Significance
The Major Facilitator Superfamily (MFS), the largest family of secondary transport proteins, catalyzes transport of a wide range of substrates. Difficulty discerning underlying mechanistic principles is due to low sequence conservation. However, a common structural feature of MFS members, suggesting that they may have arisen by intragenic multiplication, is a repeat of four three-helix bundles organized in two pseudosymmetrical domains. An alignment of these triple-helix motifs in combinatorial fashion allows detection of functionally homologous positions. Thus, substrate and H+-binding sites in distantly related symporters are located at the same relative positions. The structural organization also suggests that an ordered kinetic mechanism similar to that determined for lactose permease may be operative in other MFS symporters.
Keywords: membrane transport, sequence alignment, bioenergetics
Abstract
One fundamentally important problem for understanding the mechanism of coupling between substrate and H+ translocation with secondary active transport proteins is the identification and physical localization of residues involved in substrate and H+ binding. This information is exceptionally difficult to obtain with the Major Facilitator Superfamily (MFS) because of the broad sequence diversity of the members. The MFS is the largest and most diverse group of transporters, many of which are clinically important, and includes members from all kingdoms of life. A wide range of substrates is transported, in many instances against a concentration gradient by transduction of the energy stored in an H+ electrochemical gradient using symport mechanisms, which are discussed herein. Crystallographic structures of MFS members indicate that a deep central hydrophilic cavity surrounded by 12 mostly irregular transmembrane helices represents a common structural feature. An inverted triple-helix structural symmetry motif within the N- and C-terminal six-helix bundles suggests that the proteins may have arisen by intragenic multiplication. In the work presented here, the triple-helix motifs are aligned in combinatorial fashion so as to detect functionally homologous positions with known atomic structures of MFS members. Substrate and H+-binding sites in symporters that transport substrates, ranging from simple ions like phosphate to more complex peptides or disaccharides, are found to be in similar locations. It also appears likely that there is a homologous ordered kinetic mechanism for the H+-coupled MFS symporters.
The exceptionally diverse Major Facilitator Superfamily (MFS) includes over 10,000 sequenced members and is comprised of 74 families with members from Archaea to Homo sapiens (1–3). MFS proteins catalyze transport of a wide range of substrates, including amines, acids, amino acids, sugars, peptides, and antibiotics, in many instances by transducing the energy stored in an H+ electrochemical gradient (∆µ̃H+, interior negative and/or alkaline) into a concentration gradient of substrate by substrate/H+ symport mechanisms (4).
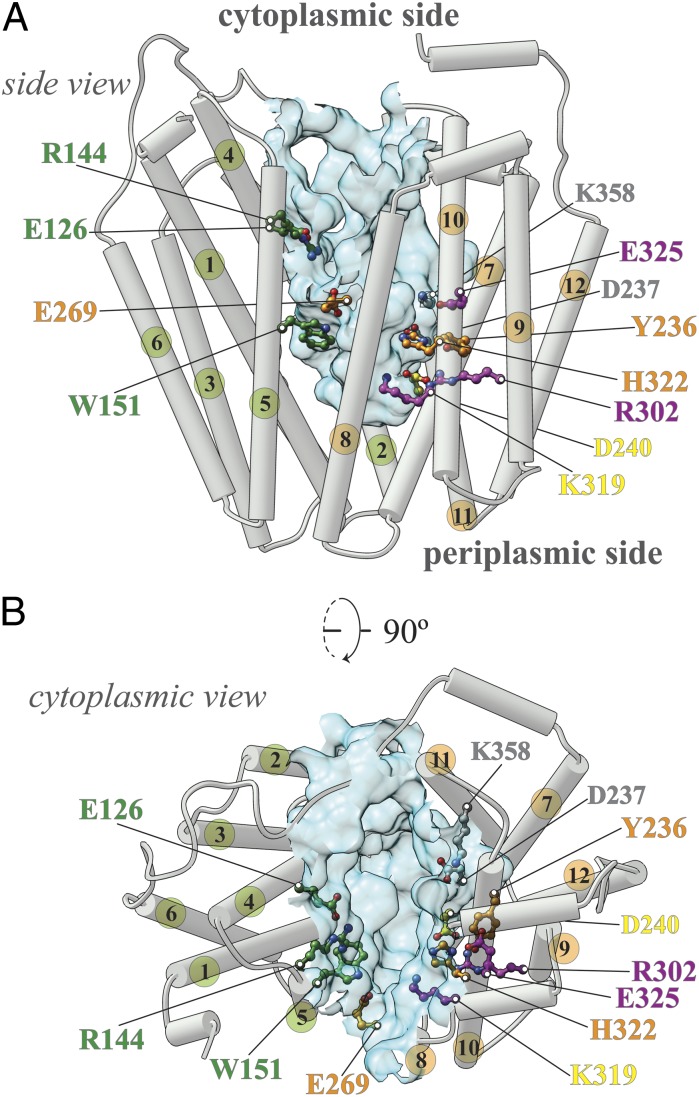
The lactose permease from Escherichia coli (LacY), a galactoside/H+ symporter, arguably the most intensively studied secondary transporter known at present (5, 6), is the paradigm of the MFS. LacY is comprised of 417 amino acid residues organized into two pseudosymmetrical six-helix bundles with the N and C termini on the cytoplasmic face of the membrane (Fig. 1). To determine which residues play an obligatory role in the mechanism and to create a library of mutants with a single-Cys residue at each position of the molecule for structure/function studies, each residue was replaced individually with Cys in a functional mutant devoid of native Cys residues (7). The great majority of the single-Cys mutants are expressed normally in the membrane and catalyze accumulation of lactose against a significant concentration gradient, thereby demonstrating that Cys replacement at most positions does not induce severe perturbations