Abstract
The pluripotent state, which is first established in the primitive ectoderm cells (PE) of blastocysts, is lost progressively and irreversibly during subsequent development1. For example, development of postimplantation epiblast from PE involves significant transcriptional and epigenetic changes, including DNA methylation and X inactivation2, which creates a robust epigenetic barrier and prevents their reversion to a PE-like state. Epiblast cells are refractory to leukaemia inhibitory factor (LIF)-STAT3 signaling, but they respond to Activin/bFGF to form self-renewing epiblast stem cells (EpiSC), which exhibit essential properties of epiblast cells3,4, that differ from embryonic stem cells (ESC) derived from PE5. Here we show reprogramming of advanced epiblast cells from E5.5 - E7.5 embryos with uniform expression of N-cadherin and inactive X chromosome, to ES-like cells (rESC) in response to LIF-STAT3 signaling. Cultured epiblast cells (cEpi) overcome the epigenetic barrier progressively as they proceed with the erasure of key properties of epiblast cells, involving DNA demethylation, X reactivation and expression of E-cadherin. The accompanying changes in the transcriptome result in a loss of phenotypic and epigenetic memory of epiblast cells. Notably, using this new approach, we report reversion of established EpiSC to rESC. Furthermore, unlike epiblast and EpiSC, rESC contribute to somatic tissues and germ cells in chimeras. This is a tractable model to investigate signaling molecule induced epigenetic reprogramming that can promote reacquisition of the fundamental pluripotent state.
Previous studies showed that epiblast cells, unlike PE, are refractory to LIF-STAT3 signaling3,4; instead they respond to Activin/bFGF to generate self-renewing EpiSC. EpiSC differ epigenetically from ESC, as they have an inactive X-chromosome and they cannot form chimeras when introduced into blastocysts. However, we set out to re-examine if postimplantation epiblast cells could undergo reprogramming to ESC-like cells in response to LIF-STAT3 signaling. We isolated epiblast tissue on embryonic day (E) E5.5 - E7.5 from transgenic embryos with an Oct4-ΔPE-green fluorescent protein (GFP) reporter6. This reporter, with the distal enhancer and lacking the proximal enhancer for Oct4, shows preferential expression in the PE, primordial germ cells (PGC) and ESC only, but not in the epiblast or EpiSC6. Notably, the distal enhancer of Oct4 is an enhanceosome representing the densest binding locus for the key pluripotency-specific transcripts in ESC7; its activation occurs only when all pluripotency-associated factors are expressed optimally as in PE and ESC; these must be lacking in the epiblast and its derivative, EpiSC.
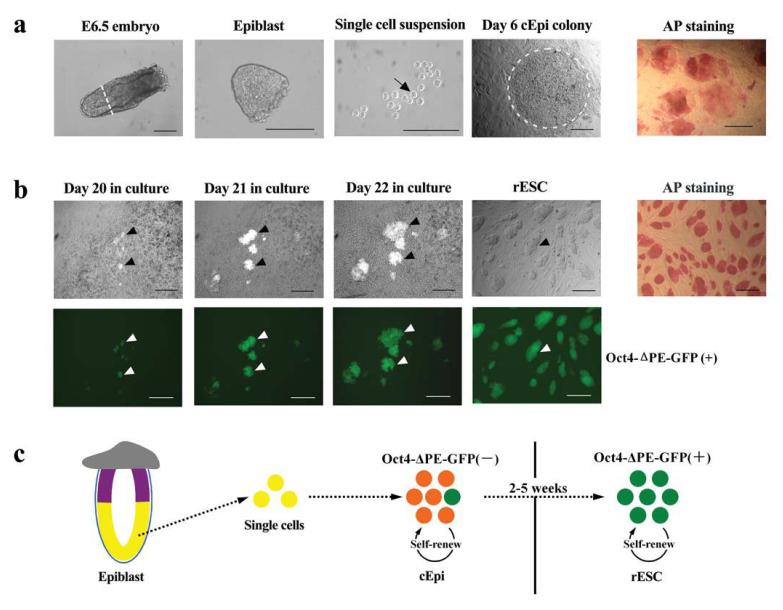
Next, for the culture of epiblast, we used LIF and fetal calf serum (FCS) on mouse embryonic fibroblasts feeder cells (MEFs), which is the standard condition used for the derivation of ESC from PE, and for reprogramming of somatic cells to induced pluripotent stem cells (iPS)5,8-11. The epiblast tissue was dissected to remove the most proximal region (the site of PGC and PGC precursors2), and the outer visceral endoderm (Fig. 1a). All the epiblast cells uniformly showed an inactive X-chromosome, and were positive for N-cadherin (see below). Notably, we then trypsinised the epiblast tissue and used single cell suspension from individual epiblasts for culture, unlike previous studies where the epiblast tissue was left intact3,4. Disruption of the epiblast, which undergoes rapid differentiation in vivo, may break the existing cell-cell interactions and permit establishment of a new signaling-induced transcriptional network in vitro. The majority of the resulting cultures revealed large colonies after about 4 - 7 days (Fig. 1a and Table 1), with many alkaline phosphatase (AP) positive cells (Fig.1a). However, all the epiblast cells at the outset, and those forming the colonies were negative for Oct4-ΔPE-GFP reporter expression, indicating that the distal enhancer was yet inactive (Fig. 1a, b). We could propagate the cEpi colonies following collagenase treatment without significant detectable morphological changes for at least 20 passages, suggesting that these cells can be maintained in LIF/FCS.
