Abstract
Protein interactions play an important role in the discovery of protein functions and pathways in biological processes. This is especially true in case of the diseases caused by the loss of specific protein-protein interactions in the organism. The accuracy of experimental results in finding protein-protein interactions, however, is rather dubious and high throughput experimental results have shown both high false positive beside false negative information for protein interaction. Computational methods have attracted tremendous attention among biologists because of the ability to predict protein-protein interactions and validate the obtained experimental results. In this study, we have reviewed several computational methods for protein-protein interaction prediction as well as describing major databases, which store both predicted and detected protein-protein interactions, and the tools used for analyzing protein interaction networks and improving protein-protein interaction reliability.
Keywords: Protein-protein interaction, Protein interaction networks, Computational prediction method, Machine learning, Networks analyzing tools, Interaction database, Gold standard dataset selection.
1. INTRODUCTION
Protein–protein interactions (PPIs) are of interest in biology because they regulate roughly all cellular processes, including metabolic cycles, DNA transcription and replication, different signaling cascades and many additional processes. Proteins carry out their cellular functions through concerted interactions with other proteins, so it is important to know the specific nature of these relationships. Indeed, the importance of understanding these interactions has prompted the development of various experimental methods used in measuring them. While the amount of genomic sequence information continues to increase exponentially, the annotation of protein sequences appears to be somewhat lagging behind, both in terms of quality and quantity. Multi-pronged, high-throughput functional genomics approaches are needed to bridge the gap between raw sequence information and the relevant biochemical and medical information. Therefore, computational methods are required for discovering interactions that are not accessible to high throughput methods. These computational predictions can then be verified by using more labor-intensive methods. A number of computational approaches for protein interaction discovery have been developed over recent years. These methods differ in feature information used for protein interaction prediction. Many studies have demonstrated that knowing the tools and being familiar with the databases is important for new research in protein-protein interaction analysis to be conducted [1-7].
The scope of this review focuses on describing these resources.
2. DATABASES
Through recent rapid advances in high-throughput technologies, massive protein-protein interaction data of various organisms have become available and are currently stored in several databases. More than 100 PPI related repositories have been published and are available online [8], these databases can be used as the major data for evaluating prediction methods. Many of these PPI data providers are independently funded and do their works in isolation and often contain redundant data from overlapping sets of publications. The issue of the integrating data from PPI disparate repositories began with the efforts of the Human Proteome Organization Proteomics Standards Initiative (HUPO-PSI) and International Molecular Exchange (IMEx) consortium and followed by publishing the ‘minimum information about a molecular interaction experiment’ (MIMIX) guidelines [9]. The HUPO-PSI has developed the PSI-MI XML format to establish a single, unified format for PPI data. Additionally, a simplified tabular format, MITAB has been developed [10]. The IMEx is an international collaboration between a group of major public interaction data providers who have agreed to share literature-curation efforts and make a nonredundant set of PPI available in a single search interface on a common website (http://www.imexconsortium.org/) [8]. IMEx defines three types of membership: Active: IMEx partner commits to producing relevant numbers of records curated to IMEx standard and providing these via a Proteomics Standards Initiative common query interface (PSICQUIC) service. Observer: Prospective IMEx consortium member. Inactive: former IMEx partner that contributed to the establishment of the IMEx curation rules. The PSICQUIC is a web service aimed at enabling users to access multiple interaction databases with a single query and standardizing the programmatic access to molecular interaction databases [11].
(Supplementary table S1 (264.3KB, pdf) ) shows almost all active databases on the PPI, but in the following we focus on the most popular repository (which has more than 1000 citations according to Google scolar at the time of writing this manuscript, May 2013) in more detail, (see Table 1) for feature by feature comparesion of these databases.
Table 1.
The Most Popular Repository (Which Have More Than 1000 Citations According to Google Scolar at the Time of Writing this Manuscript, May 2013) on the PPI, for Detailed Description About These Repository Refer to the Text
| Acronym | Organisms | Number of Interactions | Curated to IMEx/MIMIx standards | IMEx Partner | Accept submission in PSI-MI format | Availability | PSICQUIC service | Last Content Update | URL | References |
|---|---|---|---|---|---|---|---|---|---|---|
| BioGrid (IMEx partner) |
Model organisms | 279409 | IMEx | Yes (Observer) | No | Free to academic users | Yes | 2013 | http:// www.thebiogrid.org | [30-32] |
| BIND/BOND (IMEx partner) |
H. sapiens, S.cerevisiae, M. musculus, H. pylori | 198,905 | No | Yes (Inactiv) | Yes (submission by email) | Free to all users | Yes | 2004 | http://download.baderlab.org/BINDTranslation/ | [21, 33-35] |
| DIP/ LiveDIP (IMEx partner) |
Model organisms | 73268 | IMEx | Yes (Active) |
Yes (submission by email) | Free to academic users | Yes | 2005 | http://dip.doe-mbi.ucla.edu | [36-40] |
| HPRD | Human | 30,047 | No | No | Yes (submission by email) | Free to academic users | No | 2009 | http:// www.hprd.org | [24-26, 41] |
| IntAct (IMEx partner) |
Model organisms | 290,891 | IMEx/MIMIx | Yes (Active) |
Yes (submission by web-based tool) | Free to all users | Yes | 2011 | http:// www.ebi.ac.uk/intact/ | [28, 29, 42, 43] |
| MINT (IMEx partner) |
Model organisms | 241458 | IMEx | Yes (Active) |
Yes (submission by email) | Free to all users | Yes | 2011 | http://mint.bio.uniroma2.it/mint | [22, 23, 43, 44] |
BioGRID [12, 13], the General Repository for Interaction Database, is one of the most comprehensive databases of experimentally determined protein-protein interactions. It has continuously been updating the source of protein and genetic interactions from major model organisms (by the time of writing this manuscript, Feb-2012, contains 27 different organisms) compiled through comprehensive curation efforts, it comprises more than 460000 interactions and all interaction data are freely available for download in a wide variety of standardized formats, more over, this repository supplies information about the experimental methods used for interaction detection. This database does not contain information about multi-protein complexes larger than dimers and lists any interaction as pairwise interactions.
The Database of Interacting Proteins (DIPTM)[14] developed at the University of California, Los Angeles has combined data from a variety of sources to create a single, consistent set of PPI. In addition to the primary sources, DIP drives its data from a number of other databases such as Yeast Protein Database (YPD) [15], EcoCyc [16], and FlyNet [17], Kyoto Encyclopedia of Genes and Genomes (KEGG)[18]. The complete DIP dataset are freely available for download as well as specialized DIP subsets and additional data (free registration required), the database contains more than 460 organisms.
The Biomolecular Interaction Network Database (BIND) [19, 20], is a component of BOND (the Biomolecular Object Network Databank). This repository was created at the University of Toronto. It contains more than 200,000 interactions of more than 1500 organisms and it holds a large variety of interaction data including those curated by a team of curators. Although, the majority of BIND is the protein interactions data, BIND also contains many other types of interactions involving RNA, DNA, genes, complexes and small molecules. Although BIND curation stopped in 2005, BIND still remains a highly cited publicly available interaction database, because the BIND data is not available in a standard format from the official source, recently [21] a translation of BIND in the Proteomics Standard Initiative-MI (PSI-MI) 2.0 format was publicly available which makes the BIND data compatible with current software tools.
The Molecular Interaction Database (MINT) [22, 23] developed by the University of Rome Tor Vergata, interaction data and various experimental details are mined from published literature by using a literature-mining program, the MINT assistant, then expert curators establish the putative interactions. Currently MINT contains more than 230,000 interactions and more than 34,000 proteins and focused on the model organisms, this database provides confidence scores for experimentally detected PPIs, which show the reliability of the interactions. MINT is an active partner of IMEx and shares curation efforts and supports the Protein Standard Initiative (PSI) recommendation.
The Human Protein Reference Database (HPRD [24-26]) was built as a cooperative effort between Johns Hopkins University and the Institute of Bioinformatics, this resource provides a collection of human protein-protein interaction. Data are manually extracted from the literature, and each record is linked to a detailed piece of information including post-translational modifications, disease associations via OMIM for each protein in the human proteome, subcellular localizations, enzyme–substrate relationships, protein isoforms and domain architectures. This database currently contains more than 30,000 proteins and more than 39,000 protein-protein interactions.
IntAct [27-29] is a molecular interaction database that its data come from the literature or from direct data depositions, IntAct source code and data are freely available for download. Currently this resource contains more than 60,000 proteins and more than 290,000 binary interaction evidences abstracted from more than 5000 scientific publications. IntAct is an active partner of the IMEx consortium, and the majority of its protein-protein interaction data is annotated to IMEx standards. In addition to protein-protein interaction data, IntAct also includes information on DNA, RNA, and small-molecule interactions.
3. COMPUTATIONAL METHODS FOR PROTEIN-PROTEIN INTERACTION PREDICTION
In general, the available methods for predicting protein-protein interaction can be divided into four main categories: methods based on genomic context and structural information, methods that use network topology to predict protein-protein interaction, methods that detect protein-protein interaction by using text mining and literature mining (or database search) and, finally, methods based on machine learning algorithms utilizing heterogeneous genomic/proteomic features (see Table 2 for a general overview). In the following section we describe each of these methods and their application.
Table 2.
A General Overview of Computational Methods for Protein-Protein Interaction Prediction with Their References, for Description About Methods Refer to the Text
| Method | Features | References | |
|---|---|---|---|
| Methods based on genomic context and structure information | Gene fusion |
|
[54, 55] |
| Gene neighboring |
|
[46, 47, 123] | |
| Phylogenetic similarity |
|
[50-52, 124-126] | |
| Sequence and primary structure |
|
[65-74] | |
| Structure based |
|
[56-64, 127-129] | |
| Methods based on machine learning algorithms with utilizing multiple genomic/proteomic features | Decision tree and random forest |
|
[73, 121, 122, 130, 131] |
| KNN |
|
[118]. | |
| MLP |
|
[110, 111] | |
| Naive Bays |
|
[71, 74, 115-117] | |
| SVM |
|
[70, 104-106] | |
| Other methods | Using network topology for predicting protein-protein interaction |
|
[78-85] |
| Text mining methods |
|
[90-97, 99, 100] | |
3.1. Methods Based on Genomic Context and Structure Information
3.1.1. Gene Neighboring
Gene neighboring or co-localization of genes is one of the first and simplest methods for protein–protein interaction prediction methods based on the genomic context [45-47]. The main idea is that related genes are located close to one another in the genome (Fig. 1). Like many other genome-context approaches, the predictions of this method become more confident with larger numbers of genomes [48]. Contrary to prokaryotic organisms, the tendency of being located at a close genomic distance is not evident regarding related genes in eukaryotes, so a major limitation of this method is that it is not applicable in the eukaryote genomes without a doubt especially when there are no homologues in prokaryotes. While its simplicity is a benefit, this method may produce some false negative results because fails to recognize the interaction between related but distantly located genes. Another drawback of gene neighbouring method is that the choice of reference genomes can affect the performance of the method [49].