in the structure of LacY or in the symport mechanism. It is striking that fewer than 10 residues are irreplaceable for active lactose transport: Glu126 (helix IV) and Arg144 (helix V), which are critical for substrate binding, as well as Trp151 (helix V) where an aromatic side chain is essential; Glu269 (helix VIII), His322 (helix X), and Tyr236 (helix VII), which are likely involved in coupling between protonation and sugar binding; and Arg302 (helix IX) and Glu325 (helix X), which are exclusively involved in H+ translocation (5). As shown in the inward-facing crystal structures of LacY (8–11), these residues are located at the apex of a deep central hydrophilic cavity that is open to the cytoplasm only and tightly sealed on the periplasmic side (Fig.1 A and B). Although very few residues are absolutely irreplaceable, Cys substitution of 82 additional residues has a significant effect on activity, inhibiting the steady-state level of accumulation by 50–80% (7, 12).
Fig. 1.
LacY structure. (A) Side view of the inward-facing conformation of LacY (PDB ID code: 2V8N). Helices are shown as rods; the numbers of helices in the N-terminal six-helix bundle are colored green, and the numbers of C-terminal six-helix bundle are colored orange. Critical residues are indicated (green, residues involved in sugar binding exclusively; orange, residues involved in sugar affinity and H+ translocation; magenta, residues involved in H+ translocation exclusively; yellow, weakly salt-bridged residues; details are provided in the main text). The water-accessible surface of the cavity is shown as a light blue surface [calculated using the Computed Atlas of Surface Topography of proteins (CASTp) Web tool with a probe size of 1.4 Å]. (B) Cytoplasmic view; the color-coding is the same as in A.
X-ray structures give the impression that the symporters are rigid, but various biochemical and biophysical approaches provide converging evidence that LacY is in a highly dynamic state (13–15). Furthermore, binding of galactosides induces widespread conformational transitions, increasing the open probability of a hydrophilic cleft on the periplasmic side of the molecule, with closure of the cytoplasmic cavity in reciprocal fashion (6, 16). These coordinated conformational transitions are fundamental to secondary transport and represent the basis for the alternating access mechanism. Accordingly, the catalytic cycle of a transporter does not involve significant movement of sugar- and H+-binding sites relative to the membrane. Rather, the protein essentially moves around the substrate, reciprocally exposing the binding sites to either side of the membrane (i.e., alternating access in ref. 17). A wealth of biochemical and spectroscopic data demonstrates that the alternating access mechanism is operative in LacY (reviewed in refs. 6 and 16). Furthermore, X-ray structures (18–21), as well as spectroscopic findings (22), indicate that other MFS members probably function in similar fashion.
LacY is highly dynamic. On one hand, although ∼65% of the side chains are hydrophobic and buried, ∼85% of the backbone amide protons exchange with deuterium in 10–15 min at room temperature (13, 14), with 100% exchange at elevated, nondenaturing temperatures (15). On the other hand, flipping the six-helix bundles over and superposing them in structure models representing inward- or outward-open conformations yields an rmsd of about 1.2 Å. Thus, the two conformations are symmetrical, suggesting that the two domains move in rigid-body fashion during alternating access. However, the conclusion regarding rigid-body movement is incompatible with the H/D exchange data and observations that transport is blocked when helices within the N- or C-terminal helix bundles are cross-linked (23–25). Furthermore, crystal structures have been reported for other MFS transporters in an occluded conformation (20, 26) where the central cavity is inaccessible from either side of the membrane, which argues against a simple rocker-switch mechanism (27).