Figure 1.
Reprogramming epiblast cells from E6.5 embryos to generate rESC. (a) Derivation of cEpi from E6.5 epiblast. Epiblast tissue was divested of the proximal region (white line, panel 1) and visceral endoderm; a single cell suspension (black arrow) was cultured, which formed cEpi colonies. Note AP positive cells in cEpi in the last panel. (b) Derivation of rESC from cEpi. Note the appearance of clusters of Oct4-ΔPE-GFP-positive cells in cEpi colonies (black arrowheads), and corresponding white arrowheads for GFP in the panel below. Note that the rESC are uniformly AP positive. Scale bar: 100μm. (c) Schematic representation of reprogramming of epiblast through cEpi, and finally rESC.
Table 1.
Derivation of rESC lines from mouse postimplantation embryos. The embryos were isolated from E5.5 - E7.5 129/sv or ROSA129/Sv females mated with Oct4-ΔPE-GFP transgenic males. The epiblast part was dissected and digested into single cell suspension. cEpi colonies emerged after 4 - 7 days in culture, and rESC were 20 observed after 14 - 35 days.
| Transgenic lines | Stage of embryos | No. of embryos | No. of cEpi lines |
No. of rESC lines |
|---|---|---|---|---|
| Oct4-ΔPE-GFP | E5.5 | 22 | 14/22 (64%) | 6/22 (27%) |
| Oct4-ΔPE-GFP | E6.5 | 17 | 10/17 (59%) | 4/17 (24%) |
| Rosa-LacZ/Oct4-ΔPE-GFP | E6.5 | 28 | 22/28 (79%) | 10/28 (36%) |
| Oct4-ΔPE-GFP | E7.5 | 23 | 7/23 (30%) | 5/23 (22%) |
Following culture of cEpi for 14 - 35 days, we started to detect expression of Oct4-ΔPE-GFP reporter for the first time, which were seen as individual clusters of GFP-positive cells within cEpi colonies, indicating activation of the distal enhanceosome of the Oct4-ΔPE-GFP reporter and a possible reversion to an ESC-like state6. Subsequent culture of GFP-positive cells was carried out after disruption of cEpi colonies by treatment with trypsin, which is detrimental to the survival of cEpi but promotes propagation of ESC-like cells. Subsequent passaging of cells confirmed that the GFP-positive cells had started to acquire ESC-like morphology with uniform GFP expression (Fig. 1b). We call these cells, reprogrammed epiblast-ES-like cells (rESC).
The frequency of rESC derivation from E5.5 - E7.5 epiblast was not only relatively high at around 22 - 36% (Table 1), but importantly, this frequency of derivation did not diminish with increasing developmental age. This excludes a possibility that rare cells within the epiblast undergo reprogramming since, if they exist, we might expect them to diminish in number with increasing developmental age. Note also that the original epiblast cells were uniformly negative for the Oct4-ΔPE-GFP reporter but positive for N-cadherin and inactive X-chromosome. Furthermore, AP staining (a hallmark of cells undergoing reprogramming) of cEpi is more extensive and not confined to a few isolated cells (Fig. 1a), which are sustained by LIF-STAT3 under our culture procedure. It is from these cEpi colonies that clusters of GFP-positive ESC-like cells gradually emerge during subsequent culture (Fig. 1b). This is remarkable since no such ESC-like cells have previously been derived from embryos as late as E7.5; derivation of ESC has been from PE present in the inner cell mass (ICM) in blastocysts5,7,10,11 . Note that the previously described EPL cells12,13 and FAB-SC14, were derived from preimplantation or implanting blastocyst (Supplementary Table 1), and not from the advanced postimplantation epiblast cells as we report here. Furthermore, neither of them was examined for their epigenetic state nor for the status of the X chromosome.