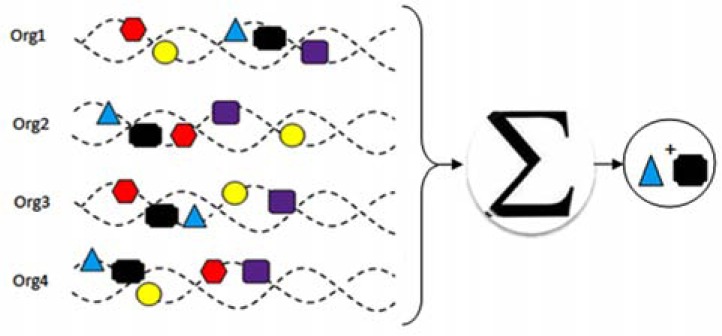
Fig. (1).
Gene neighboring method for protein–protein interaction prediction, the main idea is that related genes are located close to one another in the genome. For example the black and blue proteins predict to interact (plus sign indicates the interaction between proteins).
3.1.2. Phylogenetic Relationship
In this method, the interaction of proteins will be detected based on “phylogenic profile” similarity [50-52], phylogenetic profile for a given protein is a binary vector that reflects the presence or absence of that protein across a set of organisms (Fig 2), this method is a flexible version of the gene neighboring method which can detect some interaction that gene neighboring method fails to detect. The basic idea is that functionally related genes remain together across many distant species for playing a role in a biological process. However, this powerful method has three important drawbacks. The first is that the number and distribution of the genomes that used can influence the results dramatically [49]. The second is that it cannot be applied to essential proteins that presents in almost all organisms, and third drawback is that this method only can run on the complete genomes [48].
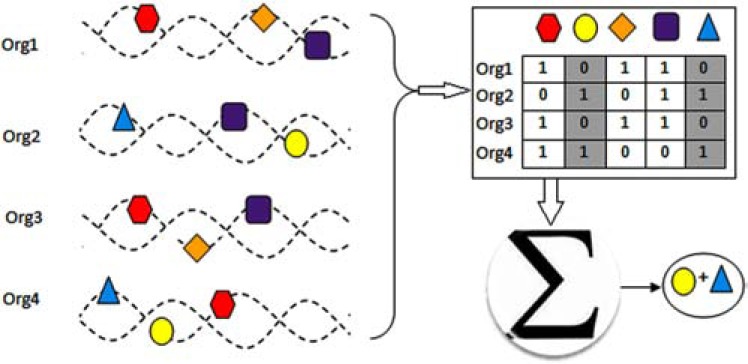
Fig. (2).
Protein-protein interaction detection based on phylogenic profile similarity. Phylogenetic profile for every protein is a binary vector that reflects the presence or absence of that protein across a set of organisms. (plus sign indicates the interaction between proteins).
3.1.3. Gene Fusion
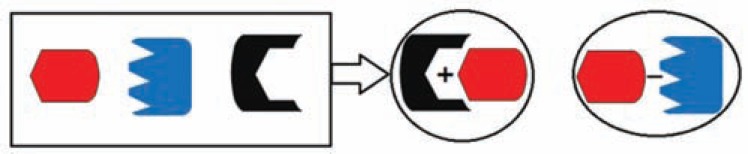
It has seen that separate related genes, probably to reduce the regulatory load of multiple interacting gene products, can be fused into a single multi-functional gene, a so-called “Rosetta Stone” protein. For example, topoisomerase II is a fusion result of Gyr A and Gyr B subunits of Escherichia coli DNA gyrase [53]. The gene fusion method uses comparative genomics and evolutionary information [54, 55] and so can be considered as complement of gene neighboring and phylogenetic profile methods (Fig 3). A major advantage of this method is its reliability, because the existing gene fusion events are very informative about functional relationship. One of the drawbacks of this method is that fusion event not abundant, especially in prokaryotes [48].
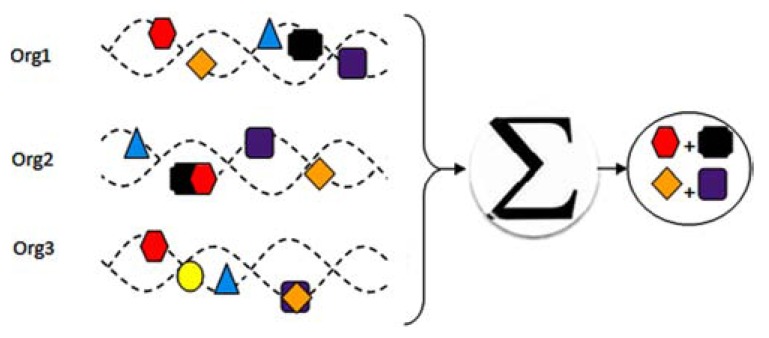
Fig. (3).
Gene fusion method for protein-protein interaction detection. In this method, the complete genome comparison must be done across multiple organisms: if two separate proteins in one organism fused in another organism then one can conclude they interact. (plus sign indicates the interaction between proteins).
3.1.4. 3D Structure Based
This method uses three-dimensional (3D) structure information to predict interactions, the information from predicted interactions also can be used for predicting interaction between new proteins that are homologous with previously predicted interacting proteins [56-63]. This method need accurate dimensional structures (Fig 4) and so cannot be used for a large number of proteins, because the number of known 3D structures in the Protein Databank (PDB) is limited, recently a genome-wide scale method based on structure information for PPI prediction that have used homology models when three dimensional structures were asbsent [64]. In comparison to other methods the results of this method have more details such as interacting residue and biophysical characteristics of the interaction.
Fig. (4).
Use three-dimensional (3D) structure information to predict interactions, this method need accurate dimensional structures.
3.1.5. Primary Structure
The information gained from protein sequences have been used in a number of Bioinformatics researches such as protein subcellular localization or protein recognition, in recent year sequence information also used for protein-protein interaction prediction [65-74]. Primary protein structure approaches predict protein-protein interaction typically based on the short conserved polypeptide such as signatures [66, 67, 69] or sequence similarity and k-let count (subsequences with length k) [70, 74-77].
3.2. Methods Based on Network Topology
Like in many real-world networks, protein-protein interaction networks in various organisms share common topological features which make these networks different from random networks. These topological features have been used as evidence to discern the difference between interactions that represent true positives and those that are false positives. These have allowed researchers to assign an improved confidence score to each interaction [78].
Analyzing the PPI networks from topological perspective is crucial for a better understanding of the underlying evolutionary mechanisms and network dynamics that shape the network. Because the network theory is a relatively new field, so for determining the significance of topological properties in a given PPI network, the properties are compared against those in random networks and then confidence scores are assigned to PPIs. Finally based on these scores some of intearctions can be eliminate and some other can be added to the network [79-85]. One problem is how to create random networks for comparison, usually the number of vertices and edges are held constant so that we can determine which properties are significant.
A random graph model is a model for generating graphs by random process that uses graph theory and probability theory. Random graph was defined in 1959 in two independent studies for the first time [86, 87]. ErdÖs-Rényi model is a model for a random graph generation, in this model the number of vertices in the random network is equals the number of vertices in the original protein-protein interaction network and the probability of an edge existing between any two vertices is equal to the edge density and is independent of other edges, so roughly speaking this model generates a network with the same number of edges and vertices. But this model is not a suitable random model for determining the significance of protein-protein interaction network properties, because many topological properties of these networks are different from protein-protein interaction networks.
Some of the most important topological concepts of protein-protein interaction networks are as follows: (in this section we consider the protein-protein interaction network as a graph G=(V, E) in which V and E are vertex set and edge set of the graph respectively, we also suppose that the number of vertices are n and the number of edges are e)
Degree: for a vertex v shows the number of its interactions and represent as deg(v)
Degree Distribution: indicate the number of vertices whit different degree.
Hub: nodes with high degree called hub which suggested having an important role in cellular processes.
k-core: A subgraph of the network where every vertex in the subgraph has degree greater than k-1 within that subgraph.
Edge Density: is the ratio of the number of network edges to the maximum possible number of edges that can be computed as equation 1:
Clustering Coefficient (CC): for a vertex such as v if Ev shows the maximum number of possible interactions between neighbors of v and Fv shows the number of neighbors of v that interact with each other then clustering coefficient for v is defined as follows:
For a network clustering coefficient define as the average clustering coefficient over all vertices:
Path length: the least number of edges needed to reach from one vertex to another called the path length between them.
Average path length: the average path length over all possible vertex pairs in the network.
Diameter: the maximum path length in the network.
Centrality: in general centrality is a structural attribute of nodes or edges that shows the importance of those nodes or edges in the network. There are many centrality measures, however, we describe three of the most popular centrality measures below:
Degree centrality: it is the simplest centrality measure, which is defined as the number of edges incidences with that node, in the normalized version the degree is divided by the maximum possible degree.
Closeness centrality: this measure is based on the distance of a node to all other nodes in the graph and is precisely defined as follows:
Betweenness centrality: betweenness centrality for a node v is defined as the number of shortest paths passing through v, this measure can be defined for an edge in the same way. A protein with high betweenness centrality value has great influence over information flows in the whole network. In spite of closeness centrality, betweenness centrality can be used for disconnected networks.
Motif: A subgraph of the network which its occurrences are significantly high (more than expected at random).
Protein-protein interaction networks of different species interestingly have many common topological features. PPI networks are also said to have a power-law degree distribution, which means there are a few nodes with many connections and many nodes with few connections and so the degree distribution of the PPI networks is heavy-tailed (power-law degree distribution)[88, 89]. Another feature in protein-protein interaction networks compared to random networks is having a high clustering coefficient: the interaction probability of the neighbors of two interacting proteins is significantly high. Unlike the high clustering, the average path length in the PPI networks is short. Protein-protein interaction networks are referred to as small-world networks because of having a high clustering coefficient and a short average path length.
In addition to being used to predict protein-protein interaction, topological properties of protein-protein interaction networks have been used to predict proteins function, finding protein complexes and finding functional modules.
3.3. Methods Based on Text Mining and Literature Mining
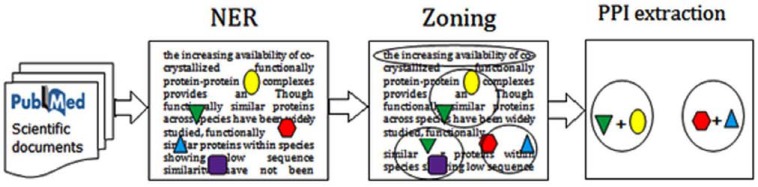
PubMed is expanding at the rate of approximately one paper every thirty seconds; this fact shows the importance of the biomedical literature mining approaches. Some methods use text mining and literature mining algorithms and use the information of co-occurrence of the proteins in the PubMed abstracts approaches for protein-protein interaction prediction [90-100]. In general, each literature mining system consists of three steps (Fig. 5):
Fig. (5).