A principal difficulty in comparing MFS proteins is their low sequence conservation. Despite the conserved fold motif and in some cases overlapping function, sequence identity ranges around 12–18% (12). However, sequence similarity is much higher because most MFS members have a hydrophobic amino acid content of 60–70%. Moreover, the inherent symmetry of the proteins with regard to helix kinks and bends provides further overlapping of residues. In addition, regions of functional similarity (e.g., substrate- or H+-binding sites) align for only very closely related MFS transporters (28, 29). In less closely related symporters, residues with similar functional roles are often at disparate locations in the sequences (12).
The X-ray structures of various MFS symporters published in the past 2 y (19–21, 26, 30–33) have increased the total number of unique sets of coordinates from three (8, 27, 34) to nine, not including coordinates for alternative conformations or mutants of the same transporter (9–11, 18, 35). With a growing number of crystallographic structures of MFS symporters, we can begin to identify functionally related residues, which are uncorrelated in the primary structures but are correlated in the tertiary structures.
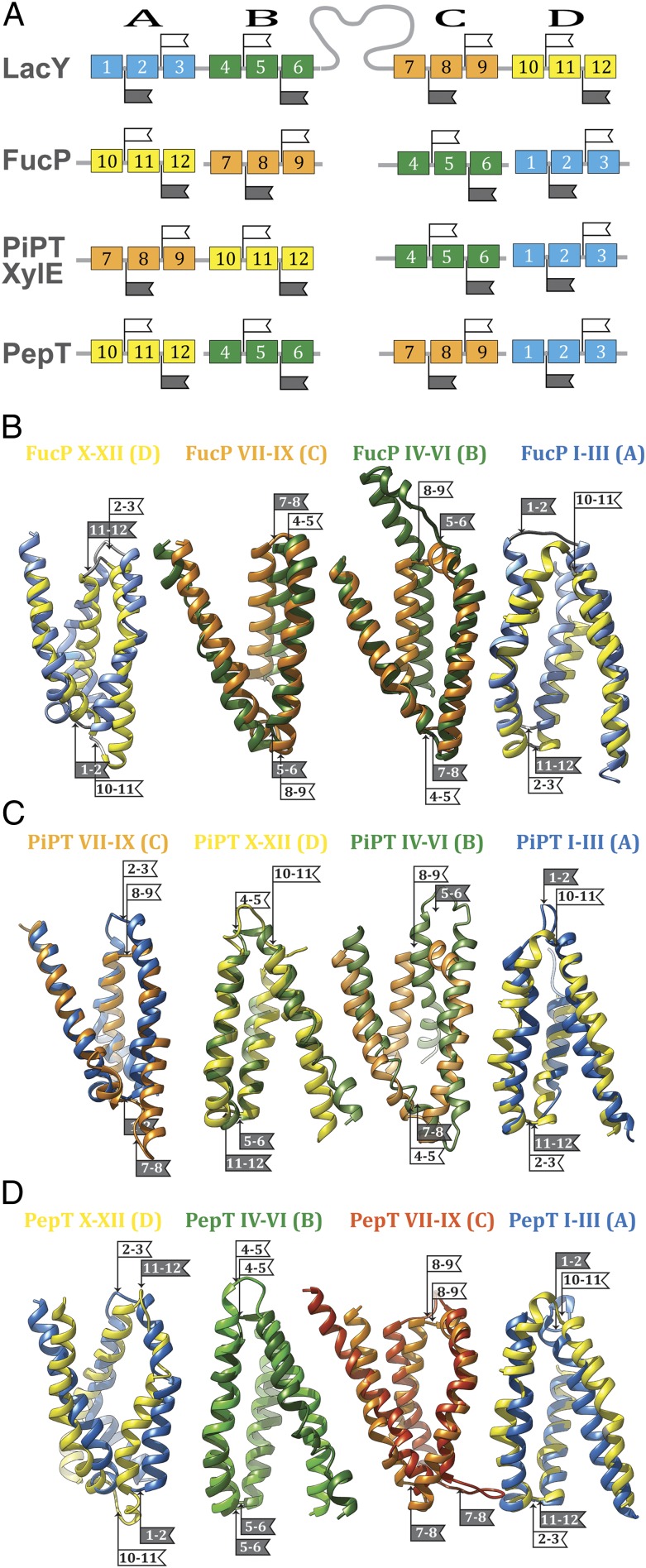
It has been suggested that the MFS transporters may have arisen by intragenic multiplication of the triple-helix motif to two pseudosymmetrical six-helix bundles (36, 37), the most common topological feature of MFS transporters. The recently presented crystallographic model of l-fucose permease (FucP) was obtained with the central cavity open to the periplasm, an outward-facing conformation opposite to that of LacY (21). It was observed that the transformation between the inward- and outward-facing conformations is obtained by an interconversion between conformations of neighboring inverted triple-helix repeats in the N- and C-terminal six-helix bundles (38). This observation prompted a comparative sequence analysis of the triple-helix motifs, screening for conservation of functional residues. An example of flexibility in design was observed within LacY and FucP, two MFS symporters (12). We permuted the order of the triplets in FucP from their natural order relative to LacY to obtain a higher order of sequence conservation (Fig. 2). The alignment was tested for conservation by comparing the 92 LacY mutants that impair function with 34 analogous functional mutations in FucP (12). In contrast to the conventional, linear sequence alignment, much stronger alignments between the sugar- and H+-binding sites in the two proteins are observed with rearrangement of the FucP triplets. We suggested that LacY and FucP might have evolved from primordial noncovalently fused helix-triplets that formed symporters. Modern symporters formed as functional mutations were being introduced and the functional segments assembled in a varying order. An alternative although less dynamic scenario is that a multispanning protein evolved first, possibly for stability reasons, and functional mutations were introduced later. In any case, using the notion of triple-helix evolution in the MFS, a more useful annotation of sequence motifs than a conventional, linear sequence alignment may be appropriate; we can test this possibility by alignment of functionally significant residues.