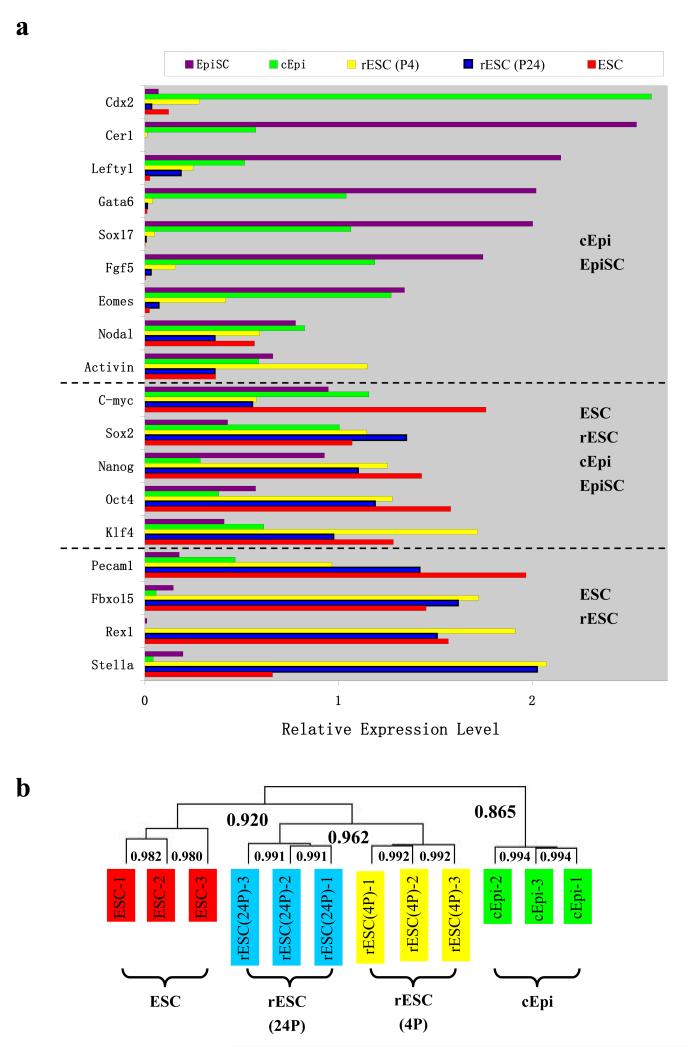
To gain insight into how epiblast cells undergo reprogramming, we examined changes in gene expression, which was evident from the progressive increase in AP staining in cEpi as they advanced towards rESC (Fig. 1a-c). Analysis of the transcriptome revealed that the cEpi were more closely related to their epiblast origin, as is the case for EpiSC3,4. Thus, cEpi showed strong expression of Eomes, Fgf5, Sox17, Gata6, Lefty1 and Cer1, but there was little expression of Stella, Pecam1, Rex1 and Fbxo15. By contrast, rESC showed the opposite transcriptome profile, with an increase in the expression of Stella, Pecam1and Rex1, and a concomitant loss of Fgf5, Eomes and Sox17 expression, which is indicative of progressive reprogramming of epiblast derived cEpi to rESC phenotype (Fig. 2a). There was also an overall and progressive increase in expression of the key pluripotency-specific genes Oct4, Sox2, Nanog, Rex1, and Fbxo15 that reached levels comparable to ESC (Fig. 2a and Supplementary Fig. 1, 2b). The advancement of reprogramming was also evident when comparing early passages rESC (passage 4: P4) with slightly higher expression levels for genes including Eomes and Lefty1, compared to their expression in P24 rESC (Fig. 2a), suggesting a progressive loss of a residual ‘memory’ of their epiblast origin (see below). Comprehensive whole-genome microarray analysis confirmed that rESC are similar to control ESC and differ from cEpi (Fig. 2b, and Supplementary Fig. 3). These overall changes in the transcriptome must account for the distal enhancer-driven activation of the Oct4-ΔPE-GFP reporter in rESC.
Figure 2.
Changes in gene expression profile. (a) Reverse transcription real-time PCR of marker genes in EpiSC, cEpi, rESC at early (P4) and late (P24) passages, and ESC. Note progressive loss of markers of epiblast detected in cEpi and EpiSC (at the top) and enhancement of expression of genes in rESC that resemble ESC (b) Whole-genome cluster analysis of transcriptomes of cEpi, rESC at early (P4) and late (P24) passages, and ESC. The labeled numbers are the corresponded Pearson correlation coefficients between different cDNA samples. Note that rESC resemble ESC and not cEpi that are more like the original epiblast cells as described above. Note also the changes between the early (P4) and late (P24) passages of rESC.
