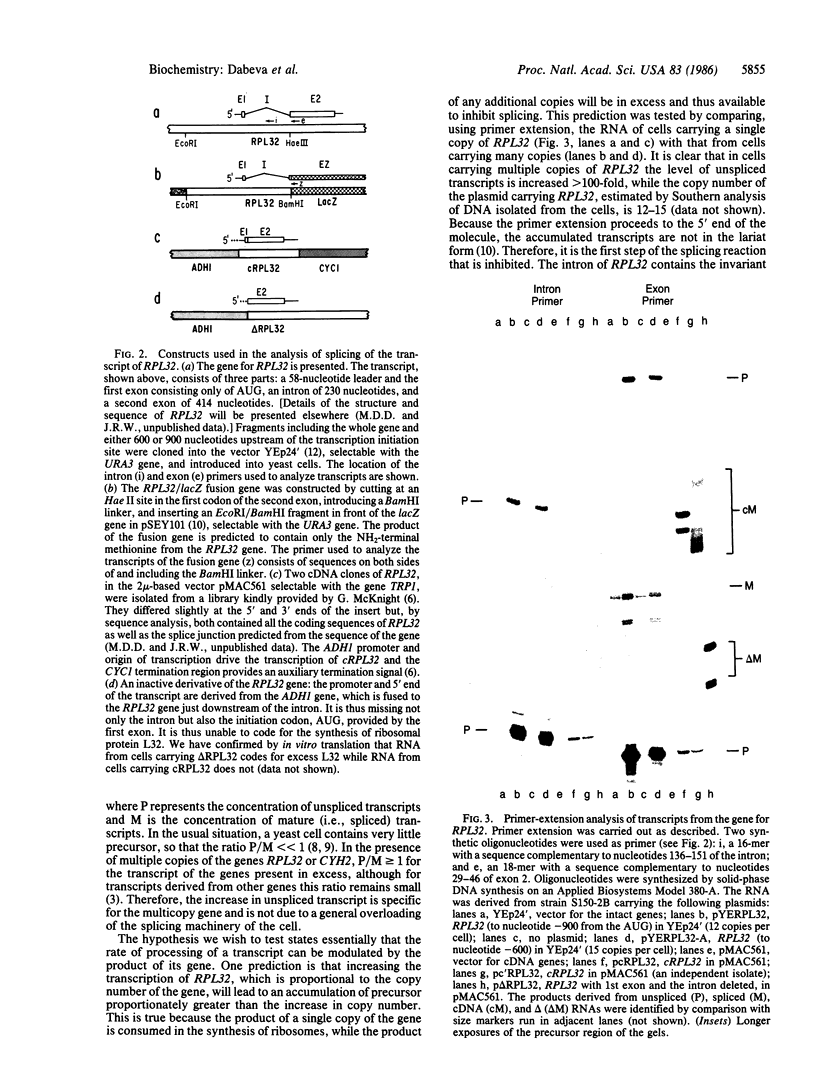
Abstract
The gene for a yeast ribosomal protein, RPL32, contains a single intron. The product of this gene appears to participate in feedback control of the splicing of the intron from the transcript. This autogenous regulation of splicing provides a striking analogy to the autogenous regulation of translation of ribosomal proteins in Escherichia coli.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abovich N., Gritz L., Tung L., Rosbash M. Effect of RP51 gene dosage alterations on ribosome synthesis in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Dec;5(12):3429–3435. doi: 10.1128/mcb.5.12.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozzoni I., Fragapane P., Annesi F., Pierandrei-Amaldi P., Amaldi F., Beccari E. Expression of two Xenopus laevis ribosomal protein genes in injected frog oocytes. A specific splicing block interferes with the L1 RNA maturation. J Mol Biol. 1984 Dec 25;180(4):987–1005. doi: 10.1016/0022-2836(84)90267-5. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Domdey H., Apostol B., Lin R. J., Newman A., Brody E., Abelson J. Lariat structures are in vivo intermediates in yeast pre-mRNA splicing. Cell. 1984 Dec;39(3 Pt 2):611–621. doi: 10.1016/0092-8674(84)90468-9. [DOI] [PubMed] [Google Scholar]
- Douglas M. G., Geller B. L., Emr S. D. Intracellular targeting and import of an F1-ATPase beta-subunit-beta-galactosidase hybrid protein into yeast mitochondria. Proc Natl Acad Sci U S A. 1984 Jul;81(13):3983–3987. doi: 10.1073/pnas.81.13.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Fried H. M., Pearson N. J., Kim C. H., Warner J. R. The genes for fifteen ribosomal proteins of Saccharomyces cerevisiae. J Biol Chem. 1981 Oct 10;256(19):10176–10183. [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Langford C. J., Gallwitz D. Evidence for an intron-contained sequence required for the splicing of yeast RNA polymerase II transcripts. Cell. 1983 Jun;33(2):519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- Leer R. J., van Raamsdonk-Duin M. M., Hagendoorn M. J., Mager W. H., Planta R. J. Structural comparison of yeast ribosomal protein genes. Nucleic Acids Res. 1984 Sep 11;12(17):6685–6700. doi: 10.1093/nar/12.17.6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight G. L., McConaughy B. L. Selection of functional cDNAs by complementation in yeast. Proc Natl Acad Sci U S A. 1983 Jul;80(14):4412–4416. doi: 10.1073/pnas.80.14.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medford R. M., Nguyen H. T., Destree A. T., Summers E., Nadal-Ginard B. A novel mechanism of alternative RNA splicing for the developmentally regulated generation of troponin T isoforms from a single gene. Cell. 1984 Sep;38(2):409–421. doi: 10.1016/0092-8674(84)90496-3. [DOI] [PubMed] [Google Scholar]
- Miller A. M. The yeast MATa1 gene contains two introns. EMBO J. 1984 May;3(5):1061–1065. doi: 10.1002/j.1460-2075.1984.tb01927.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Pearson N. J., Fried H. M., Warner J. R. Yeast use translational control to compensate for extra copies of a ribosomal protein gene. Cell. 1982 Jun;29(2):347–355. doi: 10.1016/0092-8674(82)90151-9. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Teem J. L., Rosbash M. Evidence for the biochemical role of an internal sequence in yeast nuclear mRNA introns: implications for U1 RNA and metazoan mRNA splicing. Cell. 1983 Sep;34(2):395–403. doi: 10.1016/0092-8674(83)90373-2. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Smith M., Williamson V. M., Young E. T. Nucleotide sequence of the yeast alcohol dehydrogenase II gene. J Biol Chem. 1983 Feb 25;258(4):2674–2682. [PubMed] [Google Scholar]
- Warner J. R., Mitra G., Schwindinger W. F., Studeny M., Fried H. M. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol Cell Biol. 1985 Jun;5(6):1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]