Abstract
Populations of “identical” cells are rarely truly identical. Even when in the same state of differentiation, isogenic cells may vary in expression of key signaling regulators, activate signal transduction at different thresholds, and consequently respond heterogeneously to a given stimulus. Here, we review how new experimental and analytical techniques are suited to connect these different levels of variability, quantitatively mapping the effects of cell-to-cell variability on cellular decision-making. In particular, we summarize how this helps classify signaling regulators according to the impact of their variability on biological functions. We further discuss how variability can also be leveraged to shed light on the molecular mechanisms regulating cellular signaling, from the individual cell to the population of cells as a whole.
Introduction
The existence of discrete cell states within clonal bacterial or yeast populations, or within differentiating cell populations in multicellular organisms has been appreciated for some time [1,2]. In recent years, however, observations of continuous cell-to-cell variability (CCV) in protein abundance in genetically identical eukaryotic cells sharing a common differentiation state have become ubiquitous [3]. As awareness of CCV is becoming more prevalent, the importance of understanding its origins and impact has grown, and methodologies to connect variability in gene expression, protein abundance, signaling and phenotypes have begun to be established.
Research into the origins of CCV has suggested that noise in mRNA transcript levels may be an unavoidable consequence of the transcriptional machinery [4,5]. The processes of chromatin opening and closing, and transcription initiation and termination, result in transcriptional bursts, which lead to a fundamental level of noise in mRNA production and consequently in protein abundance [5]. An emerging body of work suggests that cells have capitalized on such protein expression noise to promote evolutionarily adaptive functions. CCV in protein abundance is a precursor to the large phenotypic divergence seen in differentiating cells, wherein broad distributions of protein abundance [6–8] or differences in signaling responses [9,10] prepare cells to respond differently to a common signal, producing multiple cell types. Within the immune system in particular, naïve lymphocytes undergo differentiation into diverse cell types during most immune responses. Heterogeneity in certain receptors has been shown to prepare differentiating CD4 cells to commit to long-lived memory, or short-lived effector fates [7], and to further differentiate different types within the memory population [11]. In addition to its role in differentiation, CCV has been shown to allow a population of cells to make a graded response from decisions that are all-or-none at a single cell level, such as apoptosis or commitment to a particular differentiation type [9,12] Thus, even in an isogenic population of cells, CCV can generate subsets with distinct phenotypes based on either intrinsic differences or response to stimuli.
Additionally, and by analogy with single-celled organisms, we conjecture that CCV may serve an adaptive role in multicellular systems that must respond to uncertain external stimuli, allowing populations of variable cells to make more robust decisions than a population of homogeneous cells would. In bacteria, stochastic switching between states that confer either growth or survival benefits ensures that members of a population will survive, even in the face of sudden environmental changes. In yeast, continuous variability in certain proteins allows a spectrum of growth rate-survival tradeoffs [13]. This strategy adopted by single-celled organisms has been described as bet hedging as cells diversify their phenotype in anticipation of environmental fluctuations[1,14,15]. In multicellular organisms, the clearest analog to both of these exists in the immune system, which must react to constantly evolving pathogenic threats. To do so, the immune system must maintain cells in many discrete differentiated states, whose functional relevance has been abundantly characterized with genetic tools: Loss of particular lymphocyte subpopulations often induces susceptibility to specific pathogens or autoimmune disorders. Our recent work demonstrates the relevance of continuous variability of protein expression within individual states [16,17]. Precisely how CCV contributes to effective immune function in the face of uncertain threats remains an interesting and open question.