We next examined if LIF-STAT3 signaling is critical for the observed reprogramming of epiblast cells. We detected STAT3-phosphorylation in cEpi suggesting that these cells, like rESC and ESC respond to LIF signaling (Supplementary Fig. 4a). Notably, addition of the JAK inhibitor (Calbiochem) that prevents phosphorylation of tyrosine 705 of STAT3 to culture of E6.5 epiblast cells, initially allowed some cEpi colonies to develop, but they gradually differentiated and failed to undergo reprogramming to rESC (Supplementary Table 2). This demonstrates that LIF-STAT3 is crucial for sustaining cEpi and their reprogramming to rESC. Furthermore, culture of rESC in medium with the JAK inhibitor caused a striking reversal towards the transcriptional state of cEpi, with a decline in the expression of Stella, Pecam1 and Rex1, and a concomitant increase in the expression of Fgf5 and Eomes, whereas culture of cEpi for three days resulted in no significant changes in the transcriptional profile (Supplementary Fig. 4b). A number of STAT3 targets have been identified in ESC7, including Fbxo15, Rex1 and Stat3 itself, and of the epigenetic modifiers, Lin28, Ezh2 and Mbd3, suggesting that STAT3 has the potential to influence the transcriptional and epigenetic state of cEpi.
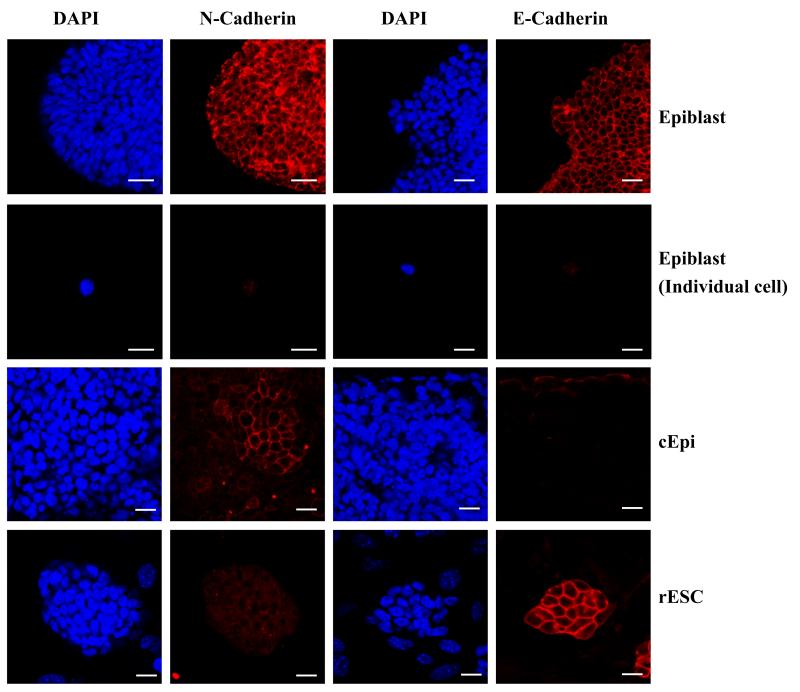
To observe the dynamic changes in cell surface properties during reprogramming of cEpi to rESC, we examined progressive changes in the expression of E-cadherin and N-cadherin in E6.5 epiblast cells (Fig. 3). Whereas expression of both E-cadherin and N-cadherin was detected in E6.5 epiblast uniformly, trypsinisation of these cells prior to culture led to the loss of these adhesion molecules. During subsequent culture, we then detected heterogeneous N-cadherin expression in cEpi. However, with further reprogramming to rESC, there was a complete loss of N-cadherin, which was replaced by uniform expression of E-cadherin. This observation is consistent with the evidence suggesting that LIF-STAT3 promotes up regulation of E-cadherin to levels detected in ESC14. Thus, changes in the cell surface property reveal the dynamic nature of the process that is promoted by LIF-STAT3 signaling.
Figure 3.
Dynamic changes in cell surface adhesion molecules; both E-cadherin and N-cadherin are detected uniformly in E6.5 epiblast, which is undetectable in single cell suspension. During culture, N-cadherin expression is heterogeneous in cEpi and eventually disappears completely and replaced by E-cadherin in rESC. Scale bar: 20μm.