Fig. 2.
Schematic alignment of the helix-triplets in consecutive order in the sequence. (A) Helix-triplets (represented by colored boxes) from FucP, PiPT, XylE, and PepT are aligned with LacY (helices 1–3, blue; helices 4–6, green; helices 7–9, orange; helices 10–12, yellow). The flags indicate the loops within symmetry motifs. Helix-triplets from LacY are aligned with FucP (B), PiPT (C), and PepT (D) (helices 1–3, blue; helices 4–6, green; helices 7–9, orange; helices 10–12, yellow). The alignments are oriented with the LacY cytoplasmic side to the top. The flags indicate the loops within symmetry motifs (white, cytoplasmic loop; gray, periplasmic loop). The numbers on the flags indicate the two helices that are connected by the respective loop. See SI Appendix, Fig. S1 for a schematic representation.
Results and Discussion
Although it is generally believed that all MFS symporters operate by an alternating access mechanism, extensive biochemical and spectroscopic evidence is available for LacY only (reviewed in refs. 6 and 16). Furthermore, as a result of intensive study over three decades, an overall mechanism for coupling in LacY has been worked out.
An Overall Mechanism for Coupling in LacY.
i) Lactose/H+ symport in the uphill or downhill energetic modes is precisely the same reaction. The difference is in the rate-limiting step. For downhill symport, deprotonation is rate-limiting; for uphill transport, deprotonation is no longer limiting, and either dissociation of sugar or a conformational change that leads to deprotonation becomes limiting.
ii) Sugar binding and dissociation—not ∆µ̃H+—are the driving force for alternating access (∆µ̃H+ has no effect whatsoever on equilibrium exchange or counterflow).
iii) LacY must be protonated to bind sugar (the pKa for sugar binding is ∼10.5).
iv) Galactoside binds by an induced-fit mechanism, which powers a transition to an occluded state.
v) Sugar dissociates first.
vi) Upon sugar dissociation, there is a conformational change that causes Arg302 (helix IX) to approximate Glu325, leading to deprotonation of LacY.
As an initial step to investigate whether a similar overall mechanism might apply to other MFS symporters, we have searched for functional homologies between individual residues in additional structurally resolved symporters.
Detection of Functionally Homologous Positions.
MFS transporters consist of four symmetrically disposed triple-helix units that can be aligned individually and examined for conservation of functionally significant residues (Table 1; Fig. 2). The combinatorial alignment of the symmetry motifs allows detection of functionally homologous positions in different MFS transporters. The procedure relies on mapping known functional markers in the helix triplets followed by inferential mapping of unknown functional positions.
Table 1.
Combinations of symmetry motifs in MFS
 |
Helix-triplets A–D from PiPT, XylE, PepT, and, for comparison, FucP are aligned to LacY. The triplets are color-coded as in Fig. 2. The values represent the similarity score and the functional similarity score according to SI Appendix, Eqs. S1 and S2 (Q/QAct). For comparison, the respective scores are also stated for the superposition of the N-terminal six helices (Nter) and the C-terminal six helices (Cter) with LacY for the respective model.
Arrangement of helix-triplets in XylE and PiPT.