Despite the appreciation of CCV in protein abundance and its clear importance to the regulation of differentiation and apoptosis in eukaryotic development, an understanding of the connections between underlying variability and heterogeneous outcomes is still developing. Observing CCV requires only the ability to measure a biological readout at the level of single cells. This can be accomplished through microscopy of live or fixed cells [4,13], flow cytometry, mass cytometry [18] or various methods of single-cell gene expression profiling [19–21]. Connecting variability to downstream effects, however, requires a variety of tools and techniques. More specifically, quantifying the impact of protein abundance on signaling responses requires different methodologies than identifying the impact of protein abundance on functional responses. As each method of single-cell analysis requires certain trade-offs, fully understanding the consequences of CCV requires the combination of multiple methods. In this review, we will focus on the combinations of methods that can connect variable protein abundance, through signaling, to functional responses. Rather than providing an exhaustive catalog of studies using cell-to-cell variability, this review will focus on a few key examples of studies demonstrating techniques to connect layers of CCV—from gene regulation to functional responses—towards the development of a more-mechanistic understanding of cellular decision-making.
Studying signaling in heterogeneous populations
Cellular signaling networks translate external environmental cues into response and change of internal cell state. There exist many layers of heterogeneity that can affect the eventual response. Studying this response with single-cell resolution requires a measurement of pathway activation, of which there are two principal categories: live cell reporters of protein activity, or antibody immunostaining for activated states of proteins, typically phospho-epitopes.
If cell signaling needs to be analyzed over a period of time, it is necessary to endow the cells with reporting capability through genetic methods. In cases where activation results in nuclear localization, such as for extracellular regulated kinase (ERK) [22], signal transducer and activator of transcription 5A/5B (STAT5) [23], or nuclear factor of activated T-cells (NFAT) [24], microscopy of live cells expressing fluorescently tagged proteins reveals activation through the proxy of localization. In other cases, direct enzymatic activity can be evaluated, such as with Förster resonance energy transfer (FRET) reporters of ERK kinase activity [25], or cleavageactivated fluorescence reporters of caspase activity [12,26]. The principal benefit of analyzing signaling in live cells is the ability to track a single cell over time. This is essential in cases where the phenomenon being studied involves an oscillatory or transient signal [27] and is also key in the ability to track the connection of signaling and phenotype (discussed in more detail below). The drawback of such live-cell methods is the complexity of constructing systems to measure multiple signals simultaneously, and the difficulty of implementation in primary cells.
In cases where time-evolution is not as critical, end-point fixed-cell methods provide the ability to measure multiple phospho-signals and regulatory proteins simultaneously through antibody staining [28–30]. Flow cytometry and microscopy of fixed cells both provide well-established methods to measure protein and phosphoprotein levels simultaneously. Applying automated methods of image processing can allow quantitative analysis of large numbers of cells [31,32], allowing the analysis of outliers or small populations. Though, in our experience, the dynamic range of measurements made by microscopy are not as large as those made by flow cytometry the added protein localization information, ability to work with adherent cells, and ability to identify physical phenotypes makes this a valuable tool in the study of cell-to-cell variability [20,31]. Flow cytometry provides the added ability to quantify simultaneously the abundance and modification of a large number of proteins. This allows the possibility to examine mixed populations of cells (such as splenocytes), identify subpopulations of interest through expression of surface markers, and still have the ability to measure the abundance of regulatory proteins and phosphoproteins. Modern fluorescence-based cytometers have the ability to measure more than 16 channels simultaneously. However, due to spectral spillover, the number of channels that can be measured with quantitative precision at any one time is limited to one per laser, typically no more than five. Recently, the introduction of mass cytometry [18] has greatly expanded the number of simultaneous parameters that can be assessed. The potential to simultaneously assess variability in entire signaling networks with single-cell resolution is an exciting prospect.
Connecting cell-to-cell variability in protein abundance to variability in signal sensitivity
Connecting CCV in signaling components to differences in signaling response requires the simultaneous measurement of protein abundance and pathway activation. To do this in live cells, over time, requires either complicated genetic intervention or restricted scope [32], and has rarely been attempted. Instead, most work has utilized flow cytometry of fixed cells to provide a snapshot of the population, as flow cytometry provides the ability to measure the abundance and activation (through phosphorylation) of multiple signaling regulators simultaneously.
