Abstract
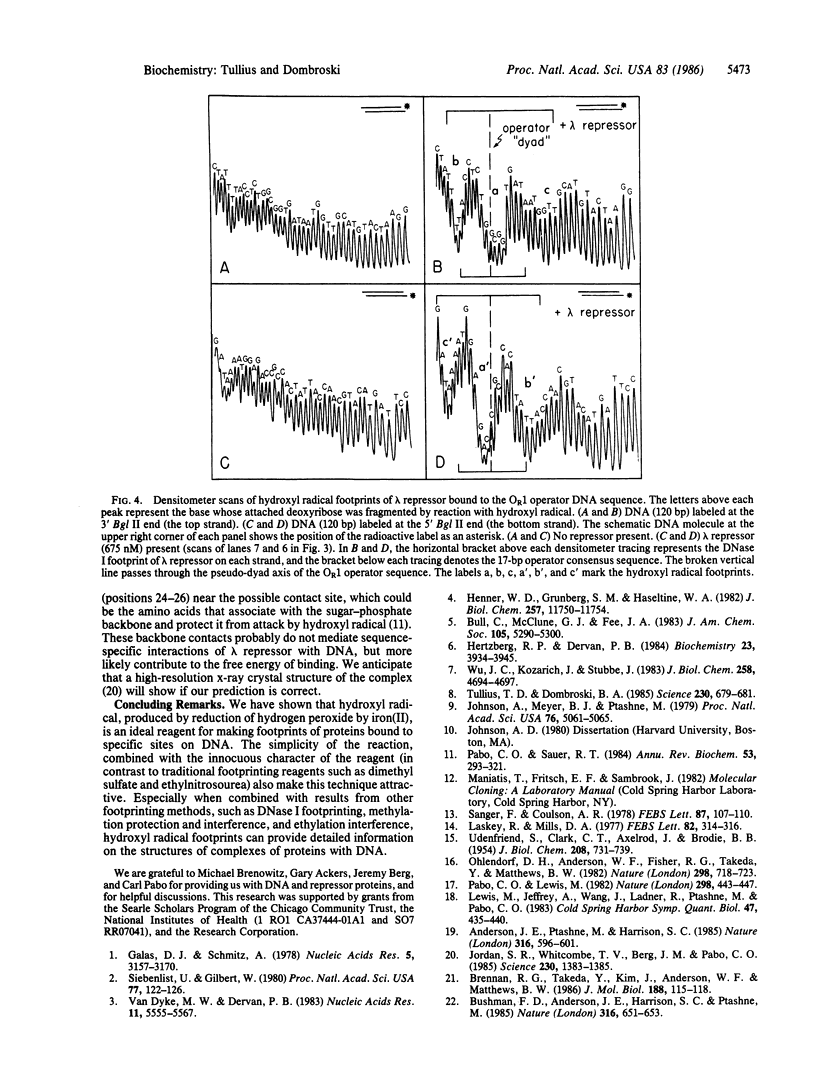
A method has been developed for making "footprints" of proteins bound to DNA. The hydroxyl radical, generated by reduction of hydrogen peroxide by iron(II), is the reagent used to cut the DNA. Hydroxyl radical breaks the backbone of DNA with almost no sequence dependence, so all backbone positions may be monitored for contact with protein. In addition to defining the DNA sequence in contact with the protein, hydroxyl radical footprints embody structural information about the DNA-protein complex. For example, hydroxyl radical footprints of the bacteriophage lambda repressor and Cro protein show directly that these proteins are bound to only one side of the DNA helix. Additional contacts of lambda repressor and Cro protein with DNA, not observed by other chemical footprinting methods, are revealed by hydroxyl radical footprinting.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. E., Ptashne M., Harrison S. C. A phage repressor-operator complex at 7 A resolution. Nature. 1985 Aug 15;316(6029):596–601. doi: 10.1038/316596a0. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Takeda Y., Kim J., Anderson W. F., Matthews B. W. Crystallization of a complex of cro repressor with a 17 base-pair operator. J Mol Biol. 1986 Mar 5;188(1):115–118. doi: 10.1016/0022-2836(86)90488-2. [DOI] [PubMed] [Google Scholar]
- Bushman F. D., Anderson J. E., Harrison S. C., Ptashne M. Ethylation interference and X-ray crystallography identify similar interactions between 434 repressor and operator. Nature. 1985 Aug 15;316(6029):651–653. doi: 10.1038/316651a0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner W. D., Grunberg S. M., Haseltine W. A. Sites and structure of gamma radiation-induced DNA strand breaks. J Biol Chem. 1982 Oct 10;257(19):11750–11754. [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Johnson A. D., Meyer B. J., Ptashne M. Interactions between DNA-bound repressors govern regulation by the lambda phage repressor. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5061–5065. doi: 10.1073/pnas.76.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan S. R., Whitcombe T. V., Berg J. M., Pabo C. O. Systematic variation in DNA length yields highly ordered repressor-operator cocrystals. Science. 1985 Dec 20;230(4732):1383–1385. doi: 10.1126/science.3906896. [DOI] [PubMed] [Google Scholar]
- Laskey R. A., Mills A. D. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977 Oct 15;82(2):314–316. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- Lewis M., Jeffrey A., Wang J., Ladner R., Ptashne M., Pabo C. O. Structure of the operator-binding domain of bacteriophage lambda repressor: implications for DNA recognition and gene regulation. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):435–440. doi: 10.1101/sqb.1983.047.01.051. [DOI] [PubMed] [Google Scholar]
- Ohlendorf D. H., Anderson W. F., Fisher R. G., Takeda Y., Matthews B. W. The molecular basis of DNA-protein recognition inferred from the structure of cro repressor. Nature. 1982 Aug 19;298(5876):718–723. doi: 10.1038/298718a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982 Jul 29;298(5873):443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Siebenlist U., Gilbert W. Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc Natl Acad Sci U S A. 1980 Jan;77(1):122–126. doi: 10.1073/pnas.77.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- UDENFRIEND S., CLARK C. T., AXELROD J., BRODIE B. B. Ascorbic acid in aromatic hydroxylation. I. A model system for aromatic hydroxylation. J Biol Chem. 1954 Jun;208(2):731–739. [PubMed] [Google Scholar]
- Van Dyke M. W., Dervan P. B. Methidiumpropyl-EDTA.Fe(II) and DNase I footprinting report different small molecule binding site sizes on DNA. Nucleic Acids Res. 1983 Aug 25;11(16):5555–5567. doi: 10.1093/nar/11.16.5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. C., Kozarich J. W., Stubbe J. The mechanism of free base formation from DNA by bleomycin. A proposal based on site specific tritium release from Poly(dA.dU). J Biol Chem. 1983 Apr 25;258(8):4694–4697. [PubMed] [Google Scholar]