (transcription, translation, and proteolysis) by RssB and the two-component system ArcB/ArcA in a "three-component system" that coordinates rpoS transcription and σ38 proteolysis and, thereby, maintains low σ38 levels in cells growing exponentially. In this system, it has been suggested that the sensor histidine kinase ArcB monitors both the oxygen and energy supplies [47]. In addition, this histidine-kinase phosphorylates ArcA (their cognate two-component transcriptional repressor regulator for RpoS) and RssB (a factor targeting σ38 for proteolysis) [47]. Both phosphorylated ArcA and RssB keep σ38 levels low and are limiting factors in σ38 proteolysis. Acetyl phosphate is an accessory component that contributes to RssB and, possibly, ArcA phosphorylation [48]. ClpXP is a protease that requires the exposure of σ38 to phosphorylate RssB [46, 49]. Given that σ38 is protected against degradation when it is bound to the RNAP core, its concentration affects its own rate of degradation [47]. Once the cell enters in stationary phase or some other stress conditions are present, σ38concentration is increased above that of non-phosphorylated RssB, thus allowing σ38 to bind to the RNAP core and transcribe its target genes [46]. In this case, the negative feedback circuit is formed directly between σ38 and RssB because the first factor transcribes the second, assuring the production of the anti-σ RssB and the control of σ38 when the environmental stress that triggered its liberation disappears (Fig. 2).
REGULATION OF σ32 BY DnaK
A temperature of 37°C or higher compromises E. coli homeostasis fundamentally due to thermal denaturation of folded proteins. In reaction to an upshift in temperature, E. coli employs the heat shock response to increase the activity of transcription directed by σ32 factor, inducing the up-regulation of over 120 products of this sigmulon. Some proteins produced under heat-shock conditions act as chaperones, such as GroEL and DnaK, which prevent protein turnover by maintaining proper folding, and proteases, such as Lon and Clp, which facilitate protein turnover (Fig. 2).
The dnaK gene is co-transcribed with dnaJ from three heat-inducible promoters, which are recognized in vitro by a RNAP containing σ32 but not by a RNAP in complex with σ70 [50]. An intracellular balance of molecular concentrations of chaperones in response to environmental conditions has significance in cellular homeostasis. The overexpression of dnaK does not have a deleterious effect on the chaperone system function, cell growth, survival and morphology [51], and also confers freeze tolerance [52].
σ32, the σ factor responsible for responding to heat shock, is maintained at low concentrations during exponential cell growth [53]. During optimal growth conditions, DnaJ (Hsp40 homolog) binds to σ32 (in the presence of ATP), presenting it to the ATP-bound DnaK (Hsp70 homolog). In this way, DnaK competes with the RNAP core to bind σ32. The complex of DnaJ-σ32-DnaK-ADP is formed after ATP hydrolysis. Then, the chaperone GrpE presents σ32 to the free FtsH protease for degradation [19, 54]. Under stress conditions, cytoplasmic proteins become denatured, acting as substrates for the DnaJ/DnaK/GrpE chaperones complex, releasing active σ32 and allowing the transcription of heat shock genes [30, 55]. This free σ32 can re-associate with the RNAP core to transcribe σ32 dependent promoters. As for σ38, σ32also takes part in the transcription of the components of its anti-σ system, ensuring that it is repressed when the environmental condition inducing its liberation (i.e., any condition responsible for cytoplasmic misfolded proteins) disappears (Fig. 2).
REGULATION OF σ28 BY FlgM
The flagellar genes of E. coli are organized into a regulatory hierarchy involving three classes of promoters, which groups genes according to their corresponding hierarchical level: early, middle and late. These classes of promoters are differentially expressed just prior the flagellum assembly in discrete intervals of the cell cycle.
σ28 activates the late genes involved in flagellum assembly. σ28 is initially expressed by an FlhDC-controlled class 2 promoter and is responsible for class 3 transcription of the flagellar regulon. In this stage, the proteins composing the flagellar filament, motility control and chemotaxis are expressed. In the late transcription hierarchical class 3, σ28 expression is also increased by transcription at an additional class 3 promoter [56, 57]. In contrast, the anti-σ gene flgM is transcribed from two promoters. The P1 promoter, which is regulated by σ70, is situated in the flgA-flgB intergenic region, and flgM is co-transcribed with flgA and flgN. The second promoter, P2, is situated upstream of flgM and is transcribed in a σ28-dependent fashion [57].