Our lab has made extensive use of these aspects of flow cytometry to derive deeper understanding of the underlying regulatory networks through the development of CCV analysis (CCVA), a computational method to correlate variability in protein expression with variability in signaling sensitivity using flow cytometric data (Figure 1). By binning cells according to variation in the abundance of regulatory proteins of interest, we are able to analyze cell-to-cell variation in stimulus sensitivity, bypassing the fact that measurements of dose-responses at the single cell level are technically impossible (cells often respond to stimuli by adapting—tuning their responsiveness according to past stimuli). We first demonstrated that T cells with identical T cell receptors (TCRs) can vary in their ability to trigger a phospho-ERK response by three orders of magnitude, due to heterogeneity of two key proteins: a co-receptor (CD8) and a negative regulator (Shp-1), but not in the levels of ERK itself. Similarly, we have used this methodology to demonstrate how CCV regulates heterogeneity in sensitivity to interleukin 2 (IL-2) [16] and other members of the γc cytokine family [17]. In all of these cases, the observed correlations between protein and signaling variability were used to refine and extend models of cellular signaling (discussed below). Furthermore, because such CCV is widespread, we have created a program enabling CCVA to be applied without specialized computational knowledge [33]. Overall, correlating CCV in protein abundance with heterogeneity in signaling responses is quite amenable because it requires simply adding a step of computational processing following the commonly used technique of flow cytometry.
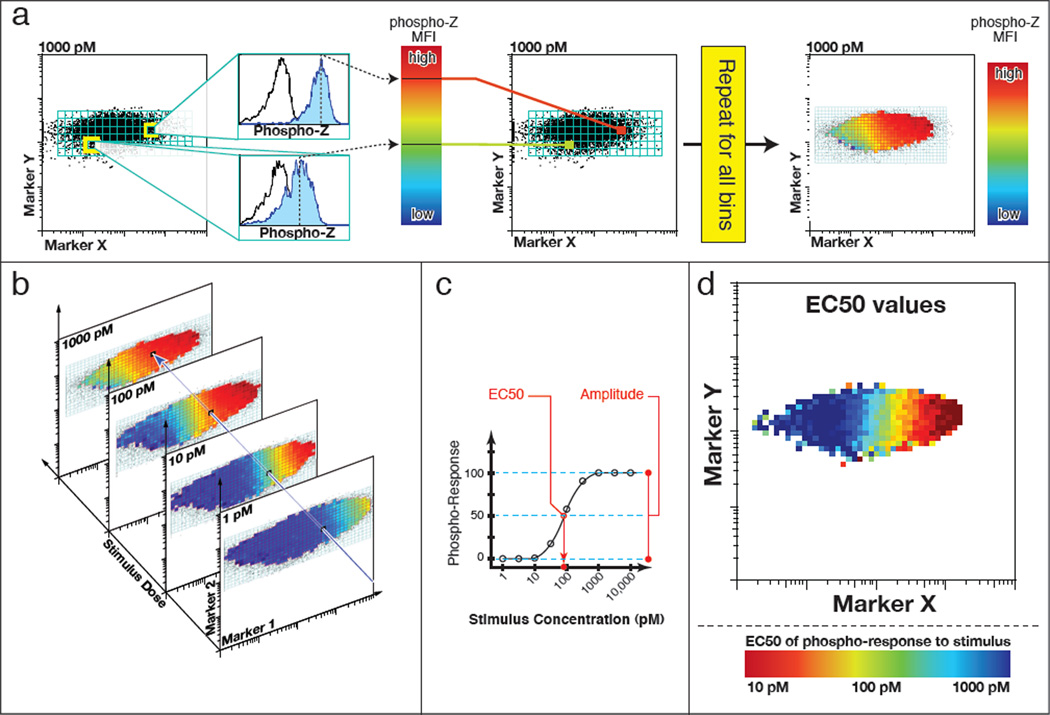
Figure 1. Cell-to-cell variability analysis (CCVA) computational processing.