The schematic text mining approaches for protein-protein interaction prediction, In general, each literature mining system consists of three steps: Named Entity Recognition or NER step, it does the identification task of protein. Zoning step, in this step the text split into basic building blocks and extract sentences from the text. PPI step that uses various algorithms to infer protein-protein interaction.
Named Entity Recognition or NER step, it does the identification task of protein names which is a crucial step for further analyzing. Zoning step, in which the text is split into basic building blocks and sentences are extracted from the text. Protein–protein interaction extraction step that uses various algorithms to infer protein-protein interaction. Current biomedical literature mining approaches for detecting protein-protein interactions can be divided into three categories:
Computational natural language processing (NLP) and linguistics-based methods, which define a grammar and use parsers to detect protein-protein interaction. Rule-based methods, these methods infer protein-protein interaction using a set of context specific rules or patterns. Machine learning approaches which don't need rules or grammar but some classifiers learn the pattern that enables them to identify protein-protein interaction from a training set.
This automated data mining results may not be as reliable as manually curated data, but the fast growth of published biomedical literature can make these methods more confident. (Supplementary Table 2 (264.3KB, pdf) ) shows literature and text mining tools for protein-protein interaction.
3.4. Methods Based on Machine Learning Algorithms with Utilizing Heterogeneous Genomic/Proteomic Features
Some other methods use heterogeneous biological data such as gene expression, codon usage [71, 101], k-let count (subsequences with length k) [70, 74-77] and physicochemical properties of amino acids [77, 102] to learn a model for predicting PPI. These methods integrate biological data sources provided by high-throughput technologies to feature vectors and use machine learning approaches to learn and predict PPI from these feature vectors (some of these methods mentioned previously as another method specially as sequence based methods, but now we focus on machine learning algorithms that were used in those studies). Generally speaking, a machine learning algorithm (classifier) for protein-protein interaction prediction uses a set of various features (or descriptors) of the proteins or protein pairs with known interaction and non-interaction as learning set to learn which proteins interact and which do not interact, and then the algorithm can classify new protein pairs to interacting or non-interacting classes. There are particular machine learning algorithms used to address protein-protein interaction prediction problem, we shall briefly describe these methods in the following context.
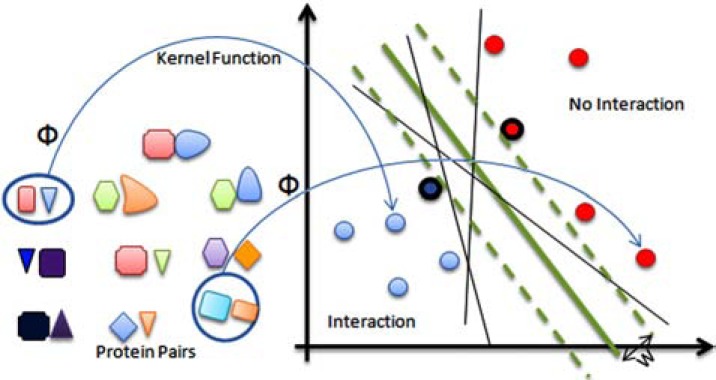
Support vector machines (SVM) or kernel machines are widely used in bioinformatics and computational biology for classifying biological data [103] as well as protein-protein interaction prediction [70, 104-106]. The support vector machine (SVM) classifier is underpinned by the idea of maximizing the margins. Intuitively, the margin for an object is related to the certainty of its classification (see Fig. 6). Objects for which the assigned label is correct and highly certain will have large margins and objects with uncertain classification are likely to have small margins [107]. An SVM can be trained using a labeled training dataset, each data marked as belonging to one of two classes, to build a model that could predict class labels for new examples. SVM is extremely powerful and can classify problems with arbitrary complexity, but it is complex and has large memory requirements also it is a little slow to train and evaluate. Another drawback of this classifier is that the parameters can greatly impact the results [103]. For more details, we refer the interested reader to [108, 109].
Fig. (6).
Schematic view of SVM classifier: At the first step, using a function that is called “kernel function” (and is denoted by Ф) the protein pairs are transformed into points in a new space (presumably making the classification easier in this space). Then, the best separating hyperplane (separating line in this figure) is selected as the boundary of two classes, in this example each three thin black lines and the thick green line are separating lines. The margin for a separating hyperplane is the shortest distance from that hyperplane to the closest positive or negative example; in this figure the margin of thick green line is denoted by ‘w’. The best separating hyperplane is the one with the maximum margin; in this example the thick green line is the best separating line (closest points to the best separating hyperplane are called support vectors, the support vectors are circled in the figure).
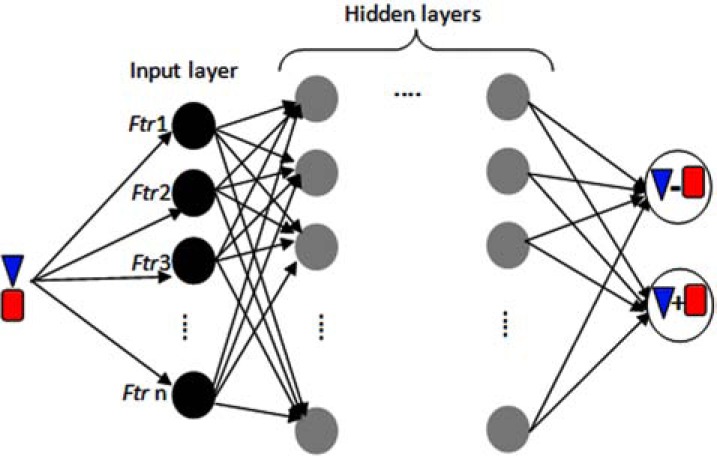
Artificial neural networks (ANNs or simply NNs) originated from the idea to model mathematically human intellectual abilities by biologically plausible engineering designs. One of the most popular NN models is the multilayer perceptron (MLP) [107], MLP is a tool used for modeling PPI with good performance [110, 111]. However, MLP has been criticised as being a black-box classifier because it is difficult to know what the model parameters mean [112]. An MLP is a feedforward artificial neural network, which consists of multiple layers and each layer is fully connected to the next layer with weighted edges. Typically there are three layers: input layer, hidden layer (intermediate layer) and output layer, each node at the hidden and output layer is a neuron with an activation function, this node contains the processing units of an MLP. The weights of the edges are optimized and adjusted on the training dataset to minimize classification error using a supervised learning approach. (Fig. 7) shows a schematic representation of a MLP for protein-protein interaction prediction. For more details, we refer the interested reader to [113].
Fig. (7).
A Multilayer Perceptron (MLP), which consists of multiple layers and each layer is fully connected to the next layer with weighted edges. Typically there are three layers: input layer, hidden layer (intermediate layer) and output layer, the features (feature abbreviated as ftr in the figure) of every protein pairs is delivered to the input layer for classifying. Each node at the hidden and output layer is a neuron with an activation function, these nodes are the processing units of an MLP. Neuron at output layer classify input protein pair into interacting (which is denoted by a ‘+’ sign between two proteins) or non-interacting (which is denoted by a ‘-’ sign between two proteins) class. The weights of the edges are optimized and adjusted on the training dataset to minimize classification error using a supervised learning approach.
Naïve Bayes is a probabilistic classifier that is based on Bayes' theorem and it is a popular algorithm owing to its simplicity (the source of simplicity is the assumption that the independent variables are statistically independent.), computational efficiency and easy to interpret. In spite of the simplicity of this classifier, it turns out that Naïve Bayse works quite well in problems involving normal distributions, which are very common in real-world problems. Naive Bayes classifiers can be trained efficiently on a small training dataset in a supervised learning approach using maximum likelihood, but in the more complex classification problem it may work not well [114]. This method has been widly used in PPI prediction problem [71, 74, 115-117].
K-Nearest neighbors (K-NN) is one of the simplest machine learning classifiers that is a prototype method for classifying objects, which assign labels to each object based on the K closest objects (parameter K must be set by user) in the feature space according to majority vote, in contrast to other statistical methods, K-NN requires no explicit training (because the choice of K is very crucial in this method, optimizing K can be considered as a kind of learning). In spite of its simplicity to implement, when a large data set or numerous features are used the computational cost and memory requirement grows rapidly. This method has been used (not widely) in PPI prediction problem [118]. For more details, we refer the interested reader to [119].
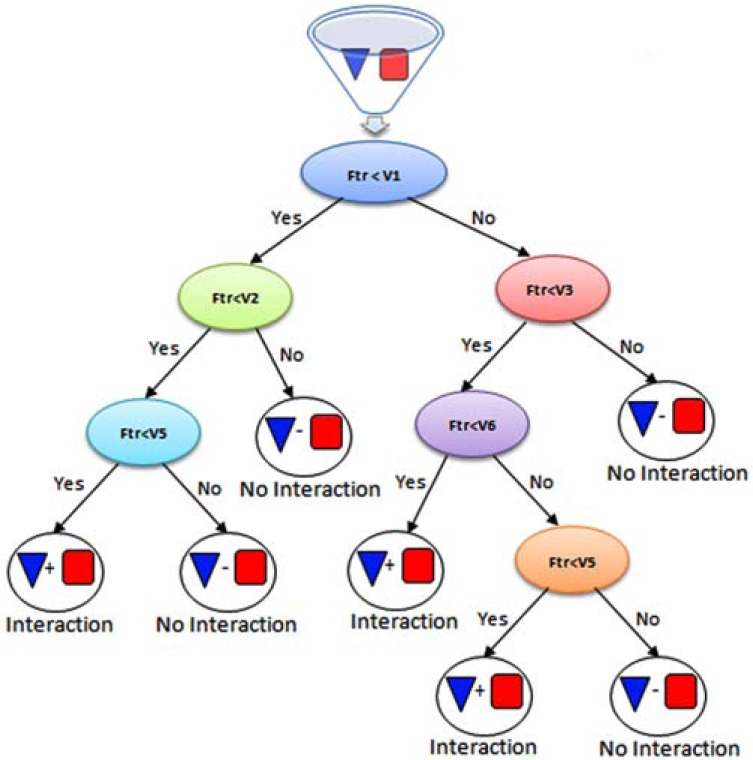
Decision tree or classification tree is a popular machine learning classifier, which has great applications in bioinformatics and computational biology and has shown to be one of the best classifier for protein-protein interaction prediction. In these trees, internal node test features, each branch correspond to feature value and finally leaves assigns a class label (Fig. 8). In the training phase, training dataset is partitioned into the subsets according to the feature values and this process is recursively done on the subsets until splitting no effect on the classification. Concerning various aspects of optimality, constructing the optimal decision tree is an NP-complete problem and so practical decision tree construction algorithms such as ID3, C4.5 and CART employ a heuristic search [114]. In addition it is efficient from computational cost and memory requirements points of view. This classifier is prone to overfitting and in some applications it may not have good generalization, but compared with other classifiers such MLP the pattern in the data can be easily explained with classification trees [112].