To avoid the participation of σ28 during assembly of the basal body of the flagellum, σ28activity needs to be inhibited. This process is carried out by the anti-σ factor FlgM [58]. In this manner, FlgM inhibits the σ28-dependent transcription of genes whose products are needed late in assembly, when the flagellar basal motor structure, the hook-basal body (HBB), is constructed [59]. With the assembly of the HBB, a type III secretion system is formed that is capable of secreting both the flagellar filament subunits and, interestingly, FlgM. This constitutes the other function of σ28, which facilitates the secretion of FlgM through the HBB, acting as an FlgM type III secretion chaperone. This results in the release of σ28, which allows σ28 to bind to a RNAP core and initiate transcription of the structural class 3 genes necessary to complete the flagellum [59].
Free σ28 is a proteolysis substrate just like other σ factors. The Lon protease is responsible for σ28 proteolysis. When σ28 is in excess, its proteolysis increases the robustness and precision of this complex control system by rapidly reestablishing a 1:1 stoichiometry between σ28 and FlgM. This contributes to limiting the up-regulation of flagellar biosynthesis, and FlgM has a protective function against σ28 degradation [60]. When the flagellar filament is assembled completely, the anti-σ FlgM produced from the transcription regulated by σ28 increases and then sequesters σ28 to suppress its function and proteolysis, forming a negative feedback regulatory circuit (Fig. 2).
REGULATION OF σ24 BY RseA
Under certain stresses (e.g., heat or osmotic shock) misfolded envelope proteins are generated. As a consequence, bacteria such as E. coli have developed signaling pathways that sense and respond to these envelope stresses. These pathways trigger the expression of genes encoding a wide variety of proteins that are able to repair envelope (e.g., periplasmic folding chaperons, proteases, and biosynthesis enzymes for lipid A). These pathways are known as extracytoplasmic (EC) or envelope stress responses. One of the EC systems monitoring envelope stress is the σ24 sigmulon, where σ factor is induced when the stress damages the periplasmic proteins. This is the only known EC response that is essential for viability in E. coli K-12 [61], whose misfolded-proteins in the cell envelope direct the expression of genes to restore envelope integrity [62].
The EC stress signal is transduced across the inner membrane to the cytoplasm via the inner membrane protein RseA, which is the anti-σ factor responsible for inhibiting σ24, and it is also known as an EC anti-σ factor. In general, the EC anti-σ factors are inner membrane proteins with at least one transmembrane domain. Additionally, it has been observed that the cytoplasmic N-terminal domain of EC anti-σ factors is conserved and possibly interacts directly with their cognate σ factor. On the other hand, little homology is observed in their transmembrane C-terminal domains (except between E. coli RseA and H. influenzae MclA), which is consistent with the possibility that these anti-σ factors may respond to different extracytoplasmic signals [20].
The rseA gene, which encodes the anti-σ24 factor, is transcribed from three promoters. Two of these promoters, P1 and P2, are located upstream the gene rpoE; while the third promoter, P3, is located inside the coding region of rpoE. No obvious σ70 consensus promoter sequence is found upstream of promoter P1 [63]. However, promoters P2 and P3 have the traditional conserved motifs sequences of σ24-dependent transcription regulation [64]. Therefore, the levels of σ24 factor partially depend on the levels of its cognate anti-σ factor and vice-versa.
RseA anchors to the inner membrane (single pass) and contains a binding site for σ24. Under unstressed conditions, biochemical analyses and crystallographic structures have shown that the periplasmic domain of RseA interacts with RseB (protecting RseA from proteolysis), while the RseA cytoplasmic domain is sandwiched between two domains of σ24. This interaction occludes the two main binding regions of σ24 for the RNAP core, repressing σ24 activity because it is sequestered at the inner membrane by the anti-σ factor RseA [65].
During envelope stress, which interferes with the folding of outer membrane proteins (OMPs) and includes heat shock, overexpression of OMP genes, and mutations in genes encoding the chaperones required for OMPs folding, E. coli employs a proteolytic cascade to transduce signals related to protein folding in the periplasm to the transcriptional apparatus in the cytoplasm. RseA is degraded in response to misfolded OMPs by the sequential action of two inner membrane proteases, DegS and YaeL (RseP). The C-termini of misfolded OMPs bind to the PDZ domains (a common structural domain found in cellular signaling proteins) of the trimeric DegS protease, activating cleavage of the periplasmic domain of RseA [66, 67]. The partially degraded RseA is now a substrate for YaeL. YaeL cleaves RseA further, releasing the cytoplasmic domain of RseA bound to σ24. The cascade activates the cytoplasmic ClpXP protease and SspB chaperone, which degrades the remaining domain of RseA in the final step of the proteolytic cascade, thus allowing the release of σ24 [68, 69]. Then, σ24 binds to the RNAP core and is ready to direct transcription of its sigmulon to cope with the environmental condition produced by the envelope stress [70]. The regulatory nucleotide ppGpp, which mediates the stringent response, and the RNAP-binding transcription protein DksA together activate σ24-dependent transcription, once σ24 is released by RseA, through subverting the dominant housekeeping σ70. In fact, σ24 activity is induced upon entry into stationary phase in E. coli by the alarmone ppGpp and not via the RseA-dependent stress signaling pathway [71]. When periplasmic stress disappears, the σ24-RseA complex is restored by the RseA transcription for σ24, which replaces the anti-σ RseA that was degraded by proteolysis (Fig. 2).