(a) The first step in CCVA is binning across abundance of two markers of interest (X, and Y in our example). In each bin, for each dose of stimulus, the mean fluorescence intensity of a phospho-response is calculated (blue histogram), a histogram of an untreated control histogram is shown for comparison (open histogram). (b) The second stage is performed within each bin, across all doses of stimulus, wherein (c) a curve is fit describing the phospho-response as a function of varied doses of stimulus. This curve has two principal output parameters: EC50 and amplitude. The EC50, which describes the dose at which half of the maximum phospho-response stimulation is reached, provides a measure of sensitivity. The amplitude, which is the difference between the lower and higher response plateaus, measures the maximal responsiveness of the cells. The output of this analysis is a map of EC50 (d) and amplitude (not shown). In this example, Marker X has a strong correlation with sensitivity, with EC50s varying from 1 nM to 1 pM. Abundance of Marker Y, however, does not strongly influence stimulus sensitivity.
Connecting Cell-to-Cell Variability in Signaling Response to Phenotypic Variability
Developing an understanding of how CCV in signaling translates into differential decisions, such as differentiation or death, presents methodological challenges since signaling is best measured with fixed-cell methods, while outcomes must be observed at later time points. Thus, connecting CCV in signaling with CCV in phenotype requires either connection of multiple snapshots of signaling, or the application of additional methods, such as computational modeling or biochemistry.
Chen et al [31] examined the connection between ERK and Akt signaling and neuronal differentiation in PC12 cells responding to EGF or NGF (epidermal and neural growth factors, respectively). Signaling through phospho-ERK (ppERK) and phospho-Akt (pAkt), and the choice between differentiation and proliferation were measured using microscopy of fixed cells, 24 hours following treatment with either growth factor. A clear map could be derived, whereby the combination of ppERK and pAkt levels identified regions of propensity towards either differentiation or proliferation. While this map provided a correlation between variable signaling and heterogeneous differentiation, discerning the cause-effect direction required intervention, both chemical (inhibitors) and genetic (siRNA). Through an siRNA screen, the group was able to identify a number of regulators that shifted the map of ppERK and pAkt, and identify a novel regulatory crosstalk between ppERK and pAkt pathways downstream of NGF stimulation. It would be interesting to further examine whether CCV in key regulatory proteins of the ERK and Akt signaling cascades maps out the ppERK-pAkt responsiveness, and how the variability of this map is regulated.
Bridging CCV in Protein abundance to divergence in phenotype
Linking variability in protein expression to heterogeneous cellular fates entails the complete connection of protein abundance, signaling regulation, and eventual phenotype, and presents the possibility of fully understanding the implications of CCV.
Biochemical models can provide the link between variability in protein abundance and variability in phenotype by simulating signaling. This requires that the signaling cascades be well characterized, restricting the freedom with which a model can be fit to a given set of experimental results. As a prime example of the utility of models in such a study, Spencer et al [12] used an established model of TNF-related apoptosis-inducing ligand (TRAIL) signaling [12] to connect variability in apoptotic regulators, such as Bcl and Bid, with heterogeneity in the timing of apoptosis. The measured variation in apoptotic regulators was small (coefficients of variation ranging from 0.21 – 0.28), and no single protein’s abundance predicted the time to death. However, a model of the collective, uncorrelated variation could predict the full range of times to death seen in the population of cells given low doses of TRAIL. This result presents an interesting conundrum: given that such small variability in a handful of proteins can lead to a wide divergence in a life-or-death decision, how do cells ensure that they respond appropriately to an apoptotic signal? The answer to this question becomes more clear when the multicellular organism, rather than the individual cell, is considered the unit of evolutionary selection: low doses of TRAIL, the variability in the system converts an all-or-none response at the individual level into a graded response at the population level, allowing the population as a whole to commit only partially in response to an uncertain signal, providing a benefit similar to that seen in bet hedging.