Fig. (8).
Decision tree or classification tree, in these trees internal nodes tests features (in each internal node corresponding feature is compared with a value), each branch corresponds to features value and finally leaves assigns a class label (positive or negative for interacting and non-interacting respectively).
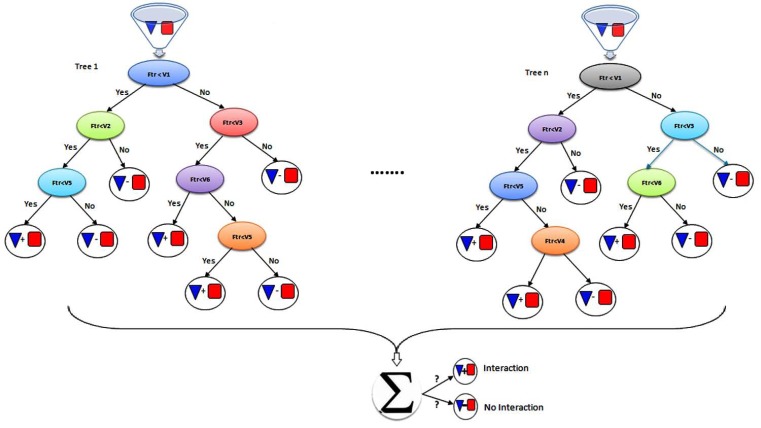
Random forest (RF) algorithm is a classification method that consists of many decision trees (Fig. 9), in training phase each tree is constructed based on random feature vectors sampled from a data set independently and for every node in a tree, a small fraction of the variables are randomly selected and then each classification tree is completely grown. To classify a new object, put the input vector down each of the trees in the forest, and finally according to the majority voting one class is assigned to the object. The RF is a practical classifier when there are a large dataset and large number of features and no need to feature selection or feature deletion, also it can rank features according to importance for classification. In addition RF can be used for recovering missing data, but in some databases containing noisy data RF may be overfit [114]. Decision trees and random forest are widely used in bioinformatics and computational biology for classifying biological data [120] especially for PPI prediction [73, 121, 122].
Fig. (9).
The random forest (RF) classifier, which consists of many decision trees. To classify a new object, put the input vector down each of the trees in the forest, and finally according to the majority voting one class assign to the protein pair.
4. RESULTS ASSESSMENT
The need for gold standard data sets, which contain both positive and negative interactions, to evaluate performance of methods for PPI prediction is a critical problem in protein-protein interaction prediction. Each available database of protein-protein interaction contains positive interactions which may also include many false positives (interactions that are not biologically real and are produced due to tools biases and errors). The most complex part is in selecting negative examples (non-interacting proteins), benchmark data can affect the performance results and may lead to overestimating the prediction performance [132, 133]. Recently two web based systems have been developed for constructing benchmark protein-protein interaction data [134, 135] and some high quality PPI data sets have been published for model organisms [136-138]but researches about constructing gold standard datasets for PPI prediction had conflicting results [132, 133, 139] and it seems there is need for more efforts. After constructing a gold standard dataset, the prediction performance could be assessed with different measures based on four following basic parameters:
TP (True Positive or hit): the number of interactions predicted correctly.
TN (True Negative or correct rejection): the number of non-interactions predicted correctly.
FP (False Positive or type I error): the number of non-interactions predicted incorrectly as interaction.
FN (False Negative or type II error): the number of interactions predicted incorrectly as non-interaction.
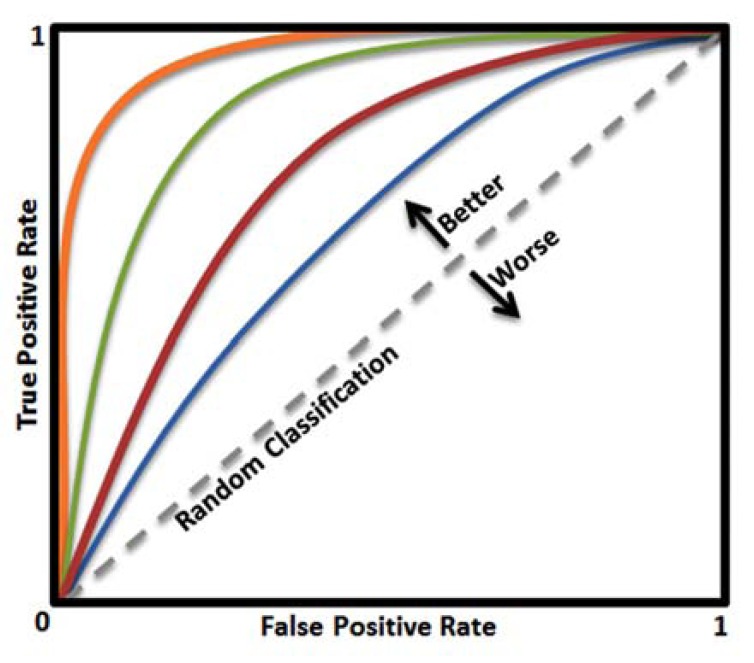
(Table 3) lists the important measures for evaluating prediction methods. In addition to these measures, one popular graphical tool for assessing the classification performance is ROC (receiver operating characteristic) curve which plots sensitivity (true positive rate) vs. one minus the specificity (true negative rate), which each of them changes between 0 and 1. The ROC curve shows the tradeoff between sensitivity and specificity (see Fig. 10), and the closer the curve follows the left-hand border and then the top border of the ROC space, the more accurate the classifier, and the closer the curve comes to the diagonal of the ROC space, the less accurate the classifier and closer to the random classifier. The area under the ROC curve (AUC or "Area Under Curve"), is another measure of classification accuracy, the closer the AUC to one the more accurate the classification. It is argued that reporting accuracy and precision can be misleading but AUC has proved to be a reliable performance measure for imbalanced problems like PPI prediction [140, 141] there are many tools for evaluating and visualizing the performance of classifiers [142, 143].
Table 3.
Important Measures to Evaluate Prediction Methods with Brief Descriptions
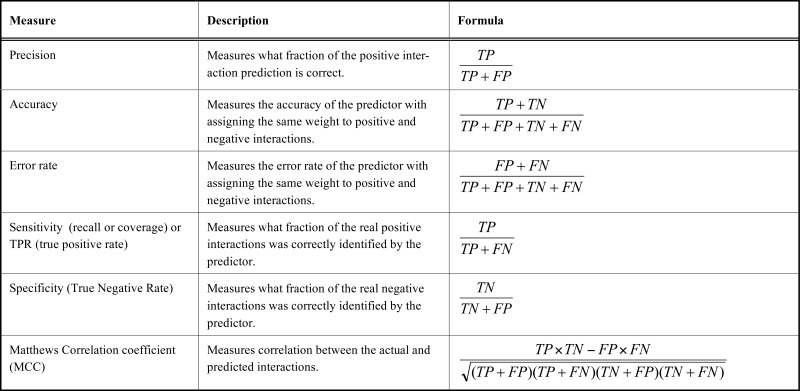 |
Fig. (10).
The ROC curve is a plot of the true positive rate against the false positive rate, it shows the tradeoff between sensitivity and specificity. The closer the curve follows the left-hand border and then the top border of the ROC space, the more accurate the classifier, and the closer the curve comes to the diagonal of the ROC space, the less accurate the classifier and closer to the random classifier. If two ROC curves do not intersect, the upper one dominates the other (in this example the orange curve is the best and the blue curve is the worst).
5. TOOLS FOR ANALYZING AND VISUALIZING PROTEIN-PROTEIN INTERACTION
After constructing the protein-protein interaction, researchers need to visualize and analyze the networks. In recent years, many tools and software tools have been developed for this purpose; (Table 4) briefly discusses some of the popular tools used for the analysis and visualization of biological networks. In the following section, some of the most popular instances of these tools are described in detail.
Table 4.