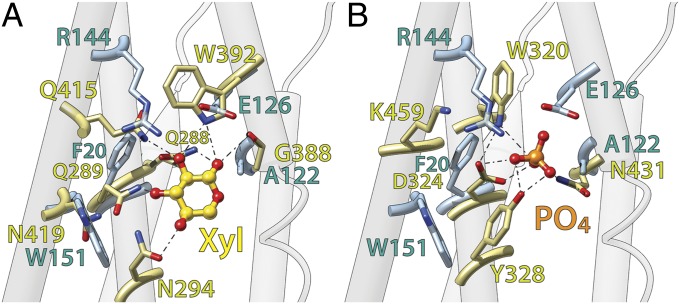
The recently solved structures for XylE and PiPT (20, 26) in the substrate-bound state allow identification of residues involved in substrate binding (Fig. 3). Although many substrate-binding residues in LacY are located in the N-terminal six-helix bundle (e.g., Arg144, Trp151, and Glu126), in XylE and PiPT, residues from the C-terminal six-helix bundle predominate. The previous study (12) comparing LacY with FucP revealed that due to the different order of the symmetry motifs, functionally equivalent residues can be located at different positions in the protein. Superposition of the C-terminal six-helix bundle from XylE or PiPT on the N-terminal bundle of LacY results in spatial alignment of substrate-binding residues in these three symporters (Fig. 3 A and B). However, an alignment of XylE and PiPT with FucP can be achieved only if symmetry motif D is superposed on motif C in FucP and, respectively, symmetry motif C is superposed on motif D in FucP (Fig. 2; Table 1). The N-terminal six-helix bundles from XylE, PiPT, and FucP superpose on each other without changing the order of symmetry motifs A and B. However, superposition on LacY succeeds only when symmetry motif C from LacY is superposed on symmetry motifs B from XylE, PiPT, and FucP, and symmetry motif D from LacY with the symmetry motifs A from the other three symporters (Fig. 2; Table 1).
Fig. 3.
Overall architecture of the substrate binding sites compared with LacY. (A) C-terminal six-helix bundle of XylE (colored in light olive) is superposed on the N-teminal six-helix bundle of LacY (side chains shown colored in light blue). The ligand of XylE, xylose, is shown as a gold ball-and-stick model. (B) C-terminal six-helix bundle of PiPT (colored in light olive) is superposed on the N-teminal six-helix bundle of LacY (side chains shown colored in light blue). The ligand of PiPT, phosphate, is shown as an orange ball-and-stick model. Oxygen and nitrogen atoms are colored red and blue respectively. The superposition of XylE and PiPT is provided in SI Appendix, Fig. S2.
Altered orientation of helix-triplets.
In FucP, the orientation of all four motifs is inverted with respect to LacY; all cytoplasmic loops superpose on periplasmic loops and periplasmic loops on their cytoplasmic counterparts in these two symporters (12). Unlike FucP, in XylE and PiPT, only motifs A and B are inverted with respect to motifs C and D in LacY, thereby placing the C terminus of the first six-helix bundle (motifs A and B) and the N terminus of the following six-helix bundle (motifs C and D) on opposite sides of the protein in the alignment of the structure motifs (Fig. 2). This is not the case in the native XylE or PiPT; rather, in these proteins, the N-terminal six-helix bundle is rotated 180° with respect to the membrane plane relative to LacY. This is compatible with the hypothesis that the symporters evolved by intragenic duplication and fusion of the helix-triplets (12, 36, 38). With single substrate and H+ binding-sites (5), the primordial MFS symporters may have exhibited substantial flexibility with regard to the orientation of the helix bundles to each other. This is not surprising considering that substrate-binding sites are generally located at the approximate middle of these symporters. Therefore, inversion of triple-helix motifs may be well tolerated. In this context, although the antiporter EmrE is not an MFS protein, it is a dimer with the binding site placed at the dimer interface in the middle of the complex. Moreover, it has been demonstrated (39) that the EmrE dimer is functional in both parallel and antiparallel configurations.
Arrangement and orientation of helix-triplets in PepT.