Building better computational models using heterogeneity
We posit here that cell-to-cell variability analysis is a useful method to develop and test quantitative models of signaling. Biochemically explicit models, as illustrated in key studies of TCR signaling, TRAIL-induced apoptosis and cytokine response [10,12,16,34] have been proposed to account for the average response of cells to external stimuli. The natural variability seen in cell populations provides the opportunity to both characterize individual proteins’ roles in signaling networks and quantitatively constrain models of signaling.
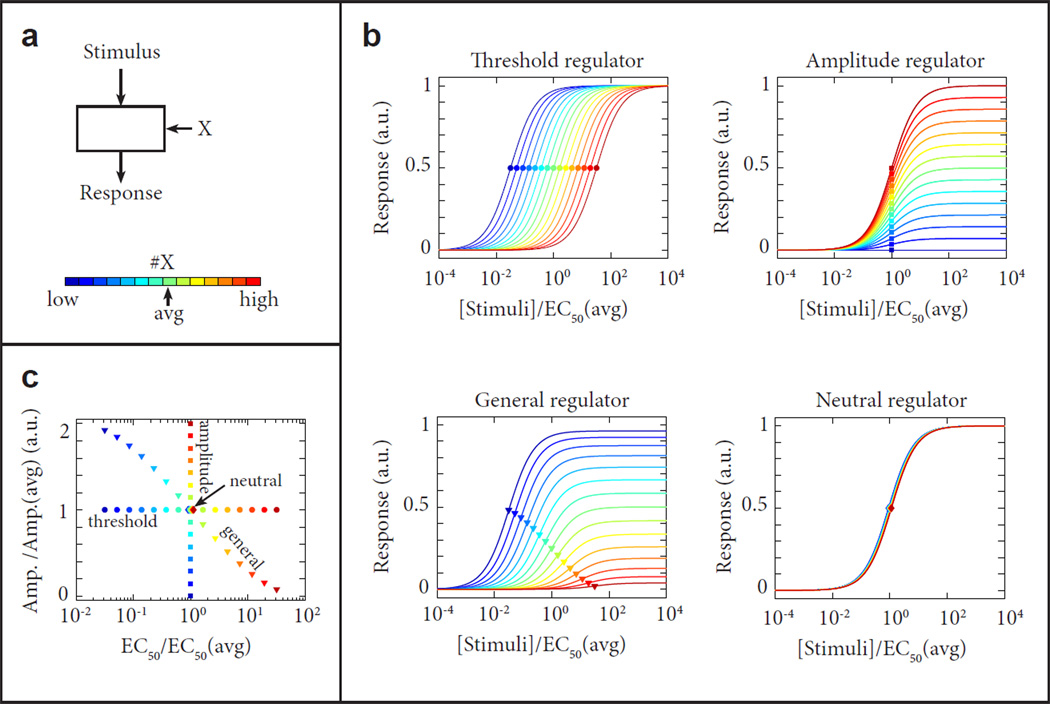
To better understand the structure of networks, their component signaling regulators’ effects can be classified along the dimensions of positive, neutral or negative, and threshold or amplitude, based on the effect that variable abundance of these regulators have on the signaling response (see figure 2). In our experience, neutral regulators are the most common: the abundance of a given protein does not impact the input/output relationship, a property known as robustness, common in enzymatic systems in which the substrate is not limiting. Threshold and amplitude regulators are defined by their impact on parameters characterizing the dose-response to stimuli, namely EC50 and amplitude, respectively. For example, in the case of TCR signaling, CCVA was used to identify a proximal signaling component (CD8) as a threshold regulator, a feedback phosphatase (Shp-1) as an amplitude regulator and the enzymatic substrate ERK as a neutral regulator, in agreement with model predictions [10]. In the case of the IL-2 receptor, IL- 2Rα was identified as a threshold regulator of STAT5 phosphorylation in response to IL-2, whereas il2rb was identified as an amplitude regulator [16], in concordance with a model whereby preformation of the IL-2 receptor regulates sensitivity to IL-2 [33].