The Most Popular Available Tools for Analyzing and Visualizing Protein-Protein Interaction Networks with Brief Descrip-tions and References. OS Column Shows Operating System(s) Including Linux (Lin), Macintosh (Mac) and Windows (Win) that the Tools Can Run on it (Them). The Popularity of Tools Range from One Star to Five Stars Based on the Numbers of Corresponding Publications’ Citations (According to Google Scholar at the Time of Writing this Manuscript, May 2013): One Star Means Less than 50 Citations; Two Stars Mean Between 50 and 100 Citations; Three Stars Mean Be-tween 100 and 200 Citations; Four Stars Mean Between 200 and 500 Citations, and Four Stars Mean Greater Than 500 Ci-tations
| Acronym | Description | OS | Availability | Popularity | Stand-alone/Web-based/Plug-in | URL and References |
|---|---|---|---|---|---|---|
| APID (Cancer Research Center) | (Agile Protein Interaction DataAnalyzer) it's an interactive web-tool that allow exploration and analysis of protein-protein interaction | Lin/Mac/Win | Free | *** | Web-based | http://bioinfow.dep.usal.es/apid/index.htm [147] |
| BiNoM | It developed to facilitate the manipulation of biological networks represented in standard systems biology formats (SBML, SBGN, BioPAX) and to carry out studies on the network structure. | Lin/Mac/Win | Free | ** | Cytoscape plug-in | https://binom.curie.fr/ [148] |
| BioLayout | BioLayout is a tool for visualization and clustering of biological networks in both 3D and 2D, it is compatible with Cytoscape. It also includes analytical approaches to microarray data analysis. | Lin/Mac/Win | Free | * | Stand-alone | http://www.biolayout.org. [149, 150] |
| Cerebral | It enhances Cytoscape's functionality by using extra annotation provided by the user to both automatically generate a more pathway-like representation of a network and to provide an environment for the visualization, comparison, and clustering of expression data from multiple conditions. | Lin/Mac/Win | Free | * | Cytoscape plug-in | http://www.pathogenomics.ca/cerebral/ [151] |
| Cytoscape | A powerful interactive open source network visualization tool | Lin/Mac/Win | Free | ***** | Stand-alone | http://cytoscapeweb.cytoscape.org/ [144, 146] |
| InterProSurf (University of Texas Medical Branch) | wWeb server for predicting the functional sites on a protein surface | Lin/Mac/Win | Free | * | Web-based | http://curie.utmb.edu/prosurf.html [152] |
| InterViewer (Inha University) | Produces a molecular interaction network of good quality without computing force between every pair of nodes | Win | Free | * | Stand-alone | http://interviewer.inha.ac.kr/ [153] |
| iSPOT (Universita di Roma) | SPOT (Sequence Prediction Of Target) infer the peptide binding specificity of any member of a family of protein binding domains. | Lin/Mac/Win | Free | * | Web-based | http://cbm.bio.uniroma2.it/ispot/[154] |
| MCODE | It finds clusters (highly interconnected regions) in a network. It can also be used to lay out any graph that requires stratification according to some characteristic and thus can be used by researchers in a variety of fields | Lin/Mac/Win | Free | ***** | Cytoscape plug-in | http://baderlab.org/Software/MCODE [155] |
| Medusa | An powerful interactive tool for visualization and clustering analysis of biological networks. | Lin/Mac/Win | Free | * | Stand-alone | https://sites.google.com/site/medusa3visualization[156] |
| meta-PPISP (Florida State University) | A web server for protein-protein interaction site prediction (this tool is built on three individual web servers: cons-PPISP, PINUP, and Promate). | Lin/Mac/Win | Free | ** | Web-based | http://pipe.scs.fsu.edu/meta-ppisp.html [157] |
| NAViGaTOR (University of Toronto) |
Software package for visualizing and analyzing protein-protein interaction networks | Lin/Mac/Win | Free | ** | Stand-alone | http://ophid.utoronto.ca/navigator/ [158] |
| NOXclass (Max-Planck-Institut fur Informatik) | A classifier identifying protein-protein interaction types implemented using a SVM algorithm. | Lin/Mac/Win | Free | *** | Web-based | http://noxclass.bioinf.mpi-inf.mpg.de/ [159] |
| Osprey | a A tool for visualization and manipulation of complex interaction networks | Lin/Mac/Win | Free (registeration is needed) | **** | Stand-alone | http://biodata.mshri.on.ca/osprey/servlet/Index [160] |
| Pajek | is a standalone application, iIt can use for analyzing large networks with up to million of nodes and vertices. | Win | Free | ***** | Stand-alone | http://vlado.fmf.uni-lj.si/pub/networks/pajek/ [161, 162] |
| PathBLAST (Whitehead Institute) | It ssearches the protein-protein interaction network of the target organism to extract all protein interaction pathways that align with a pathway query. | Lin/Mac/Win | Free | **** | Web-based | http://www.pathblast.org/ [163] |
| SCOWLP (TU Dresden) | Structural classification of protein binding regions for atomic comparative analysis of protein interactions allow individual and comparative analysis of protein interactions. | Lin/Mac/Win | Free | * | Web-based | http://www.scowlp.org/scowlp/ [164, 165] |
| UVCLUSTER | Iterative cluster analysis of protein interaction data. | Lin/Win | Free for academic users (fulfillment of software license agreement is needed) | *** | Stand-alone | http://www.uv.es/genomica/UVCLUSTER/ [166] |
| VisANT | It designed specifically for the integrative visual data-mining of multi-scale Bio-Network/Pathways. It also can find the over-represented GO terms in network modules. | Lin/Mac/Win | Free | ** | Cytoscape plug-in | http://visant.bu.edu/ [167] |
Cytoscape [144-146] is a free software package, which is one of the most popular protein-protein interaction visualization and data integration tools. It has many interesting features such as custom node graphics and attribute equations, with these features the user can project images onto nodes and use spreadsheet-like functionality capability for more enhanced network visualization (supplementary Fig. S1 (264.3KB, pdf) ). This software provides complex network searches, filtering operations and many other analysis options.
Medusa is a powerful Java standalone application for visualization of large-scale biological networks in 2D, it also implements various clustering algorithms: k-Means, spectral, predefined clustering and affinity propagation. It is very interactive and uses a variety of layout and methods (Grid, random, circular, hierarchical, fruchterman-reingold, spring embedding, distance geometry and parallel coordinates) for more intuitive visualizations. It also supports varieties of graphs such as weighted and unweighted multi-edged directed and undirected graphs. Medusa has some other interesting features: it is compatible with many other tools, have up to10 types of connections, have search functionality, the user can collapse/expand nodes and provides color schemes. This software allows users to load an arbitrary image as a background for more descriptive visualizations (supplementary Fig. S2 (264.3KB, pdf) ).
NAViGaTOR is a graphing tool for the 2D and 3D visualization of biological networks which has been implemented in Java and is freely available for researchers, it can be installed on Windows, Mac, Linux and Unix. (Supplementary Fig. S3 (264.3KB, pdf) ) shows NAViGaTOR interface and one rendered network. This software uses hardware acceleration to facilitate the visualization of large networks. It supports some popular data interchange formats, such as PSI-MI, BioPAX and GML makes it compatible with other tools. NAViGaTOR includes many functions for network analysis and visualizing options and allows the user to generate high quality images, it also can be extended through an application programming interface (API).
6. FUTURE DIRECTION AND CONCERNS: EVOLUTION OF PROTEIN-PROTEIN INTERACTION NETWORKS
Protein-protein interaction network is highly dynamic [168] and studying the evolution of protein-protein interaction networks is one of the central problems of systems biology, the results of such researches are crucial for a better understanding of the evolution of living systems and could be used for protein interaction and function prediction.
Statistics Versus Comparative Approaches
Generally, it is possible to categorize studies on protein-protein interaction network evolution in two ways: those based on a statistical and mathematical models, and those based on a comparative network analysis. In approaches based on statistical and mathematical models after analyzing protein-protein interaction networks (by focusing on the topological features) mathematical and statistical models of evolving networks is produced and then by tuning parameters we proceed to reproducing properties observed in experimentally produced networks. In approaches based on comparative network analysis protein-protein interaction networks of species with different levels of complexity are analyzed and then by comparing networks we try to find the evolutionary processes that generally shaped these networks [169-171]. In both of these approaches, three main evolutionary events are considered as the main processes that have shaped the structure of the protein-protein interaction network.
Addition of New Nodes
Gene duplication is an important evolutionary mechanism that naturally increases the number of proteins in the protein-protein interaction networks. A gene duplication event therefore corresponds to the addition of a node and with links identical to the original node, followed by the divergence of some of the initially redundant links between the two duplicate nodes [172]. After gene duplication, a protein product that has the ability to bind strongly to its partner will be better able to explore mutations that allow it to co-evolve, or to dimerize with other, existing, homologues using the ancestral binding mode. This duplicating effect should, therefore lead to an enhanced ability to create homologous interacting pairs of proteins, and could have played a role in the early emergence of protein-protein interaction networks. This should allow for an increased resistance to environmental change, or adaptability However, single gene duplications may lead to an immediate stoichiometric imbalance, which would therefore tend to be counter selected [173]. In smaller gene family sizes, especially for genes encoding protein complex components, there is less potential for paralogs to evolve new interaction partners through mutation and selection.
Addition and Elimination of Edges
There are two reasons for the loss and gain of new interactions, namely neofunctionalization and subfunctionalization. After gene duplication, the second copy of the gene is relatively free from selective pressure and is able to diverge and accumulate mutations faster than a functional single-copy gene because these mutations often have no deleterious effects [174, 175]. If these mutations are solely degenerative in nature then this will lead to a nonfunctional gene product, but if instead they are innovative, then this can lead to neofunctionalization and the acquisition of novel features. In another possible trajectory, some of the functions of the original gene are assigned to the new copy and both copies accumulate degenerative mutations leading to a differentiation of function and division of labor (i.e. subfunctionalization). The result of these processes is the divergence at interaction pattern of the original gene and its copy (ie. the addition and elimination of edges).
Elimination of Nodes
After gene duplication the mutation that takes place on the new copy of a gene could convert it to a nonfunctional gene, consequently it is deleted from the network because it does not have any interaction (gene lost).
ACKNOWLEDGEMENTS
We appreciate great help from all LBB member during the course of study. Part of this work was supported by Iranian INSF (insf.gov.ir). We would like to thank Dr. Bidkhori for his critical reading the manuscript.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIALS
Supplementary material is available on the publisher’s web site along with the published article.