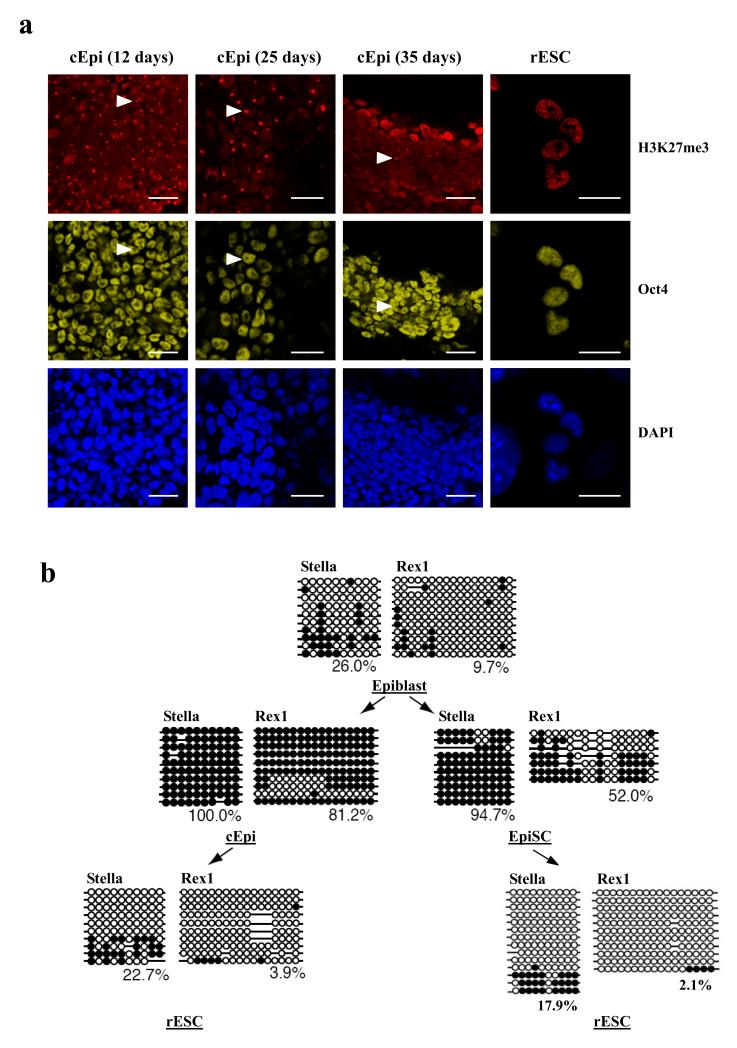
Next, we examined epigenetic changes in epiblast during reprogramming to rESC. Notably, reactivation of the late-replicating inactive X chromosome in the epiblast15, would indicate a major epigenetic change16, since we found that all of the E6.5 epiblast cells (96/96 cells; Supplementary Fig 5a) had the characteristic accumulation of histone H3 lysine 27 trimethylation (H3K27me3) epigenetic mark, which is diagnostic for the inactive X17,18. Furthermore, after culture of epiblast cells for 12 days, nearly all of the cEpi (99/100 cells) still had inactivated X-chromosome, but after 25 days, there was evidence for gradual X reactivation since only 62% of the cells (37/60 cells) had the H3K27me3 ‘spot’ (Fig. 4a). The number of cells with the inactive X declined further to only 9% after 35 days of culture (7/80 cells). These observations show continuing epigenetic reprogramming of epiblast cells through cEpi phenotype towards reversion to rESC, where none of the cells had the characteristic H3K27me3 spot. Reactivation of the X chromosome, a hallmark of epigenetic reprogramming, is also seen following somatic nuclear transplantation into oocytes, in mouse iPS cells, as well as in ESC-somatic cell hybrids19-23.
Figure 4.
Epigenetics changes during reprogramming of epiblast cells. (a) Female cEpi exhibit uniform accumulation of H3K27me3 associated with the inactive X (white arrowhead), which is gradually lost from individual cells during culture and finally lost in rESC. Scale bar: 20 μm (b) Changes in DNA methylation of Stella and Rex1 during reprogramming of epiblast. Although Stella and Rex1 are repressed in epiblast cells, these loci are initially unmethylated; they undergo DNA methylation transiently in cEpi and stably in EpiSC. Reprogramming to form rESC results in the loss of DNA methylation.