Figure 2. Identification and classification of signaling regulators.
(a) Schematic diagram of a signaling module. The input-output relationship must be evaluated as a function of the abundance of a regulatory component X. (b) For component X, deviations from the average abundance can have different effects on the input-output relationship of the signaling module. Based on such effect, regulatory components can be identified and classified. Using their effect on the relative EC50 of the response (threshold regulators), its amplitude (amplitude regulators), or both (general regulators). The input-output relationship can also be largely unaffected by changes in the abundance of a component X, in which case the component is identified as a neutral regulator. Examples of threshold, amplitude, general and neutral regulators are given here. (c) Diagram of the Relative Amplitude and EC50 for the deviation from the average response when the abundance of component X varies. These deviations classify the regulators as in (b) as threshold, amplitude, general and neutral are shown.
Alternatively, the rich experimental observables generated by analysis of variable populations of cells can be used to better constrain or refine mathematical models of signaling [24], or to uncover connections that would be invisible if observed at an average level [33]. As an example, Tay et al. observed heterogeneous responses to TNF stimulation through single cell tracking of NFκb localization dynamics. To account for this heterogeneity, the researchers constructed a model of NFκb regulation that relied on stochasticity of transcription to create cell-to-cell variability in the response to TNF. We propose that experimental measurement of CCV in TNF response regulators would add quantitative observations to further test the proposed mechanism for the generation of heterogeneous responses in this system. Such an analysis was performed for a model of TRAIL signaling, using quantitative single cell measurements to experimentally validate the model’s parameter sensitivity [35,36]. Similarly, in the context of the signaling response to common gamma cytokines among T lymphocytes, this stringency led to reconsideration [33] of a previous model of IL-2 signaling [16]. CCVA revealed an inhibitory effect of the IL-2 receptor alpha chain (IL-2Rα) abundance on IL-7 sensitivity. As IL-2Rα is directly involved only in IL-2 signaling, this necessitated an expansion of the model for IL-2 receptor formation to allow competition for the common gamma chain, a signaling chain shared with the receptors for IL-7 and other cytokines [33,37]. In this case, the greater stringency placed on the model by CCVA necessitated an increased number of parameters than would have been statistically valid without the inclusion of variability.
Leveraging cell-to-cell variability to interpret mutational sensitivity
With the proliferation of whole-genome sequencing of tumors, the number of potentially oncogenic mutations has multiplied, while the development of novel methods to characterize the functional significance of these mutations has lagged. Some of these mutations imply dominant effects on phenotypes, allowing genetic approaches such as siRNA knockdown to serve as sufficient validation. For more subtle cases, in which mutations marginally alter the enzymatic activities or the binding affinities of proteins, more subtle methods such as CCVA may prove better suited. CCVA has the ability to identify the nodes of the signaling network that have significant analog or digital effects (see Figure 2), for which marginal increase in abundance may serve as a suitable analogy to the effects of a marginal increase in enzymatic activity.
This analogy could work in reverse as well: The natural heterogeneity of abundance in protein expression could help identify outlier subpopulations whose extremes of signal regulator expression produce the same overactive signaling that results from oncogenic transformation [18,38]. Single-cell resolution of cellular responses could identify how outlier subpopulations with aberrant phenotypes are kept in check in healthy individuals, or contribute to oncogenesis when these checks fail [39].