REFERENCES
- 1.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions Part I Experimental techniques and databases. Plos comput biol. 2007;3(3):e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions.Part II. Computational methods to predict protein and domain interaction partners. . Plos Comput Biol. 2007;3(4):e43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theofilatos K, Dimitrakopoulos C, Tsakalidis A, Likothanassis ST, Papadimitriou S, Mavroudi S. Computational Approaches for the Prediction of Protein-Protein Interactions. A Survey Curr Bioinform. 2011;6(4):398–414. [Google Scholar]
- 4.Tuncbag N, Kar G, Keskin O, Gursoy A, Nussinov R. A survey of available tools and web servers for analysis of protein-protein interactions and interfaces. Brief. Bioinform. 2009;10(3):217–32. doi: 10.1093/bib/bbp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiaoli L, Min W, Chee-Keong K, See-Kiong N. Computational approaches for detecting protein complexes from protein interaction networks: a survey. BMC genomics. 2010;11 doi: 10.1186/1471-2164-11-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol. Biotechnol. 2008;38(1):1–17. doi: 10.1007/s12033-007-0069-2. [DOI] [PubMed] [Google Scholar]
- 7.Raman K. Construction and analysis of protein-protein interaction networks. Autom Exp. 2010;2 (1):2. doi: 10.1186/1759-4499-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman F, Cesareni G. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9(4):345–350. doi: 10.1038/nmeth.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P. The minimum information required for reporting a molecular interaction experiment (MIMIx). Nat Biotechnol. 2007;25(8):894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 10.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn A, Vinod N, Bader G, Xenarios I, Wojcik J, Sherman D. Broadening the horizon-level 2 5 of the HUPO-PSI format for molecular interactions. BMC. Biology. 2007;5(1):44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aranda B, Blankenburg H, Kerrien S, Brinkman F S, Ceol A, Chautard E, Dana J M, De Las Rivas J, Dumousseau M, Galeota E. PSICQUIC and PSISCORE: accessing and scoring molecular interactions. Nat Methods. 2011;8(7):528–529. doi: 10.1038/nmeth.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitkreutz B, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner D, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID inter- action database 2008 update. Nucleic Acids Res. 2008;36:D637– D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark C, Breitkreutz B, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID a general repository for interaction datasets. Nucleic Acids Res. 2006;34(Database issue):D535–9. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xenarios I, Salwinski L, Duan X, Higney P, Kim S, Eisenberg D. DIP the Database of Interacting Proteins a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30:303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges PE, Payne WE, Garrels JI. l The Yeast Protein Database (YPD) a curated proteome database for Saccharomyces cerevisiae. Nucleic Acids Res. 1998;26(1):68–72. doi: 10.1093/nar/26.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keseler I, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen I, Peralta-Gil M, Karp P. l EcoCyc a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33 (Database Issue):D334–337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian F, Shah P K, Liu X, Negre N, Chen J, Karpenko O, White KP, Grossman RL. Flynet a genomic resource for Drosophila melanogaster transcriptional regulatory networks. Bioinformatics. 2009;25(22):3001–4. doi: 10.1093/bioinformatics/btp469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database Issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bader G, Betel D, Hogue C. BIND the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31(1):248 –250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bader GD, Hogue CW. BIND--a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16(5):465–77. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 21.Isserlin R, El-Badrawi RA, Bader GD. The Biomolecular Interaction Network Database in PSI-MI. 25 Database Database (Oxford) 2011;baq037 doi: 10.1093/database/baq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatr-aryamontri A, Ceol A, Palazzi L, Nardelli G, Schneider M, Castagnoli L, Cesareni G. MINT the Molecular INTeraction database. Nucleic Acids Res. 2007;35(Database Issue):D572–574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ceol A, Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT the molecular interaction database 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, Rashmi BP, Shanker K, Padma N, Niranjan V, Harsha HC, Talreja N, Vrushabendra BM, Ramya MA, Yatish AJ, Joy M, Shivashankar H N, Kavitha MP, Menezes M, Choudhury DR, Ghosh N, Saravana R, Chandran S, Mohan S, Jonnalagadda CK, Prasad CK, Kumar-Sinha C, Deshpande KS, Pandey A. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32(Database issue):D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan T M, Menon S, Hanumanthu G, Gupta M, Upendran S, Gupta S, Mahesh M, Jacob B, Mathew P, Chatterjee P, Arun K S, Sharma S, Chandrika K N: Deshpande, N. Palvankar, K. Raghavnath, R. Krishnakanth, R. Karathia, H. Rekha, B. Nayak, R. Vishnupriya, G. Kumar, H. G. Nagini, M. Kumar, G. S. Jose, R. Deepthi, P. Mohan, S. S. Gandhi, T. K. Harsha, H. C.: Deshpande K S, Sarker M, Prasad T S, Pandey A. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34 (Database issue):D411–4. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keshava Prasad T S, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla: D. Raju, R. Shafreen, B. Venugopal, A. Balakrishnan, L. Marimuthu, A. Banerjee, S. Somanathan: D S, Sebastian A, Rani S, Ray S, Harrys Kishore C J, Kanth S, Ahmed M, Kashyap M K, Mohmood R, Ramachandra Y L, Krishna V, Rahiman B A, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37 (Database issue):D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hermjakob L, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A, Margalit H, Armstrong J, Bairoch A, Cesareni1 G, Sherman D, Apweiler R. IntAct an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452 – D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R. IntAct - open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561 – D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian A, Kerrien S, Khadake J. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stark C, Breitkreutz B J, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone M S, Nixon J, Van Auken K, Wang X, Shi X, Reguly T, Rust J M, Winter A, Dolinski K, Tyers M. The BioGRID Interaction Database 2011 update. Nucleic Acids Res. 2011;39(Database issue):D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breitkreutz B J, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner D H, Bahler J, Wood V, Dolinski K, Tyers M. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36 (Database issue):D637–40. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stark C, Breitkreutz B, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willis R C, Hogue C W, Baxevanis.Andreas D. Searching viewing and visualizing data in the Biomolecular Interaction Network Database (BIND). Current protocols in bioinformatics . 2006;Chapter 8:Unit 8 9. doi: 10.1002/0471250953.bi0809s12. [DOI] [PubMed] [Google Scholar]
- 34.Bader G D, Betel D, Hogue C W. BIND the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31(1):248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bader G D, Donaldson I, Wolting C, Ouellette B F, Pawson T, Hogue C W. BIND--The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29(1):242–5. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xenarios I, Salwinski L, Duan X J, Higney P, Kim S M, Eisenberg D. DIP the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30(1):303–5. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salwinski L, Miller C, Smith A, Pettit F, Bowie J, Eisenberg D. The Database of Interacting Proteins 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xenarios I, Rice D W, Salwinski L, Baron M K, Marcotte E M, Eisenberg D. DIP the database of interacting proteins. Nucleic Acids Res. 2000;28(1):289–91. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xenarios I, Fernandez E, Salwinski L, Duan XJ, Thompson M J, Marcotte E M, Eisenberg D. DIP The Database of Interacting Proteins 2001 update. Nucleic Acids Res. 2001;29(1):239–41. doi: 10.1093/nar/29.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Duan X J, Xenarios I, Eisenberg D. Describing biological protein interactions in terms of protein states and state transitions: the LiveDIP database. Mol Cell Proteomics. 2002;1(2):104–16. doi: 10.1074/mcp.m100026-mcp200. [DOI] [PubMed] [Google Scholar]
- 41.Peri S, Navarro J D, Amanchy R, Kristiansen T Z, Jonnalagadda C K, Surendranath V, Niranjan V, Muthusamy B, Gandhi T K, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar H N, Rashmi B P, Ramya M A, Zhao Z, Chandrika KN, Padma N, Harsha HC, Yatish AJ, Kavitha MP, Menezes M, Choudhury DR, Suresh S, Ghosh N, Saravana R, Chandran S, Krishna S, Joy M, Anand SK, Madavan V, Joseph A, Wong GW, Schiemann W P, Constantinescu S N, Huang L, Khosravi-Far R, Steen H, Tewari M, Ghaffari S, Blobe GC, Dang CV, Garcia J G, Pevsner J, Jensen O N, Roepstorff P, Deshpande K S, Chinnaiyan A M, Hamosh A, Chakravarti A, Pandey A. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13(10):2363–71. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A. Int Act an open source molecular interaction database. Nucleic Acids Res. 2004; (32 Database):D452. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chatr-aryamontri A, Kerrien S, Khadake J, Orchard S, Ceol A, Licata L, Castagnoli L, Costa S: Derow, C. Huntley, R. Aranda, B. Leroy, C. Thorneycroft: D, Apweiler R, Cesareni G, Hermjakob H. MINT and IntAct contribute to the Second BioCreative challenge: serving the text-mining community with high quality molecular interaction data. Genome Biol. 2008;9 (Suppl 2 ):S5. doi: 10.1186/gb-2008-9-s2-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, Cesareni G. MINT A Molecular INTeraction Database. Febs Letters. 2002;513(1):135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- 45.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23(9):324–8. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 46.Tamames J, Casari G, Ouzounis C, Valencia A. Conserved clusters of functionally related genes in two bacterial genomes. J Mol Evol. 1997;44(1):66–73. doi: 10.1007/pl00006122. [DOI] [PubMed] [Google Scholar]
- 47.Overbeek R F, M.; D'Souza M, Pusch G D, Maltsev N. Use of contiguity on the chromosome to predict functional coupling. In Silico Biol. 1999;1(2):93–108. [PubMed] [Google Scholar]
- 48.Panchenko A, Przytycka T. Protein-protein interactions and networks identification computer analysis and prediction. Springer. 2008;9 [Google Scholar]
- 49.Muley V Y, Ranjan A. Effect of reference genome selection on the performance of computational methods for genome-wide protein-protein interaction prediction. Plos one. 2012;7(7):e42057. doi: 10.1371/journal.pone.0042057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcotte E, Pellegrini M, Ng HL, Rice D, Yeates T, Eisenberg D. Detecting Protein Function and Protein-Protein Interactions from Genome Sequences. Science. 1999;285(5428):751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 51.Juan D, Pazos F, Valencia A. High-confidence prediction of global interactomes based on genome-wide coevolutionary networks. Proc Natl Acad Sci USA. 2008;105(3):934–939. doi: 10.1073/pnas.0709671105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guimaraes K, Jothi R, Zotenko E, Przytycka T. Predicting domain-domain interactions using a parsimony approach. Genome Biol. 2006;7(11):R104. doi: 10.1186/gb-2006-7-11-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54(1):665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- 54.Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg DA. combined algorithm for genome-wide prediction of protein function. Nature. 1999;402(6757):83–6. doi: 10.1038/47048. [DOI] [PubMed] [Google Scholar]