Next, we examined epigenetic changes with respect to DNA methylation of the promoter region of two pluripotency genes, Stella and Rex1. Both these genes, (and others such as Pecam1) are repressed in the epiblast but active in PE and ESC24. Initially, the promoter regions of Stella and Rex1 were unmethylated in the epiblast, but they may be poised for enduring repression by DNA methylation (Fig. 4b). Indeed, Stella and Rex1 loci undergo DNA methylation during the derivation of EpiSC and remain repressed thereafter (see below). This is also the case in cEpi, where both Stella and Rex1 loci became methylated, albeit transiently (Fig. 4b), since with continuing culture of cEpi, they became demethylated. This represents an important epigenetic reprogramming event towards rESC, leading to the changes in the transcriptome described above. Consistently, we also found that rESC derived from epiblast cells with the Stella-GFP reporter, showed activation of this reporter (Supplementary Fig. 2a). Once established, the rESC epigenotype was stable and heritable. The epigenetic and transcriptional changes in rESC distinguish them from EpiSC derived in response to Activin/bFGF signaling, which retain key properties of epiblast cells, including inactive X and with some key genes such as Stella, Pecam1 and Rex1 in a repressed state. These findings are relevant for human ESC (hESC), which resemble EpiSC, and not ESC or rESC.
Although very unlikely, we wanted to exclude a possibility that rESC could have originated from PGC in epiblast cultures. First, the epiblast tissue used in our experiments was dissected away from the most proximal region to exclude PGC precursors in E6.5, and PGC from E7.5 embryos. Importantly, there is no further allocation to PGC from epiblast after E7.5 (Ref. 25). Second, PGC could not survive in the culture conditions used to propagate cEpi, since they require FGF2 and stem cell factor (SCF) for proliferation, as well as for dedifferentiation into pluripotent embryonic germ cells (EGC). It is known that ESC largely retain the parent of origin dependent DNA methylation of imprinted genes, which are fully or partially erased in EGC derived from E8.5 PGC26,27. We found that the imprinting status in rESC was maintained as in ESC and EpiSC. This was confirmed with analysis of methylation of three imprinted genes in rESC derived from E6.5 cEpi, E7.5 cEpi, and E6.5 EpiSC (see below) (Supplementary Fig. 5c). The cumulative evidence makes it unlikely that rESC could originate from PGC.
In view of our findings demonstrating reversion of the postimplantation epiblast cells, we went onto to test if EpiSC, which resembles epiblast when cultured in Activin/bFGF over a long period, could undergo reversion to rESC in our culture with LIF. We chose two different EpiSC lines; one with the Oct4-ΔPE-GFP reporter (Passage 20), and the other with an X-GFP reporter28. EpiSC with the X-GFP reporter were FACS sorted to establish lines where the reporter was repressed because of its location on the inactive X chromosome (Passage23) (Supplementary Fig. 5b). Hence, neither of these EpiSC lines showed GFP expression suggesting their stable repression in culture containing Activin/bFGF, which have AP positive cells (Supplementary Fig. 6a). Following their culture with LIF on feeder cells for 10 to 20 days, we detected GFP-positive cells, from which we established rESC as before. Expression of the X-GFP reporter indicates reactivation of the inactive X. Furthermore, we also found that Stella and Rex1 that are stably repressed in EpiSC by DNA methylation, became de-repressed in rESC following DNA demethylation of their promoter regions (Fig. 4b).
In two previous studies, reversion of EpiSC in response to changes in culture conditions proved impossible without the introduction of the exogenous Klf4 or c-Myc transcription factor29,30. However, in our culture procedure containing LIF-FCS on feeder cells are evidently conducive for the initial survival of EpiSC before their reversion to rESC without requiring exogenous transcription factors. We had previously shown that ESC in LIF-FCS exist in a metastable state and fluctuate between ESC and epiblast-like states but without proceeding completely to the EpiSC-like state. The presence of feeders also apparently helps to promote a shift towards ESC-like state24.