REGULATION OF σ19 BY FecR
The other EC signaling pathway is the σ19 (FecI) sigmulon of E. coli. Two signals are necessary to activate the expression of fecI: iron starvation and the presence of ferric-citrate in the environment. Whereas iron starvation is sensed by the Fur protein, iron citrate is bound by the FecA receptor in the outer membrane [72].
A σ19 deletion mutation is totally devoid of ferric citrate transport system expression because expression of fecIR mRNA requires σ70 and the transcription of the fecABCDE transport genes is regulated by σ19, which responds to ferric citrate via the anti-σ system FecA-FecR. fecI and fecR co-transcription is inhibited by the iron-loaded Fur repressor, which in turn transcribes the fec transport genes at a low level [73].
Ferric citrate serves as an iron source for E. coli [74]; however, the transport of this ferric chelator into the periplasm requires energy and a functional complex of three inner membrane proteins: TonB, ExbB, and ExbD. A physical interaction between the periplasmic portion of TonB and FecA results in the transduction of energy from the proton gradient across the inner membrane [72, 75]. This energetic cost may be responsible for the low position of ferric citrate in the hierarchy of iron sources.
FecA (an outer membrane protein), which recognizes ferric citrate, serves a dual role: It is 1) an iron transporter of ferric citrate across the outer membrane that operates with the fec transport gene operon, and 2) an anti-σ sensor that regulates the transcription of the ferric citrate transport genes fecABCDE in the cytoplasm. Both processes are independent and require energy transduction by the inner membrane TonB-ExbB-ExbD complex [75].
FecA, the anti-σ sensor, elicits a signaling cascade from the cell surface to the cytoplasm [29]. This signaling flux involves a series of conformational changes of the proteins FecA (anti-σ sensor), FecR (anti-σ factor), and FecI (EC σ factor), respectively.
It has been proposed that σ19 binds to its anti-σ factor, FecR; thus, FecR sequesters σ19 until FecR is inactivated by undergoing a structural change in responseto the induction signal of FecA (iron-citrate).
Activated FecA interacts in the periplasmic space with the C-terminus of FecR (anchored in the cytoplasmic membrane). Then, the signal is transferred across the cytoplasmic membrane into the cytoplasm via FecR N-terminus [76]. Thus, FecR releases the σ19 factor, allowing activation of the σ19 factor that directs the RNAP core to induce transcription of the fecABCDE transport genes [77]. σ19 contributes to the transcription of the anti-σ sensor FecA. When ferric citrate becomes exhausted or a different iron source becomes available, FecR is reverted to its prior conformation by FecA, and FecR becomes able to sequester σ19 thus inhibiting the transcription of its corresponding genes (Fig. 2).
DOES σ54 HAVE AN ANTI-σ FACTOR?
The second family of σ factors in E. coli is exclusively represented by σ54, which directs the recognition of distinct promoter motifs located at positions -24 and -12 relative to the transcriptional start site. Although initially identified for their role in nitrogen assimilation, σ54 is utilized in different physiological processes and can remedy certain conditions that make nitrogen assimilation very difficult [78]. σ54 does not have primary sequence similarity to σ70 proteins and regulates transcription by a different mechanism. σ54 can not spontaneously isomerize (melt) DNA to form open promoter complexes. This step of transcription strictly depends on mechanotranscriptional activators (also known as bacterial enhancer binding proteins, or bEBPs) that utilize ATP hydrolysis to drive conformational changes for this transition [79]. Twelve bEBPs have been described in E. coli [28, 78, 79]. Apparently, σ54 factor does not have an anti-σ factor. The σ54 impairing to transcribe genes without the help of σ54-dependent transcriptional activators (bEBPs) may explain the ostensible absence of an anti-σ factor.
SYSTEMS-LEVEL LESSONS FROM σ/ANTI-σ CIRCUITS
Biological systems are, both intrinsically and extrinsically, noisy. Therefore, a mechanism enabling the cell to maintain a stable internal state is required. For maintain homeostasis, organisms need to detected changes in their environment and respond accordingly in a variety of manners. Negative feedback circuits have evolved in organisms as a common tool for maintaining homeostasis and filtering noise, thus providing a robust response. A feedback mechanism occurs when an organism senses an internal or external environmental perturbation and changes the level or activity of a substance, thus starting a regulatory cascade that eventually reverses the initial perturbation. While negative feedback circuits are ubiquitous in biological regulation, certain design principles emerge as lessons from the evolution of feedback in the anti-σ control.