- 55.Enright A, Iliopoulos I, Kyrpides N, Ouzounis C. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402 (6757):86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- 56.Edwards AM, Kus B, Jansen R, Greenbaum D, Greenblatt J, Gerstein M. Bridging structural biology and genomics: assessing protein interaction data with known complexes. Trends Genet. 2002;18(10):529–36. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- 57.Aloy P, Russell R. InterPreTS protein Interaction Prediction through Tertiary Structure. Bioinformatics. 2003;19(1):161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 58.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci U S A. 2002;99(9):5896–901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hue M, Riffle M, Vert JP, Noble WS. Large-scale prediction of protein-protein interactions from structures. BMC bioinformatics. 2010;11(1):144. doi: 10.1186/1471-2105-11-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith GR, Sternberg MJ. Prediction of protein-protein interactions by docking methods. Curr. Opin. Struct. Biol. 2002;12(1):28–35. doi: 10.1016/s0959-440x(02)00285-3. [DOI] [PubMed] [Google Scholar]
- 61.Singh R, Park D, Xu J, Hosur R, Berger B. Struct2 Net a web service to predict protein-protein interactions using a structure-based approach. N ucleic Acids Res. 2010;38 ( suppl 2):W508–W515. doi: 10.1093/nar/gkq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin A C, Bork P, Superti-Furga G, Serrano L, Russell R B. Structure-based assembly of protein complexes in yeast. Science. 2004;303(5666):2026–9. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 63.Wass M N, Fuentes G, Pons C, Pazos F, Valencia A. Towards the prediction of protein interaction partners using physical docking. Mol Syst Biol. 2011;7(1) doi: 10.1038/msb.2011.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490 (7421):556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bock JR, Gough DA. Predicting protein--protein interactions from primary structure. Bioinformatics. 2001;17(5):455–60. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- 66.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or interologs. Genome Res. 2001;11(12):2120–6. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. J Mol Biol. 2001;311(4):681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- 68.Gomez S, Noble W, Rzhetsky A. Learning to predict protein-protein interactions from protein sequences. Bioinformatics. 2003;19(15):1875–1881. doi: 10.1093/bioinformatics/btg352. [DOI] [PubMed] [Google Scholar]
- 69.Pitre S, Dehne F, Chan A, Cheetham J, Duong A, Emili A, Gebbia M, Greenblatt J, Jessulat M, Krogan N. PIPE a protein-protein interaction prediction engine based on the re-occurring short polypeptide sequences between known interacting protein pairs. BMC bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Y, Yu L, Wen Z, Li M. Using support vector machine combined with auto covariance to predict proteinprotein interactions from protein sequences. Nucleic Acids Res. 2008;36(9):3025–3030. doi: 10.1093/nar/gkn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Najafabadi H S, Salavati R. Sequence-based prediction of protein-protein interactions by means of codon usage. Genome Biol. 2008;9(5):R87. doi: 10.1186/gb-2008-9-5-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Y, Li M, Pu X, Li G, Guang X, Xiong W, Li J. PRED_PPI a server for predicting protein-protein interactions based on sequence data with probability assignment. BMC Research Notes. 2010;3(1):145. doi: 10.1186/1756-0500-3-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia JF, Han K, Huang DS. Sequence-based prediction of protein-protein interactions by means of rotation forest and autocorrelation descriptor. Protein Pept Lett. 2010;17(1):137. doi: 10.2174/092986610789909403. [DOI] [PubMed] [Google Scholar]
- 74.Liu CH, Li KC, Yuan S. Human protein–protein interaction prediction by a novel sequence-based co-evolution method: co-evolutionary divergence. Bioinformatics. 2013;29(1):92–98. doi: 10.1093/bioinformatics/bts620. [DOI] [PubMed] [Google Scholar]
- 75.Ren X, Wang YC, Wang Y, Zhang XS, Deng NY. Improving accuracy of protein-protein interaction prediction by considering the converse problem for sequence representation. BMC bioinformatics. 2011;12(1):409. doi: 10.1186/1471-2105-12-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu CY, Chou LC, Chang DT. Predicting protein-protein interactions in unbalanced data using the primary structure of proteins. BMC bioinformatics. 2010;11(1):167. doi: 10.1186/1471-2105-11-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, Li Y, Jiang H. Predicting protein–protein interactions based only on sequences information. Proc Natl Acad Sci USA. 2007;104(11):4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goldberg DS, Roth FP. Assessing experimentally derived interactions in a small world. Proc. Natl. Acad. Sci. USA. 2003;100(8):4372–6. doi: 10.1073/pnas.0735871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pei P, a Z. A In Atopological measurement for weighted protein interaction network Proceedings of 16th. IEEE Computational Systems Bioinformatics Conference. 2005:268–278. doi: 10.1109/csb.2005.8. [DOI] [PubMed] [Google Scholar]
- 80.Dyer MD, Murali TM, Sobral BW. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23(13):i159–66. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 81.Saito R, Suzuki H, Hayashizaki Y. Construction of reliable protein-protein interaction networks with a new interaction generality measure. Bioinformatic. 2003;19(6):756–63. doi: 10.1093/bioinformatics/btg070. [DOI] [PubMed] [Google Scholar]
- 82.Chen J, Hsu W, Lee ML, Ng SK. Increasing confidence of protein interactomes using network topological metrics. Bioinformatics. 2006;22(16):1998–2004. doi: 10.1093/bioinformatics/btl335. [DOI] [PubMed] [Google Scholar]
- 83.Saito R, Suzuki H, Hayashizaki Y. Interaction generality a measurement to assess the reliability of a protein-protein interaction. Nucleic Acids Res. 2002;30(5):1163–8. doi: 10.1093/nar/30.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen PY, Deane CM, Reinert G. Predicting and validating protein interactions using network structure. PLoS Comput Biol. 2008;4(7):e1000118. doi: 10.1371/journal.pcbi.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wuchty S. Topology and weights in a protein domain interaction network–a novel way to predict protein interactions. BMC. genomics. 2006;7(1):122. doi: 10.1186/1471-2164-7-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Erdös P, Rényi A. On random graphs. Publ Math. 1959:290–297. [Google Scholar]
- 87.Gilbert E N. Random Graphs. Ann Math Stud. 1959;30:1141–1144. [Google Scholar]
- 88.Wagner A. The yeast protein interaction network evolves rapidly and contains few redundant duplicate genes. Mol Biol Evol. 2001;18 (7):1283–92. doi: 10.1093/oxfordjournals.molbev.a003913. [DOI] [PubMed] [Google Scholar]
- 89.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–2. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 90.Jaeger S, Gaudan S, Leser U, Rebholz-Schuhmann D. Integrating protein-protein interactions and text mining for protein function prediction. BMC bioinformatics. 2008;9 (Suppl 8 ):S2. doi: 10.1186/1471-2105-9-S8-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oyama T, Kitano K, Satou K, Ito T. Extraction of knowledge on protein-protein interaction by association rule discovery. Bioinformatics. 2002;18(5):705–14. doi: 10.1093/bioinformatics/18.5.705. [DOI] [PubMed] [Google Scholar]
- 92.Skusa A, Ruegg A, Kohler J. Extraction of biological interaction networks from scientific literature. Brief. Bioinform. 2005;6(3):263–76. doi: 10.1093/bib/6.3.263. [DOI] [PubMed] [Google Scholar]
- 93.Ono T, Hishigaki H, Tanigami A, Takagi T. Automated extraction of information on protein-protein interactions from the biological literature. Bioinformatics. 2001;17(2):155–61. doi: 10.1093/bioinformatics/17.2.155. [DOI] [PubMed] [Google Scholar]
- 94.Huang M, Zhu X, Hao Y, Payan D, Qu K, Li M. Discovering patterns to extract protein-protein interactions from full texts. Bioinformatics. 2004;20:3604–12. doi: 10.1093/bioinformatics/bth451. [DOI] [PubMed] [Google Scholar]
- 95.Hao Y, Zhu X, Huang M, Li M. Discovering patterns to extract protein-protein interactions from the literature. Part II Bioinformatics. 2005;21(15):3294–300. doi: 10.1093/bioinformatics/bti493. [DOI] [PubMed] [Google Scholar]
- 96.Marcotte EM, Xenarios I, Eisenberg D. Mining literature for protein-protein interactions. Bioinformatics. 2001;17(4):359–63. doi: 10.1093/bioinformatics/17.4.359. [DOI] [PubMed] [Google Scholar]
- 97.Bunescu R, Ge R, Kate RJ, Marcotte EM, Mooney RJ, Ramani AK, Wong YW. Comparative experiments on learning information extractors for proteins and their interactions. Artif Intell Med. 2005;33(2):139–55. doi: 10.1016/j.artmed.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 98.Kim S, Shin S, Lee I, Kim S, Sriram R, Zhang B. PIE an online prediction system for protein-protein interactions from text. Nucleic Acids Res. 2008;36 (Web Server issue):W411–5. doi: 10.1093/nar/gkn281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donaldson I, Martin J, De Bruijn B, Wolting C, Lay V, Tuekam B, Zhang S, Baskin B, Bader G D, Michalickova K, Pawson T, Hogue C W. PreBIND and Textomy--mining the biomedical literature for protein-protein interactions using a support vector machine. BMC bioinformatics. 2003;4:11. doi: 10.1186/1471-2105-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Blaschke Christian A M A. In Automatic extraction of biological information from scientific text protein protein interactions Proceedings of the seventh International conference on intelligent systems for molecular. biology. 1999: 60–67. [PubMed] [Google Scholar]
- 101.Zhou Y, Zhou YS, He F, Song J, Zhang Z. Can simple codon pair usage predict protein–protein interaction?. Molecular BioSystems. 2012;8(5):1396–1404. doi: 10.1039/c2mb05427b. [DOI] [PubMed] [Google Scholar]
- 102.Zhang SW, Cheng YM, Luo L, Pan Q. In Prediction of Protein-Protein Interaction using Distance Frequency of Amino Acids grouped with their physicochemical properties Bio-Inspired Computing. Theories and Applications (BIC-TA) 2011. Sixth International Conference on IEEE. 2011:70–74. [Google Scholar]
- 103.Ben Hur A, Ong CS, Sonnenburg S, Schölkopf B, Rätsch G. Support Vector Machines and Kernels for Computational Biology. PLoS. comput. biol. 2008;4(10):e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lo S, Cai C, Chen Y, Chung M. Effect of training datasets on support vector machine prediction of protein-protein interactions. Proteomics. 2005;5(4):876 – 884. doi: 10.1002/pmic.200401118. [DOI] [PubMed] [Google Scholar]
- 105.Dohkan S, Koike A, Takagi T. Improving the Performance of an SVM-Based Method for Predicting Protein-Protein Interactions. In. Silico Biol. 2006;6:515–529. [PubMed] [Google Scholar]
- 106.Rashid M, Ramasamy S, PS Raghava G. A simple approach for predicting protein-protein interactions. Curr.Pro.Pept. Sci. 2010;11(7):589–600. doi: 10.2174/138920310794109120. [DOI] [PubMed] [Google Scholar]
- 107.Kuncheva LI. Combining Pattern Classifiers Methods and Algorithms (Kuncheva LI 2004)[book review]. IEEE Transactions on Neural Networks. 2007;18(3):964–964. [Google Scholar]
- 108.Schölkopf B, Smola AJ. Learning with kernels. The MIT Press. 2002 [Google Scholar]
- 109.Bishop C M, Nasrabadi N M. Vol. 1. New York: springer; 2006. Pattern recognition and machine learning. [Google Scholar]
- 110.P.Fariselli A Z M, Finelli P, Martelli R. Casadio In A neural network method to improve prediction of proteinprotein interaction sites in heterocomplexes Proceedings of the 13th IEEE. Workshop on Neural Networks for Signal Processing. 2003:33–41. [Google Scholar]
- 111.Z. Ma C Z, L. Lu Y Ma, P. Sun Y. Cui In Predicting protein-protein interactions based on BP neural network, Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine. Workshops. 2007:3–7. [Google Scholar]
- 112.Yang Z R. Machine learning approaches to bioinformatics. World Scientific Publishing Company. 2010;4 [Google Scholar]
- 113.Bishop C. Pattern Recognition and Machine Learning. 2006 [Google Scholar]
- 114.Witten I H, Frank E, Hall M A. Data Mining Practical Machine Learning Tools and Techniques: Practical Machine Learning Tools and Techniques. Morgan Kaufmann. 2011 [Google Scholar]
- 115.Lu L J, Xia Y, Paccanaro A, Yu H, Gerstein M. Assessing the limits of genomic data integration for predicting protein networks. Genome Res. 2005;15(7):945–53. doi: 10.1101/gr.3610305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan N, Chung S, Emili A, Snyder M, Greenblatt J, Gerstein M. A Bayesian Networks Approach for Predicting Protein-Protein Interactions from Genomic Data. Science. 2003;302(5644):449 – 453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 117.Lin X, Chen XW. Heterogeneous data integration by tree-augmented naïve Bayes for protein–protein interactions prediction. Proteomics. 2013;13(2):261–268. doi: 10.1002/pmic.201200326. [DOI] [PubMed] [Google Scholar]