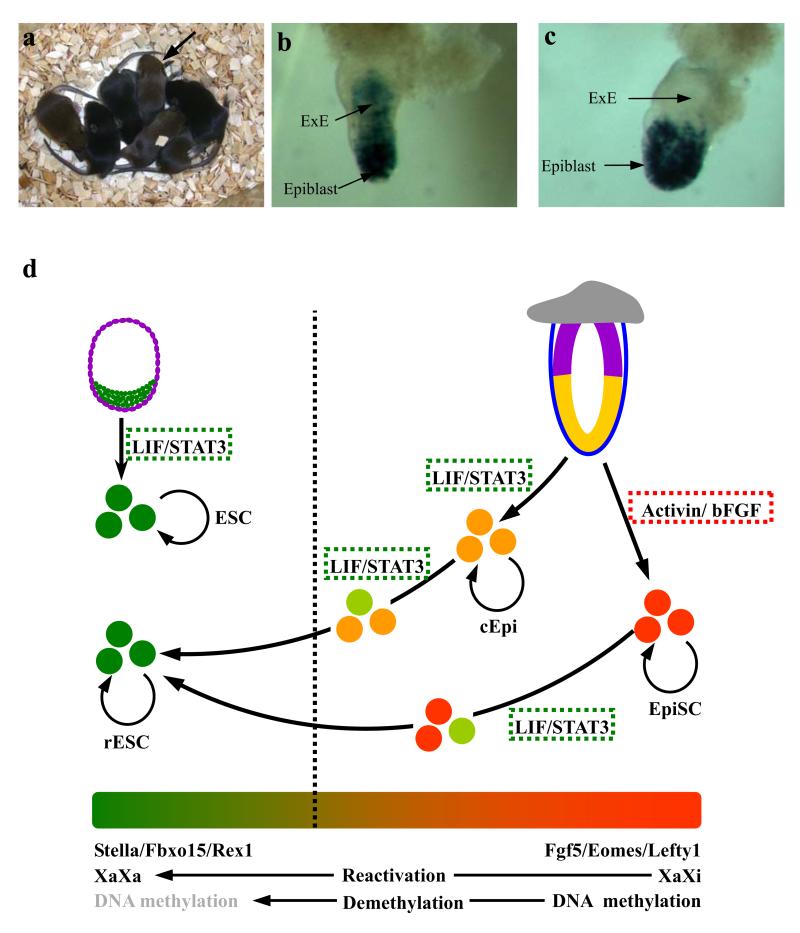
Finally, we tested the developmental potential of rESC derived from EpiSC and postimplantation epiblast in chimeric embryos, by introducing them into normal blastocysts. Using rESC from epiblast with the ROSA-lacZ reporter, we detected an extensive contribution in developing embryos (Supplementary Fig. 7), as well as in adults with germ line transmission, a key property of pluripotency (Fig. 5a, Supplementary Fig. 7 and Supplementary Table 3). While the early passage rESC from epiblast also contributed to the extraembryonic ectoderm (ExE; a derivative of the trophectoderm lineage) in E6.5 embryos (Fig. 5b), this was not the case with late passage rESC (Supplementary Table 5 and Fig. 5c). This suggests transient retention of the residual memory in rESC of their epiblast origin, a characteristic shared with EpiSC that are prone to differentiation into trophectoderm cells in vitro3,4 (Supplementary Table 5). With progressive changes in their transcriptome and the epigenetic state, rESC undergo reprogramming and erase all the key characteristics of their origin from epiblast cells. Most significant was also the observation that the rESC derived from EpiSC could participate in chimeras and contribute to the germ line in E13.5 embryos (Supplementary Fig. 6b and Supplementary Table 4). It has not proved possible to generate chimeras with EpiSC3,4, indicating that functionally, rESC derived from EpiSC have undergone a stable reversion to an ESC-like state.
Figure 5.
Contribution of rESC to chimeras. (a) Progeny derived from chimera with rESC showing germline transmission indicated by black arrow. (b) E6.5 chimera with ROSA-lacZ rESC: Note contribution of early passage rESC both to the epiblast and to the extraembryonic ectoderm (ExE), a derivative of trophectoderm cells. (c) E6.5 chimera derived with ROSA-lacZ ESC; Note contribution predominantly to the epiblast. (d) Schematic representation of reprogramming of cEpi and EpiSC to rESC. Note the epigenetic and transcriptional changes during reprogramming of cEpi and EpiSC.
In conclusion, we demonstrate that postimplantation epiblast cells and established EpiSC can overcome a robust epigenetic barrier and undergo reversion to rESC. Reprogramming of epiblast in response to LIF-STAT3 differs from the response to Activin/bFGF when epiblast cells develop into self-renewing EpiSC. However, EpiSC, unlike rESC, retain significant properties of the original postimplantation epiblast cells, transcriptionally and epigenetically. This distinction is crucial and of practical significance because hESC resemble EpiSC3,4,31. Our study of epiblast derived cEpi and rESC provides a novel and tractable experimental model for critical insights into the signal-induced epigenetic reprogramming, including erasure of DNA methylation and X-reactivation. A major barrier to reprogramming of somatic cells to iPS show many similar features to those seen during reversion of epiblast to rESC9,19,32,33. Notably, specification of PGC is also accompanied by epigenetic reprogramming that brings the germ cell lineage epigenetically closer to PE (Ref. 34,35; Hayashi et al., unpublished). Thus, the underlying mechanisms involved in PGC specification and during the experimental reversion of epiblast to rESC provide novel insights on epigenetic reprogramming that are of wide interest and relevant to human diseases, stem cells and regenerative medicine.