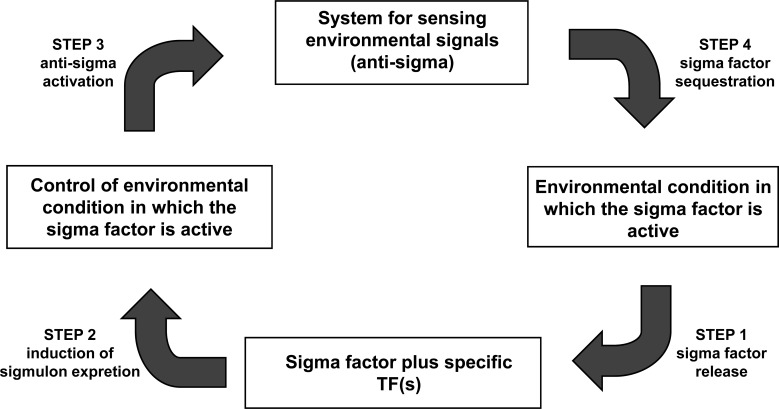
Through this review, we surveyed the different regulatory systems formed by the σ and anti-σ factors in E. coli. Following an integrative approach, we were not only interested in describing their biological differences, but also their systems-level similarities. (Fig. 2) shows the intricate cellular complexity of the σ/anti-σ circuitry. Despite this complexity, a small set of common mechanisms could be outlined. First, the anti-σ factor releases its cognate σ factor in response to a specific environmental signal (step 1, Fig. 3). It has been reported that the latter causes a rapid increase in the cognate σ factor concentration [1-3], supporting the ultrasensitive response discussed in the Introduction. Second, the σ factor and other transcriptional factors participate, directly or indirectly, in the transcription of their correspondent anti-σ factor (step 2, Fig. 3). Third, the anti-σ factor plays one of two different roles: 1) it is kept inactive by other factors or adaptors (antagonistically by anti-anti-σ factorsor anti-adaptors, and cooperatively by co-anti-σ factors, or post-translationally by proteolysis) during the presence of the specific environmental signal, or 2) it participates actively in the response to the environmental signal (step 3, Fig. 3). Finally, when this signal disappears, anti-σ factors efficiently repress their σ factors by sequestration (step 4, Fig. 3). The sequestered σ factor has two possible fates: it can be protected against proteolysis or proteolyzed. The negative feedback circuits generated by these conditions in all alternative σ factors (except σ54) and their cognate anti-σ factors interactions enable the cell to return to a stable condition similar to that before the environmental signal. An interesting exception to these principles is the housekeeping σ70 factor. As all the alternative σ factors, σ70 also transcribes its anti-σ factors (rsd and hscC). Nevertheless, there are some intriguing differences. Both anti-σ70 factors seem to be also transcribed by alternative σ factors. As previously discussed, the rsd expression has been validated in vitro to be promoted by σ70 and σ38, while the promoter region of hscC has two promoters, σ70 and σ32, which have been proposed in silico. Whether these predictions are functional requires further research. However, this evidence opens the possibility that the σ70 concentration may be indirectly controlled by entry into stationary phase and heat shock, respectively, two conditions requiring alternative σ factors. Whether these are the only environmental signals controlling the availability of σ70 remains an open question.
Fig. (3).
Conditions needed to be fulfilled to generate a negative feedback circuit of σ/anti-σ factors assuring an efficient and robust cellular response.
CONCLUSIONS
Mechanisms for coordinating the regulation of gene expression are hierarchic and complex. This coordination appears to be critical for the maintenance of a balanced gene expression profile during various environmental conditions. This is achieved by two different but complementary mechanisms. On the one hand, transcriptional regulatory protein compete with many other regulators to redirect the RNAP to transcribe a set of genes. On the other hand, σ factors show a rapid increase in their concentration and then compete with a small number of other σ factors for the limited RNAP core. The scarce RNAP core is a key aspect for the successful functioning of this regulatory mechanism through competence. Then, by activating alternative σ factors, gene expression can be efficiently and globally reprogrammed to respond to sudden environmental cues. As a consequence, the specificity that σ factors confer to the RNAP for certain promoters allows the circumvention of the transcriptional regulatory proteins currently active in a previous growth condition enabling a specific and efficient response to the new condition.