- 118.F.Browne HW H, Zheng F. Azuaje.In Supervised statistical and machine learning approaches to inferring pairwise and module-based protein interaction networks. IEEE International Conference on Bioinformatics and Bioengineering. 2007:1365–1369. [Google Scholar]
- 119.Hastie T, Tibshirani R, Friedman J, Franklin J. The elements of statistical learning: data mining, inference and prediction. Math. Intell. 2005;27(2):83–85. [Google Scholar]
- 120.Chen X, Wang M, Zhang H. The use of classification trees for bioinformatics.Wiley Interdisciplinary Reviews. J of Data Mini and Know Disc. 2011;1(1):55–63. doi: 10.1002/widm.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen XW, Liu M. Prediction of protein-protein interactions using random decision forest framework. Bioinformatics. 2005;21 (24):4394–400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- 122.Qi Y, Klein-Seetharaman J, Bar-Joseph Z. Random forest similarity for protein-protein interaction prediction from multiple sources. Pac Symp Biocomput. 2005:531–542. [PubMed] [Google Scholar]
- 123.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order a fingerprint of proteins that physically interact. Trends Biochem Sci . 1998;23 (9):324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 124.Jothi R, Kann MG, Przytycka TM. Predicting protein-protein interaction by searching evolutionary tree automorphism space. Bioinformatics. 2005;21 ( Suppl 1):i241–50. doi: 10.1093/bioinformatics/bti1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Marcotte EM, Xenarios I, Van Der Bliek AM, Eisenberg D. Localizing proteins in the cell from their phylogenetic profiles. Proc Natl Acad Sci USA. 2000;97 (22):12115–20. doi: 10.1073/pnas.220399497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pellegrini M, Marcotte E M, Thompson M J, Eisenberg D, Yeates T O. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96 (8):4285–8. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aloy P, Russell R. Interrogating protein interaction networks through structural biology. Proc Natl Sci Acad USA. 2001;98:4569–74. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Edwards A, Kus B, Jansen R, Greenbaum D, Greenblatt J, Gerstein M. Bridging structural biology and genomics: assessing protein interaction data with known complexes. TRENDS in Genetics. 2002;18 (10):529–536. doi: 10.1016/s0168-9525(02)02763-4. [DOI] [PubMed] [Google Scholar]
- 129.Aloy P, Russell RB. InterPreTS protein interaction prediction through tertiary structure. Bioinformatics. 2003;19 (1):161–2. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 130.Qi Y, Klein-Seetharaman J, Bar-Joseph Z. Random forest similarity for protein-protein interaction prediction from multiple sources. Pac Symp Biocomput. 2005:531–42. [PubMed] [Google Scholar]
- 131.Chen XW, Liu M. Prediction of protein-protein interactions using random decision forest framework. Bioinformatics. 2005;21 (24):4394– 4400. doi: 10.1093/bioinformatics/bti721. [DOI] [PubMed] [Google Scholar]
- 132.Yu J, Guo M, Needham C, Huang Y, Cai L, Westhead D. Simple sequence-based kernels do not predict protein-protein interactions. Bioinformatics. 2010;26(20):2610–14. doi: 10.1093/bioinformatics/btq483. [DOI] [PubMed] [Google Scholar]
- 133.Park Y, Marcotte EM. Revisiting the negative example sampling problem for predicting protein–protein interactions. Bioinformatics. 2011;27 (21):3024–3028. doi: 10.1093/bioinformatics/btr514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Browne F, Wang H, Zheng H, Azuaje F. GRIP A web-based system for constructing Gold Standard datasets for protein-protein interaction prediction. Source Code Biol Med. 2009;4:2. doi: 10.1186/1751-0473-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chen XW, Jeong JC, Dermyer P. KUPS constructing datasets of interacting and non-interacting protein pairs with associated attributions. Nucleic Acids Res. 2011;39(Database issue):D750–4. doi: 10.1093/nar/gkq943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Dreze M, Carvunis A-R, Charloteaux B, Galli M, Pevzner S J, Tasan M, Ahn YY, Balumuri P, Barabási AL, Bautista V. Evidence for network evolution in an Arabidopsis interactome map. Science. 2011;333(6042):601–607. doi: 10.1126/science.1203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Simonis N, Rual JF, Carvunis AR, Tasan M, Lemmens I, Hirozane-Kishikawa T, Hao T, Sahalie JM, Venkatesan K, Gebreab F. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2008;6(1):47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322(5898):104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Park Y, Marcotte E M. Flaws in evaluation schemes for pair-input computational predictions. Nat. Methods. 2012;9(12):1134–1136. doi: 10.1038/nmeth.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Weng C G, Poon J. In A new evaluation measure for imbalanced datasets, Proceedings of the 7th Australasian Data Mining Conference-Volume 87. Australian Computer Society Inc. 2008:27–32. [Google Scholar]
- 141.Fawcett T. ROC graphs Notes and practical considerations for researchers. ML. 2004;31:1–38. [Google Scholar]
- 142.Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR visualizing classifier performance in R. Bioinformatics. 2005;21 (20):3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 143.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, Müller M. pROC an open-source package for R and S+ to analyze and compare ROC curves. BMC bioinformatics. 2011;12 (1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shannon P, Markiel A, Ozier O, Baliga N, Wang J, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lopes C T, Franz M, Kazi F, Donaldson S L, Morris Q, Bader G D. Cytoscape Web an interactive web-based network browser. Bioinformatic. 2010;26(18):2347–8. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Smoot M E, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2 8 new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–2. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Prieto C, De Las Rivas J. APID Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006;34 (Web Server issue):W298–302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zinovyev A, Viara E, Calzone L, Barillot E. BiNoM a Cytoscape plugin for manipulating and analyzing biological networks. Bioinformatics. 2008;24(6):876–877. doi: 10.1093/bioinformatics/btm553. [DOI] [PubMed] [Google Scholar]
- 149.Freeman T, Goldovsky L, Brosch M, van Dongen S, Maziere P, Grocock R, Freilich S, Thornton J, Enright A. Construction visualisation and clustering of transcription networks from microarray expression data. PLoS comput biol. 2007;3 (10):2032 – 2042. doi: 10.1371/journal.pcbi.0030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goldovsky L, Cases I, Enright AJ, Ouzounis CA. BioLayout(Java) versatile network visualisation of structural and functional relationships. Appl .Bioinformatics. 2005;4 (1):71–4. doi: 10.2165/00822942-200504010-00009. [DOI] [PubMed] [Google Scholar]
- 151.Barsky A, Munzner T, Gardy J, Kincaid R. Cerebral Visualizing multiple experimental conditions on a graph with biological context. IEEE Transactions on Visualization and Computer Graphics. 2008;14 (6):1253–1260. doi: 10.1109/TVCG.2008.117. [DOI] [PubMed] [Google Scholar]
- 152.Negi SS, Schein CH, Oezguen N, Power TD, Braun W. InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatic. 2007;23 (24):3397–9. doi: 10.1093/bioinformatics/btm474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ju B H, Park B, Park J H, Han K. Visualization and analysis of protein interactions. Bioinformatics. 2003;19(2):317–8. doi: 10.1093/bioinformatics/19.2.317. [DOI] [PubMed] [Google Scholar]
- 154.Brannetti B, Helmer-Citterich M. iSPOT A web tool to infer the interaction specificity of families of protein modules. Nucleic Acids Res. 2003;31 (13):3709–11. doi: 10.1093/nar/gkg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pavlopoulos GA, Hooper SD, Sifrim A, Schneider R, Aerts J. Medusa A tool for exploring and clustering biological networks. BMC Res Notes. 2011;4(1):384. doi: 10.1186/1756-0500-4-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Qin S, Zhou H X. meta-PPISP a meta web server for protein-protein interaction site prediction. Bioinformatics. 2007;23(24):3386–7. doi: 10.1093/bioinformatics/btm434. [DOI] [PubMed] [Google Scholar]
- 158.Brown K R, Otasek D, Ali M, McGuffin M J, Xie W, Devani B, Toch I L, Jurisica I. NAViGaTOR Network Analysis, Visualization and Graphing Toronto. Bioinformatics. 2009;25 (24):3327–9. doi: 10.1093/bioinformatics/btp595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zhu H, Domingues F S, Sommer I, Lengauer T. NOXclass prediction of protein-protein interaction types. BMC bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Breitkreutz B J, Stark C, Tyers M. Osprey a network visualization system. Genome Biol. 2003;4 (3):R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Batagelj V, Mrvar A. Pajek - Program for Large Network Analysis. Connections. 1998;21:47–57. [Google Scholar]
- 162.Batagelj V, Mrvar A. Pajek analysis and visualization of large networks. Springer. 2004 [Google Scholar]
- 163.Kelley BP, Yuan B, Lewitter F, Sharan R, Stockwell B R, Ideker T. PathBLAST a tool for alignment of protein interaction networks. Nucleic Acids Res. 2004;32 (Web Server issue):W83–8. doi: 10.1093/nar/gkh411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Teyra J, Doms A, Schroeder M, Pisabarro MT. SCOWLP a web-based database for detailed characterization and visualization of protein interfaces. BMC bioinformatics. 2006;7:104. doi: 10.1186/1471-2105-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Teyra J, Samsonov SA, Schreiber S, Pisabarro MT. SCOWLP update 3D classification of protein-protein -peptide, -saccharide and -nucleic acid interactions and structure-based binding inferences across folds. BMC bioinformatics. 2011;12:398. doi: 10.1186/1471-2105-12-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Arnau V, Mars S, Marin I. Iterative cluster analysis of protein interaction data. Bioinformatics. 2005;21 (3):364–78. doi: 10.1093/bioinformatics/bti021. [DOI] [PubMed] [Google Scholar]
- 167.Hu Z, Hung J-H, Wang Y, Chang YC, Huang CL, Huyck M, DeLisi C. VisANT 3 5 multi-scale network visualization analysis and inference based on the gene ontology. Nucleic Acids Res. 2009;37 ( suppl 2):W115–W121. doi: 10.1093/nar/gkp406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhang Y, Gao P, Yuan JS. Plant protein-protein interaction network and interactome. Curr genomics. 2010;11 (1):40. doi: 10.2174/138920210790218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Peterson GJ, Presse S, Peterson KS, Dill KA. Simulated evolution of protein-protein interaction networks with realistic topology. PloS one. 2012;7(6):e39052. doi: 10.1371/journal.pone.0039052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Przulj N, Kuchaiev O, Stevanovic A, Hayes W. Geometric evolutionary dynamics of protein interaction networks. Pac Symp Biocomput. 2010:178–89. doi: 10.1142/9789814295291_0020. [DOI] [PubMed] [Google Scholar]
- 171.D'Antonio M, Ciccarelli FD. Modification of gene duplicability during the evolution of protein interaction network. PLoS comput biol. 2011;7(4):e1002029. doi: 10.1371/journal.pcbi.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pastor-Satorras R, Smith E, Sole R. Evolving protein interaction networks through gene duplication. J Theor Biol. 2003;222 (2):199–210. doi: 10.1016/s0022-5193(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 173.Mintseris J, Weng Z. Structure function and evolution of transient and obligate protein–protein interactions. Proc Nat Acad Sci USA. 2005;102(31):10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Evlampiev K, Isambert H. Conservation and topology of protein interaction networks under duplication-divergence evolution. Proc Nat Acad Sci USA. 2008;105 (29):9863–8. doi: 10.1073/pnas.0804119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Bagowski CP, Bruins W, te Velthuis AJ. The nature of protein domain evolution shaping the interaction network. Curr genomics. 2010;11 (5):368. doi: 10.2174/138920210791616725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.