Conclusion
In this review, we have presented studies that connect different layers of cell-to-cell variability with the goal of explaining how continuous distributions of proteins, through divergent signaling, lead to heterogeneous phenotypes. In our view, CCV appears to be a default characteristic of protein expression across a population, an inherent feature of the eukaryotic protein expression machinery. Specific mechanisms to limit CCV arise only when variability would be deleterious. In other cases, CCV in “neutral regulators” may remain unregulated, since it has no effect on overall phenotype. Most interestingly, however, CCV for threshold and amplitude regulators may provide a preexisting mechanism to endow populations of cells with phenotypic variability that diversifies their response to environmental cues. Hence cells can rely on CCV to adapt to varied stimuli without having to evolve specific regulatory pathways. The studies presented in this review have begun to address this larger question of how CCV contributes to evolutionarily adaptive developmental or bet-hedging mechanisms. These studies used single-cell methods to connect two layers of variability, and diverse methods such as modeling and/or biochemistry to create a fuller picture of the significance of variability to the function of the cell. Systems allowing direct connections between all of these layers will be highly valuable to forge a mechanistic understanding of how such complexes processes as differentiation and apoptosis are regulated at both cellular and population levels.
Highlights.
Cell-to-cell variability is ubiquitous in genetically identical cells.
Variability exists at multiple levels: genes, proteins, signaling and phenotypes.
New methods have been developed to connect different levels of variability.
Variability provides insight into cells’ function, individually and in populations.
Computational modeling maps out variability that in turns helps improve models.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- 2.Niepel M, Spencer SL, Sorger PK. Non-genetic cell-to-cell variability and the consequences for pharmacology. Current opinion in chemical biology. 2009;13:556–561. doi: 10.1016/j.cbpa.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelkmans L. Using cell-to-cell variability--a new era in molecular biology. Science. 2012;336:425–426. doi: 10.1126/science.1222161. [DOI] [PubMed] [Google Scholar]
- 4.Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. 2008;135:216–226. doi: 10.1016/j.cell.2008.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai L, Friedman N, Xie XS. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- 6.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008;453:544–547. doi: 10.1038/nature06965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Yuan TL, Wulf G, Burga L, Cantley LC. Cell-to-cell variability in PI3K protein level regulates PI3K-AKT pathway activity in cell populations. Current biology. 2011;21:173–183. doi: 10.1016/j.cub.2010.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen J-Y, Lin J-R, Cimprich Ka, Meyer T. A two-dimensional ERK-AKT signaling code for an NGF-triggered cell-fate decision. Molecular cell. 2012;45:196–209. doi: 10.1016/j.molcel.2011.11.023. •• This work uses microscopy and extensive biochemistry to understand the mechanisms by which heterogeneity in ERK and AKT signaling leads to heterogeneity in the decision to differentiate or proliferate.
- 10.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suel GM, Kulkarni RP, Dworkin J, Garcia-Ojalvo J, Elowitz MB. Tunability and noise dependence in differentiation dynamics. 2007;315:1716–1719. doi: 10.1126/science.1137455. [DOI] [PubMed] [Google Scholar]
- 15.Caǧatay T, Turcotte M, Elowitz MB, Garcia-Ojalvo J, Süel GM. Architecture-dependent noise discriminates functionally analogous differentiation circuits. Cell. 2009;139:512–522. doi: 10.1016/j.cell.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 16.Feinerman O, Jentsch G, Tkach KE, Coward JW, Hathorn MM, Sneddon MW, Emonet T, Smith KA, Altan-Bonnet G. Single-cell quantification of IL-2 response by effector and regulatory T cells reveals critical plasticity in immune response. Molecular systems biology. 2010;6:437. doi: 10.1038/msb.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cotari J, Voisinne G, Even Dar O, Karabacak V, Altan-Bonnet G. Dissecting common γ chain cytokine family signaling in T cells using cell-to-cell variability analysis. Science Signaling. 2013 doi: 10.1126/scisignal.2003240. Pre-publication. •• This work provides a full explanation of cell-to-cell variability analysis (CCVA), and an example of using this technique to drive computational modeling of signal transduction.
- 18.Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14:1938–1947. doi: 10.1101/gr.2890204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kolitz SE, Lauffenburger Da. Measurement and Modeling of Signaling at the Single-Cell Level. Biochemistry. 2012 doi: 10.1021/bi300846p. •• This review presents a comprehensive treatment of the technical aspects of single-cell analysis, including measurement and modeling.