The availability of E. coli σ factors is regulated at multiple levels by specific signal transduction pathways. These pathways are activated in response to specific stresses or environmental conditions. Several interactions play a prominent role in this regulatory process. These interactions operate synergistically and shape complex regulatory networks and cascades, constituting the regulatory mechanism establishing the gene expression profile. Thus, the cell can have a wide range of gene expression profiles that allow cells to cope with different requirements, such as sudden environmental changes or developmental programs. Anti-σ systems evolved in E. coli as a hierarchy of negative feedback interactions, allowing the cell to control the gene expression in response to specific environmental conditions in a way resembling compartmentalization by partitioning the gene expression states into a set of discrete functional units. When these environmental conditions disappear, the system relieves gene expression thereby completing the cycle.
To allow the efficient response of some σ factors, it is important that they are present in the cell at basal levels at all times. This may be achieved by σ70, whose promoter is present in the upstream region of each alternative σ factor (Fig. 1). Nevertheless, σ activity has to be strictly controlled by preventing its binding to the RNAP core. This also have the desired effect of silencing the genes not required under a certain biological context. Anti-σ factors facilitate this requirement by operating as a system that senses the environment and sequesters the σ factor when the gene expression driven by them is superfluous given the biological context.
SUPPLEMENTARY MATERIALS
Supplementary material is available on the publisher’s web site along with the published article.
ACKNOWLEDGEMENTS
This work was supported by the Mexican Council of Science and Technology, CONACYT (Consejo Nacional de Ciencia y Tecnología, URL: http://www.conacyt.mx), grant CB_2012_183795, and by Research Stimulus Fund Program PROMEP (Programa de Mejoramiento del Profesorado) from SEP (Secretaría de Educación Pública de México), grant PROMEP/103.5/11/132. We apologize to all researchers of this field whose original contributions could not be cited due to space limitations.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
REFERENCES
- 1.Österberg S, del Peso-Santos T, Shingler V. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 2011;65:37–55. doi: 10.1146/annurev.micro.112408.134219. [DOI] [PubMed] [Google Scholar]
- 2.Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- 3.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu. Rev. Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 4.Buchler NE, Cross FR. Protein sequestration generates a flexible ultrasensitive response in a genetic network. Mol. Syst. Biol. 2009;5:272. doi: 10.1038/msb.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc. Natl. Acad. Sci. U.S.A. 2000;97(9):4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Csete ME, Doyle JC. Reverse engineering of biological complexity. Science. 2002;295(5560):1664–1669. doi: 10.1126/science.1069981. [DOI] [PubMed] [Google Scholar]
- 7.Goulian M. Robust control in bacterial regulatory circuits. Curr. Opin. Microbiol. 2004;7(2):198–202. doi: 10.1016/j.mib.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 8.Freyre-González JA, Treviño-Quintanilla LG, Valtierra-Gutiérrez IA, Gutiérrez-Ríos RM, Alonso-Pavón JA. Prokaryotic regulatory systems biology: Common principles governing the functional architectures of Bacillus subtilis and Escherichia coli unveiled by the natural decomposition approach. J. Biotechnol. 2012;161(3):278–86. doi: 10.1016/j.jbiotec.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann N, Wurm R, Wagner R. The E. coli anti-sigma factor Rsd: studies on the specificity and regulation of its expression. PLoS One. 2011;6(5):e19235. doi: 10.1371/journal.pone.0019235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piper SE, Mitchell JE, Lee DJ, Busby SJ. A global view of Escherichia coli Rsd protein and its interactions. Mol. Biosyst. 2009;5(12):1943–1947. doi: 10.1039/B904955j. [DOI] [PubMed] [Google Scholar]
- 11.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 2009;160(9):667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 12.Hengge R. The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv. Exp. Med. Biol. 2008;631:40–53. doi: 10.1007/978-0-387-78885-2_4. [DOI] [PubMed] [Google Scholar]
- 13.Arsène F, Tomoyasu T, Bukau B. The heat shock response of Escherichia coli. Int J Food Microbiol. 2000;55(1-3):3–9. doi: 10.1016/s0168-1605(00)00206-3. [DOI] [PubMed] [Google Scholar]