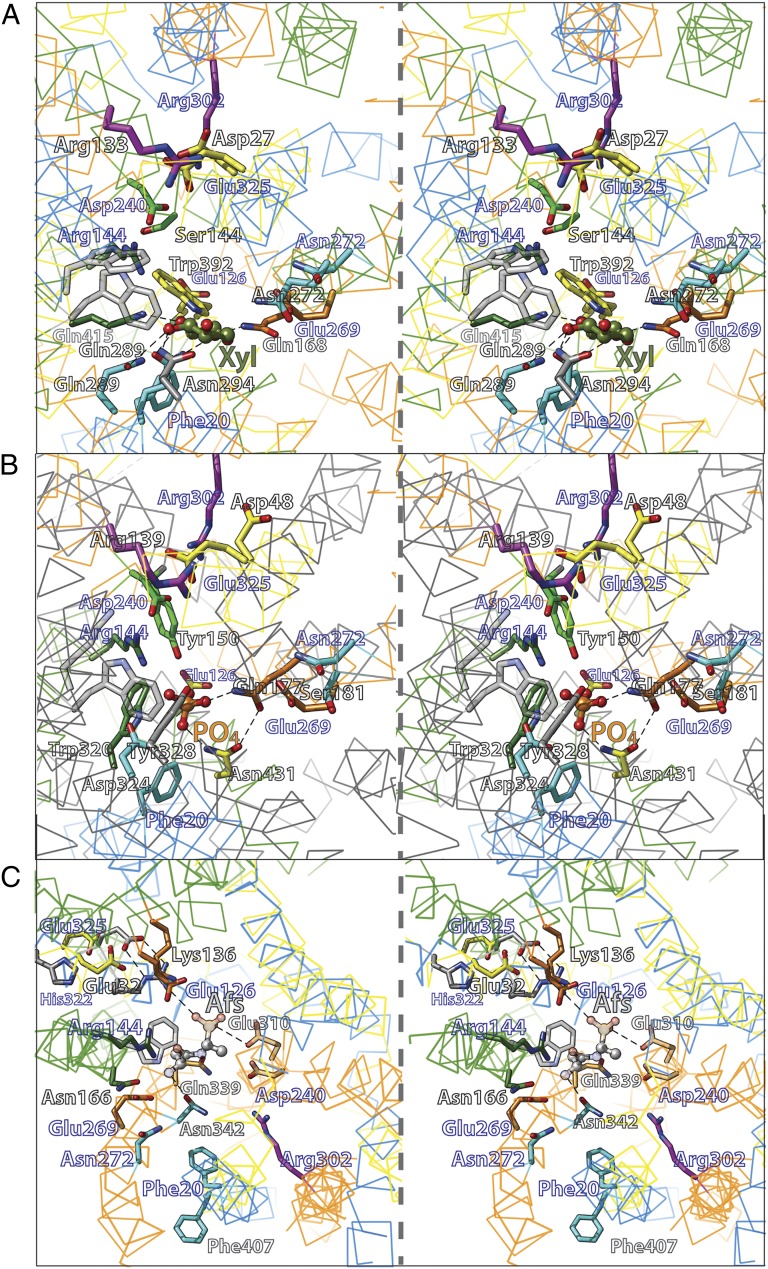
The oligopeptide/H+ symporter (PepT) (19, 30, 31, 35) provides another variation to the order of the triple-helix units because the symmetry motifs in PepT and LacY align in the consecutive order D-B-C-A to LacY A-B-C-D by using known substrate-binding residues as functional markers (Figs. 2 and 4C; Table 1). Clearly, the orientations of motifs A and D are inverted with respect to the membrane relative to LacY (Figs. 2 and 4C). Consequently, only residues at the center of PepT superimpose without inversion (Fig. 4C) (Glu310 in PepT and Asp240 in LacY). In contrast, functional positions displaced from the center are observed on the cytoplasmic side of the substrate-binding site [i.e., at a symmetry related location on the helix (Fig. 4C): Glu32 in PepT and Glu325 in LacY].
Fig. 4.
Functional alignment of XylE (A), PiPT (B), and PepT (C) to LacY helix-triplets (stereo-view). The Cα atom trace is show as wire and colored according to the helix-triplet (Fig. 2 and Table 1) except for PiPT in B. Significantly equivalent residue pairs are shown as sticks in the same color. Labels of LacY residues are indicated in blue.
Alignment of substrate-binding side chains.
In the X-ray structure of the inward-facing conformer of PepTGk from Geobacillus kaustophilus with the bound inhibitor alafosfalin (19), contacts to the inhibitor are provided primarily from motifs A and C. The positions of most side-chain interactions observed in the crystal structures of other MFS symporters are also observed in PepTGk although the side chains themselves vary, as expected. Residue Gln339 (C-terminal, helix VIII) in symmetry-motif C in PepT and corresponding to Glu269 in LacY provides contact between the N- and C-terminal halves of PepTGk by interacting with substrate ligand Arg43 and with the substrate (Figs. 4C) (N-terminal, helix I). Gln339 is located on the same face of helix VIII as the substrate ligand Asn342. The corresponding two residues, which appear to be involved in substrate binding, are observed in homologous symmetry motifs in XylE (Glu168, Gln175), PiPT (Gln177, Ser185) (Fig. 4B), FucP (Gln159, Asn162) (12), and in LacY (Glu269, Asn272) (Fig. 4). In all crystallographic structures with a bound substrate, the position corresponding to Glu269 in LacY couples the two six-helix bundles.