- 21.Beuneu H, Lemaitre F, Deguine J, Moreau HD, Bouvier I, Garcia Z, Albert ML, Bousso P. Visualizing the functional diversification of CD8+ T cell responses in lymph nodes. Immunity. 2010;33:412–423. doi: 10.1016/j.immuni.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Cohen-Saidon C, Cohen Aa, Sigal A, Liron Y, Alon U. Dynamics and variability of ERK2 response to EGF in individual living cells. Molecular cell. 2009;36:885–893. doi: 10.1016/j.molcel.2009.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Iyer J, Reich NC. Constitutive nuclear import of latent and activated STAT5a by its coiled coil domain. The FASEB Journal. 2008;22:391–400. doi: 10.1096/fj.07-8965com. [DOI] [PubMed] [Google Scholar]
- 24.Tay S, Hughey JJ, Lee TK, Lipniacki T, Quake SR, Covert MW. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomida T, Oda S, Takekawa M, Iino Y, Saito H. The Temporal Pattern of Stimulation Determines the Extent and Duration of MAPK Activation in a Caenorhabditis elegans Sensory Neuron. Science Signaling. 2012;5 doi: 10.1126/scisignal.2002983. ra76-ra76. [DOI] [PubMed] [Google Scholar]
- 26.Garrod KR, Moreau HD, Garcia Z, Lemaitre F, Bouvier I, Albert ML, Bousso P. Dissecting T cell contraction in vivo using a genetically encoded reporter of apoptosis. Cell Rep. 2012;2:1438–1447. doi: 10.1016/j.celrep.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 27.Loewer A, Lahav G. We are all individuals: causes and consequences of non-genetic heterogeneity in mammalian cells. Current opinion in genetics & development. 2011;21:753–758. doi: 10.1016/j.gde.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud O, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. 2004;118:217–228. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Krutzik PO, Clutter MR, Nolan GP. Coordinate analysis of murine immune cell surface markers and intracellular phosphoproteins by flow cytometry. J Immunol. 2005;175:2357–2365. doi: 10.4049/jimmunol.175.4.2357. [DOI] [PubMed] [Google Scholar]
- 30.Krutzik PO, Hale MB, Nolan GP. Characterization of the murine immunological signaling network with phosphospecific flow cytometry. J Immunol. 2005;175:2366–2373. doi: 10.4049/jimmunol.175.4.2366. [DOI] [PubMed] [Google Scholar]
- 31.Anderson MS, Venanzi ES, Chen Z, Berzins SP, Benoist C, Mathis D. The cellular mechanism of Aire control of T cell tolerance. Immunity. 2005;23:227–239. doi: 10.1016/j.immuni.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 32.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, et al. Dynamic proteomics of individual cancer cells in response to a drug. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 33.Cotari JW, Voisinne G, Dar OE, Karabacak V, Altan-Bonnet G. Cell-to-Cell Variability Analysis Dissects the Plasticity of Signaling of Common {gamma} Chain Cytokines in T Cells. Science Signaling. 2013;6 doi: 10.1126/scisignal.2003240. ra17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albeck JG, Burke JM, Aldridge BB, Zhang M, Lauffenburger DA, Sorger PK. Quantitative analysis of pathways controlling extrinsic apoptosis in single cells. Mol Cell. 2008;30:11–25. doi: 10.1016/j.molcel.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaudet S, Spencer SL, Chen WW, Sorger PK. Exploring the Contextual Sensitivity of Factors that Determine Cell-to-Cell Variability in Receptor-Mediated Apoptosis. PLoS Comput Biol. 2012;8:e1002482. doi: 10.1371/journal.pcbi.1002482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aldridge BB, Gaudet S, Lauffenburger DA, Sorger PK. Lyapunov exponents and phase diagrams reveal multi-factorial control over TRAIL-induced apoptosis. Mol Syst Biol. 2011;7:553. doi: 10.1038/msb.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nature reviews. Immunology. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brock A, Chang H, Huang S. Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet. 2009;10:336–342. doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]