- 14.Barembruch C, Hengge R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 2007;65(1):76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 15.Barchinger SE, Ades SE. Regulated Proteolysis: Control of the Escherichia coli σ(E)-Dependent Cell Envelope Stress Response. Subcell. Biochem. 2013;66:129–160. doi: 10.1007/978-94-007-5940-4_6. [DOI] [PubMed] [Google Scholar]
- 16.Mokdad A, Herrick DZ, Kahn AK, Andrews E, Kim M, Cafiso DS. Ligand-induced structural changes in the Escherichia coli ferric citrate transporter reveal modes for regulating protein-protein interactions. J. Mol. Biol. 2012;423(5):818–830. doi: 10.1016/j.jmb.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lonetto M, Gribskov M, Gross CA. The sigma 70 family: sequence conservation and evolutionary relationships. J. Bacteriol. 1992;174(12):3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Record MT, Reznikoff WS, Craig ML, McQuade KL, Schlax PJ. Escherichia coli RNA polymerase (E_70), promoters, and kinetics of the steps of transcription initiation. In: Neidhardt FC, Curtiss R, editors. Escherichia coliand Salmonella: Cellular and Molecular Biology. Washington D.C: ASM Press; 1996. pp. 792–821. [Google Scholar]
- 19.Hughes KT, Mathee K. The anti-sigma factors. Annu. Rev. Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 20.Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- 21.Huerta AM, Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333(2):261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 22.Jishage M, Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 1999;181(12):3768–76. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klauck E, Typas A, Hengge R. The sigmaS subunit of RNA polymerase as a signal integrator and network master regulator in the general stress response in Escherichia coli. Sci. Prog. 2007;90(Pt 2-3):103–127. doi: 10.3184/003685007X215922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jishage M, Iwata A, Ueda S, Ishihama A. Regulation of RNA polymerase sigma subunit synthesis in Escherichia coli: intracellular levels of four species of sigma subunit under various growth conditions. J. Bacteriol. 1996;178(18):5447–5451. doi: 10.1128/jb.178.18.5447-5451.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dove SL, Hochschild A. Bacterial two-hybrid analysis of interactions between region 4 of the sigma (70) subunit of RNA polymerase and the transcriptional regulators Rsd from Escherichia coli and AlgQ from Pseudomonas aeruginosa. J. Bacteriol. 2001;183:6413–6421. doi: 10.1128/JB.183.21.6413-6421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 2002;66(3):373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jishage M, Ishihama A. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 1997;179(3):959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeda H, Fujita N, Ishihama A. Competition among seven Escherichia coli sigma subunits: relative binding affinities to the core RNA polymerase. Nucleic Acids Res. 2000;28(18):3497–3503. doi: 10.1093/nar/28.18.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahren S, Schnell H, Braun V. Occurrence and regulation of the ferric citrate transport system in Escherichia coli B, Klebsiella pneumoniae, Enterobacter aerogenes, and Photorhabdus luminescens. Arch. Microbiol. 2005;184(4):175–186. doi: 10.1007/s00203-005-0035-y. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20(13):1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patikoglou GA, Westblade LF, Campbell EA, Lamour V, Lane WJ, Darst SA. Crystal structure of the Escherichia coli regulator of sigma70, Rsd, in complex with sigma70 domain 4. J. Mol. Biol. 2007;372(3):649–659. doi: 10.1016/j.jmb.2007.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jishage M, Dasgupta D, Ishihama A. Mapping of the Rsd contact site on the sigma 70 subunit of Escherichia coli RNA polymerase. J. Bacteriol. 2001;183(9):2952–2956. doi: 10.1128/JB.183.9.2952-2956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr. Opin. Microbiol. 2008;11(2):121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arifuzzaman M, Oshima T, Nakade S, Mori H. Characterization of HscC (Hsc62), homologue of Hsp70 in Escherichia coli: over-expression of HscC modulates the activity of housekeeping sigma factor sigma70. Genes Cells. 2002;7(6):553–566. doi: 10.1046/j.1365-2443.2002.00545.x. [DOI] [PubMed] [Google Scholar]
- 35.Piper SE, Mitchell JE, Lee DJ, Busby SJ. A global view of Escherichia coli Rsd protein and its interactions. Mol. Biosyst. 2009;5(12):1943–1947. doi: 10.1039/B904955j. [DOI] [PubMed] [Google Scholar]