In addition to polar substrate ligands, aromatic side chains also play an important role in substrate binding. For example, Trp151 in LacY (8, 40) and Phe308, its counterpart in FucP (41), are essential for sugar binding (22), and they align with Trp416 in XylE, Lys459 in PiPT, and Phe170 in PepTGk. Irreplaceable Arg144 in LacY (42, 43) is in close proximity and on the cytoplasmic side of Trp151, and, in FucP, Arg312 (21) is functionally important and lies in a similar position relative to Phe308 (12). Similarly, in XylE, Trp416 and Asn415 align with Trp151 and Arg144 in LacY. Asn415 is involved in a complex H-bound network with Gln289 and Asn294 that interacts with xylose (20). In the open-inward, inhibitor-bound structure of PepTGk, the position of Asn166 (Fig. 6A) corresponds to Asn415 in XylE, Arg312 in FucP, and Arg144 in LacY (Fig. 4). Although mutation of Asn166 impairs active transport and counterflow (19), in this conformation, Asn166 in PepTGk is too far from the bound inhibitor to be involved in an H-bond network analogous to XylE. However, it is possible that tilting of PepT helix V against helix I would move Asn166 into proximity with substrate and close the cavity. Such conformational movement was suggested for LacY, where helix V and I are thought to undergo a substrate-dependent, scissors-like movement at position Gly24 (helix I) and Cys154 (helix V) (10, 24, 44), and indeed these positions correspond to residues Gly41 and Ala172 in PepTGk.
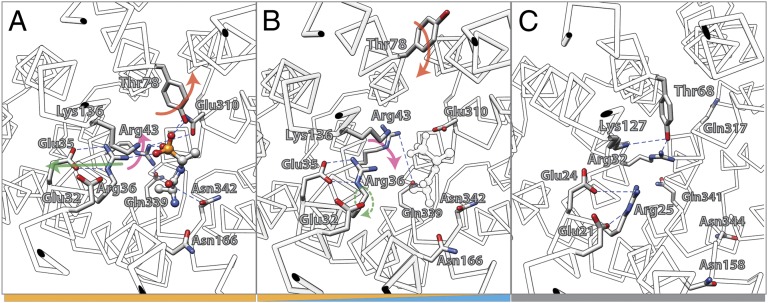
Fig. 6.
Conformational changes in the substrate binding site of PepT (see SI Appendix, Fig. S3 for stero-view version of this figure). (A) Inward-open, substrate-bound state of PepTGk (PDB ID code: 4IKZ). (B) Inward-open, substrate-free state of PepTGk (PDB ID code: 4IKV). The position of the substrate detected in the substrate-bound state is indicated as white profile. (C) Inward-occluded conformation of PepTSo (PDB ID code: 2XUT). The colors of the bars at the bottom indicate the mechanistic affiliation of the respective states with regard to the mechanistic model shown in Fig. 5.
Alignment of H+-binding side chains.
Helix X in LacY (symmetry-motif D) corresponds to helix I in PepT, XylE, PiPT (Figs. 2 and 4), and FucP (12) in symmetry motif A. Helix I carries the carboxyl side chains (Glu32 in PepT, Asp27 in XylE, Asp45 in PiPT, and Asp46 in FucP) that align with the well-characterized Glu325 in LacY. Strikingly, mutants with neutral replacements for Glu325 in LacY do not catalyze lactose/H+ symport, but counterflow and equilibrium exchange are unaffected (45). This and additional evidence (5, 46–48) indicate that Glu325 is irreplaceable and is directly involved in H+ symport. Similar findings have been reported for Asp46 in FucP (22), Glu22 in PepTSt (31), and Asp27 in XylE, supporting the conclusion that these residues are functionally homologous to Glu325 in LacY.
Residues on the same face of helix X as Glu325 play a critical role in the mechanism of LacY (5). In particular, His322, which is located one helix-turn toward the periplasm from Glu325, is another functionally irreplaceable residue (49–53). In contrast to Glu325 mutants, exchange and counterflow are blocked in His322 mutants, and replacement with Asn or Gln results in a major decrease in affinity for galactosides (52, 54). However, mutant H322R catalyzes lactose influx down a concentration gradient at a slow rate without H+ translocation (52). Although His322 is clearly involved in affinity for galactosides, it has been suggested that it is also involved in H+ translocation (54). Moreover, the notion has been put forward that structural water coordinated within a triad between His322, Tyr236, and Glu269 (two additional irreplaceable residues) may act as a cofactor for binding and galactoside/H+ symport by forming a hydronium ion intermediate during turnover (54, 55). In this regard, an analogous situation is apparent in PepTGk where Glu35, which corresponds to His322 in LacY, is not a direct substrate ligand. However, Glu35 is involved in substrate binding by positioning the guanidinium group of Arg36 with substrate (Figs. 4C and 6A) (19).
Functional Correlations.
Homology of primary amino acid sequences reflects evolution and therefore provides a guide to structure, mechanism, and function. Proteins that are related by common descent are expected to exhibit homologous structures and functions proportional to the degree of their sequence similarity. This principle provides the motivation to define protein phylogenetic relationships and correlate specific residues with function.
A kinetic mechanism for LacY.