Methods Summary
Derivation of rESC from mouse postimplantation epiblast (E5.5 - E7.5). The embryo was dissected with forceps, and the epiblast was isolated by cutting out the extraembryonic and proximal epiblast cells with glass needles. The epiblast was then treated with EGTA and trypsin, dissociated into single cells by pipetting with a hand-pulled glass capillary. The single cell suspension of epiblast cells was cultured in standard ESC medium with LIF and FCS on feeder cells. The formed colonies were called cEpi (cultured Epiblast cells), and were regularly passaged on feeder cells at 3 - 6 days interval. Following culture of cEpi for 14 - 35 days, about 10 - 50 GFP-positive cells appeared in individual cEpi colonies. When these colonies containing GFP-positive cells grew to 100 - 200 μm diameter, they were treated with trypsin, and the resulting cells were cultured to produce GFP-positive colonies. We refer to these cells as rESC.
Supplementary Material
Acknowledgments
We thank Caroline Lee for expert assistance. This work was supported by grants from the Wellcome Trust to M.A.S.
References
- 1.Gardner RL, Rossant J. Investigation of the fate of 4-5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 1979;52:141–152. [PubMed] [Google Scholar]
- 2.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Tesar PJ, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- 4.Brons IG, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- 5.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 6.Yeom YI, et al. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122:881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 7.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 10.Chung Y, et al. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–219. doi: 10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
- 11.Niwa H. How is pluripotency determined and maintained? Development. 2007;134:635–646. doi: 10.1242/dev.02787. [DOI] [PubMed] [Google Scholar]
- 12.Rathjen J, et al. Formation of a primitive ectoderm like cell population, EPL cells, from ES cells in response to biologically derived factors. J. Cell Sci. 1999;112:601–612. doi: 10.1242/jcs.112.5.601. [DOI] [PubMed] [Google Scholar]
- 13.Rathjen J, Washington JM, Bettess MD, Rathjen PD. Identification of a biological activity that supports maintenance and proliferation of pluripotent cells from the primitive ectoderm of the mouse. Biol. Reprod. 2003;69:1863–1871. doi: 10.1095/biolreprod.103.017384. [DOI] [PubMed] [Google Scholar]
- 14.Chou YF, et al. The growth factor environment defines distinct pluripotent ground states in novel blastocyst-derived stem cells. Cell. 2008;135:449–461. doi: 10.1016/j.cell.2008.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 16.Chuva de Sousa Lopes SM, et al. X chromosome activity in mouse XX primordial germ cells. PLoS Genet. 2008;4:e30. doi: 10.1371/journal.pgen.0040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 18.Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra X-tra News from the mouse X chromosome. Dev. Biol. 2006;298:344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 19.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, et al. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat. Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- 23.Bao S, et al. Initiation of epigenetic reprogramming of the X chromosome in somatic nuclei transplanted to a mouse oocyte. EMBO Rep. 2005;6:748–754. doi: 10.1038/sj.embor.7400461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3:391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohinata Y, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 26.Shovlin TC, Durcova-Hills G, Surani A, McLaren A. Heterogeneity in imprinted methylation patterns of pluripotent embryonic germ cells derived from pre-migratory mouse germ cells. Dev. Biol. 2008;313:674–681. doi: 10.1016/j.ydbio.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Meissner A. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Non-invasive sexing of preimplantation stage mammalian embryos. Nat. Genet. 1998;19:220–222. doi: 10.1038/893. [DOI] [PubMed] [Google Scholar]
- 29.Guo G, et al. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development. 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna J, et al. Metastable pluripotent states in NOD-mouse-derived ESCs. Cell Stem Cell. 2009;4:513–524. doi: 10.1016/j.stem.2009.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 32.Silva J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat. Biotechnol. 2008;26:916–924. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surani MA, Durcova-Hills G, Hajkova P, Hayashi K, Tee WW. Germ Line, Stem Cells, and Epigenetic Reprogramming. Cold Spring Harb. Symp. Quant. Biol. 2008 Nov 6; doi: 10.1101/sqb.2008.73.015. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Hayashi K, Surani MA. Resetting the epigenome beyond pluripotency in the germline. Cell Stem Cell. 2009;4:493–498. doi: 10.1016/j.stem.2009.05.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.