- 36.Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase. Proc. Natl. Acad. Sci. U.S.A. 1998;95(9):4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jishage M, Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J. Bacteriol. 1999;181(12):3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell JE, Oshima T, Piper SE, Webster CL, Westblade LF, Karimova G, Ladant D, Kolb A, Hobman JL, Busby SJ, Lee DJ. The Escherichia coli regulator of sigma 70 protein, Rsd, can up-regulate some stress-dependent promoters by sequestering sigma 70. J. Bacteriol. 2007;189(9):3489–3495. doi: 10.1128/JB.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jishage M, Kvint K, Shingler V, Nyström T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16(10):1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurie AD, Bernardo LM, Sze CC, Skarfstad E, Szalewska-Palasz A, Nystrom T, Shingler V. The role of the alarmone (p)ppGpp in sigma N competition for core RNA polymerase. J. Biol. Chem. 2003;278(3):1494–1503. doi: 10.1074/jbc.M209268200. [DOI] [PubMed] [Google Scholar]
- 41.Costanzo A, Nicoloff H, Barchinger SE, Banta AB, Gourse RL, Ades SE. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 2008;67(3):619–632. doi: 10.1111/j.1365-2958.2007.06072.x. [DOI] [PubMed] [Google Scholar]
- 42.Typas A, Hengge R. Role of the spacer between the -35 and -10 regions in sigmas promoter selectivity in Escherichia coli. Mol. Microbiol. 2006;59(3):1037–1051. doi: 10.1111/j.1365-2958.2005.04998.x. [DOI] [PubMed] [Google Scholar]
- 43.Shimada T, Makinoshima H, Ogawa Y, Miki T, Maeda M, Ishihama A. Classification and strength measurement of stationary-phase promoters by use of a newly developed promoter cloning vector. J. Bacteriol. 2004;186(21):7112–7122. doi: 10.1128/JB.186.21.7112-7122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hengge R. Proteolysis of sigmaS (RpoS) and the general stress response in Escherichia coli. Res. Microbiol. 2009;160(9):667–676. doi: 10.1016/j.resmic.2009.08.014. [DOI] [PubMed] [Google Scholar]
- 45.Ruiz N, Peterson CN, Silhavy TJ. RpoS-dependent transcriptional control of sprE: regulatory feedback loop. J. Bacteriol. 2001;183(20):5974–5981. doi: 10.1128/JB.183.20.5974-5981.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becker G, Klauck E, Hengge-Aronis R. Regulation of RpoS proteolysis in Escherichia coli: the response regulator RssB is a recognition factor that interacts with the turnover element in RpoS. Proc. Natl. Acad. Sci. U.S.A. 1999;96(11):6439–6444. doi: 10.1073/pnas.96.11.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mika F, Hengge R. A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of sigmaS (RpoS) in E. coli. Genes Dev. 2005;19(22):2770–2781. doi: 10.1101/gad.353705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouche S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol. Microbiol. 1998;27(4):787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 49.Studemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R. Sequential recognition of two distinct sites in sigma(S) by the proteolytic targeting factor RssB and ClpX. EMBO J. 2003;22(16):4111–4120. doi: 10.1093/emboj/cdg411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cowing DW, Bardwell JC, Craig EA, Woolford C, Hendrix RW, Gross CA. Consensus sequence for Escherichia coli heat shock gene promoters. Proc. Natl. Acad. Sci. U.S.A. 1985;82(9):2679–2683. doi: 10.1073/pnas.82.9.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lobacz B, Wolska KI. Escherichia coli defects caused by DnaK and DnaJ proteins overproduction. Acta Microbiol. Pol. 1997;46(4):393–397. [PubMed] [Google Scholar]
- 52.Chow KC, Tung WL. Overexpression of dnaK/dnaJ and groEL confers freeze tolerance to Escherichia coli. Biochem. Biophys. Res. Commun. 1998;253(2):502–505. doi: 10.1006/bbrc.1998.9766. [DOI] [PubMed] [Google Scholar]
- 53.Straus DB, Walter WA, Gross CA. The heat shock response of E. coli is regulated by changes in the concentration of sigma 32. Nature. 1987;329(6137):348–351. doi: 10.1038/329348a0. [DOI] [PubMed] [Google Scholar]
- 54.Obrist M, Milek S, Klauck E, Hengge R, Narberhaus F. Region 2.1 of the Escherichia coli heat-shock sigma factor RpoH (sigma32) is necessary but not sufficient for degradation by the FtsH protease. Microbiology. 2007;153:2560–2571. doi: 10.1099/mic.0.2007/007047-0. [DOI] [PubMed] [Google Scholar]
- 55.Fredriksson A, Ballesteros M, Dukan S, Nystrom T. Defense against protein carbonylation by DnaK/DnaJ and proteases of the heat shock regulon. J. Bacteriol. 2005;187(12):4207–4213. doi: 10.1128/JB.187.12.4207-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ikebe T, Iyoda S, Kutsukake K. Promoter analysis of the class 2 flagellar operons of Salmonella. Genes Genet. Syst. 1999;74(4):179–183. doi: 10.1266/ggs.74.179. [DOI] [PubMed] [Google Scholar]
- 57.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64(4):694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daughdrill GW, Chadsey MS, Karlinsey E, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat. Struct. Biol. 1997;4(4):285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 59.Aldridge PD, Karlinsey JE, Aldridge C, Birchall C, Thompson D, Yagasaki J, Hughes KT. The flagellar-specific transcription factor, sigma28, is the Type III secretion chaperone for the flagellar-specific anti-sigma28 factor FlgM. Genes Dev. 2006;20(16):2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barembruch C, Hengge R. Cellular levels and activity of the flagellar sigma factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 2007;65(1):76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- 61.De Las Peñas A, Connolly L, Gross CA. SigmaE is an essential sigma factor in Escherichia coli. J. Bacteriol. 1997;179(21):6862–6864. doi: 10.1128/jb.179.21.6862-6864.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chaba R, Grigorova IL, Flynn JM, Baker TA, Gross CA. Design principles of the proteolytic cascade governing the sigmaE-mediated envelope stress response in Escherichia coli: keys to graded, buffered, and rapid signal transduction. Genes Dev. 2007;21(1):124–136. doi: 10.1101/gad.1496707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raina S, Missiakas D, Georgopoulos C. The rpoE gene encoding the sigma E (sigma 24) heat shock sigma factor of Escherichia coli. EMBO J. 1995;14(5):1043–1055. doi: 10.1002/j.1460-2075.1995.tb07085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4(1):e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campbell EA, Tupy JL, Gruber TM, Wang S, Sharp MM, Gross CA, Darst SA. Crystal structure of Escherichia coli sigmaE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell. 2003;11(4):1067–1078. doi: 10.1016/s1097-2765(03)00148-5. [DOI] [PubMed] [Google Scholar]
- 66.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85(7):1067–1076. doi: 10.1016/s0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 67.Sohn J, Grant RA, Sauer RT. Allosteric activation of DegS, a stress sensor PDZ protease. Cell. 2007;131(3):572–583. doi: 10.1016/j.cell.2007.08.044. [DOI] [PubMed] [Google Scholar]
- 68.Grigorova IL, Chaba R, Zhong HJ, Alba BM, Rhodius V, Herman C, Gross CA. Fine-tuning of the Escherichia coli sigmaE envelope stress response relies on multiple mechanisms to inhibit signal-independent proteolysis of the transmembrane anti-sigma factor, RseA. Genes Dev. 2004;18(21):2686–2697. doi: 10.1101/gad.1238604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brooks BE, Buchanan SK. Signaling mechanisms for activation of extracytoplasmic function (ECF) sigma factors. Biochim. Biophys. Acta. 2007;1778(9):1930–1945. doi: 10.1016/j.bbamem.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flynn JM, Levchenko I, Sauer RT, Baker TA. Modulating substrate choice: the SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes Dev. 2004;18(18):2292–2301. doi: 10.1101/gad.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Costanzo A, Ades SE. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3',5'-bispyrophosphate (ppGpp) J. Bacteriol. 2006;188(13):4627–4634. doi: 10.1128/JB.01981-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogierman M, Braun V. Interactions between the outer membrane ferric citrate transporter FecA and TonB: studies of the FecA TonB box. J. Bacteriol. 2003;185(6):1870–1885. doi: 10.1128/JB.185.6.1870-1885.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ochs M, Angerer A, Enz S, Braun V. Surface signaling in transcriptional regulation of the ferric citrate transport system of Escherichia coli: mutational analysis of the alternative sigma factor FecI supports its essential role in fec transport gene transcription. Mol. Gen. Genet. 1996;250(4):455–465. doi: 10.1007/BF02174034. [DOI] [PubMed] [Google Scholar]
- 74.Frost GE, Rosenberg H. The inducible citrate-dependent iron transport system in Escherichia coli K12. Biochim. Biophys. Acta. 1973;330(1):90–101. doi: 10.1016/0005-2736(73)90287-3. [DOI] [PubMed] [Google Scholar]
- 75.Yue WW, Grizot S, Buchanan SK. Structural evidence for iron-free citrate and ferric citrate binding to the TonB-dependent outer membrane transporter FecA. J. Mol. Biol. 2003;332(2):353–368. doi: 10.1016/s0022-2836(03)00855-6. [DOI] [PubMed] [Google Scholar]
- 76.Braun V. Surface signaling: novel transcription initiation mechanism starting from the cell surface. Arch. Microbiol. 1997;167(6):325–331. doi: 10.1007/s002030050451. [DOI] [PubMed] [Google Scholar]
- 77.Harle C, Kim I, Angerer A, Braun V. Signal transfer through three compartments: transcription initiation of the Escherichia coli ferric citrate transport system from the cell surface. EMBO J. 1995;14(7):1430–1438. doi: 10.1002/j.1460-2075.1995.tb07129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reitzer L. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 2003;57:155–176. doi: 10.1146/annurev.micro.57.030502.090820. [DOI] [PubMed] [Google Scholar]
- 79.Shingler V. Signal sensory systems that impact σ54-dependent transcription. FEMS Microbiol. Rev. 2011;35(3):425–440. doi: 10.1111/j.1574-6976.2010.00255.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.