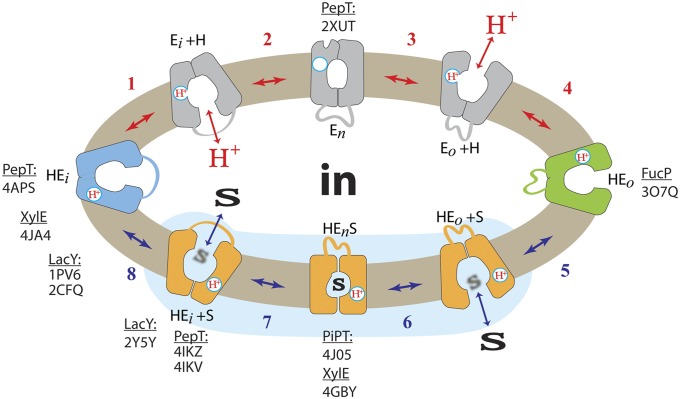
Varied experimental efforts have been undertaken with LacY to develop a kinetic scheme for lactose/H+ symport (5, 6, 46), and Fig. 5 depicts a simplified scheme. The sequence is initiated from the inward-facing conformer (HEi) in which the carboxyl group of Glu325 is protonated due to the low dielectric of the local environment, as evidenced from the X-ray structures. Because the apparent pKa of Glu325 (helix X) is ∼10.5 (54), deprotonation is likely due to the transient proximity of Arg302 (helix IX) (56) (step 1). Subsequently, the cytoplasmic cavity closes (step 2), resulting in the formation of the apo-intermediate (En). This conformer can relax back to the inward-facing conformation or open to the periplasmic side where it is reprotonated, which may involve Tyr236 (helix VII), His322 (helix X), and Glu269 (helix VIII) (55) (step 3). The protein then assumes a conformation symmetrical to the inward-facing conformer to reorient the helices for transfer of the H+ to negatively charged Glu325 and binding of sugar from the periplasmic side (HEo, steps 4 and 5). All of the specificity of LacY for substrate is directed toward the galactopyranosyl end of the substrate, and binding begins with a nonspecific hydrophobic interaction between the bottom of the galactopyranosyl ring with the hydrophobic surface of the indole ring of Trp151 (40, 57). Once the galactoside is oriented properly, formation of the binding site occurs. Arg144 (helix V) and Glu126 (helix IV), as well as Glu269 (helix VIII), interact with specific OH groups on the galactopyranosyl ring. Thus, galactopyranoside binding probably involves induced-fit (58). As the protonated galactoside-bound LacY ternary complex forms, the protein closes around the sugar to form an occluded state (HEnS, step 6). This conformer can either relax back to the outward-facing conformation or the cytoplasmic cavity opens with release of sugar to the cytoplasm (step 7). With release of sugar, the initial inward-facing conformation is restored (step 8). By reorientation of helices, Tyr236 (helix VII) is displaced from a position between Arg302 and protonated Glu325, thereby causing deprotonation of Glu325 (Ei+H+) with reinitiation of the cycle (step 1) (41). This ordered scheme was proposed originally in 1979 (59) and has been continuously refined since.
Fig. 5.
Transport cycle of LacY. Overview of the postulated steps in the transport model. Inward-facing (blue) and outward-facing (green) conformations are separated by the apo-intermediate conformational cluster (gray) or by the occluded-intermediate conformational cluster (orange). Substrate (S) and H+ are indicated. Steps are numbered consecutively: Substrate translocating transitions are indicated by blue arrows (steps 5–8) and transitions recycling the outward-open cavity are indicated by red arrows (steps 1–4). All steps are reversible (indicated by double-headed arrows). The blue-shaded area demarcates the equilibrium-exchange reaction. Examples of experimental coordinates (transporter and PDBID) associated with respective conformations are indicated.
The ordered kinetic mechanism.
There is an abundance of evidence for the ordered mechanism as depicted in Fig. 5 (reviewed in refs. 5 and 46). As stated above, the pKa for galactoside binding is ∼10.5. It is apparent that LacY is protonated at physiologic pH before substrate binding, and recent experiments supporting this conclusion more directly have been presented (60). Critically, neutral replacements for Glu325 yield mutants that are totally unable to carry out any reaction that involves net H+ transport but catalyze equilibrium exchange and counterflow as well as or better than the WT. As indicated in Fig. 5, a satisfying explanation for the behavior of Glu325 neutral replacement mutants that provides very strong evidence for the ordered mechanism shown is that the mutants can oscillate within the shaded portion of the scheme, but cannot deprotonate, and therefore cannot form the apo-intermediate and complete the cycle. In other words, the order of release in the WT must be dissociation of the galactoside first, followed by deprotonation. Because FucP, XylE, and PepT all have a carboxyl group corresponding to Glu325 in LacY that displays a similar phenotype when neutralized by mutagenesis, it is apparent that these symporters and perhaps many other MFS symporters likely operate by a similar ordered kinetic mechanism.
A common mechanistic pattern.
As discussed, the initial step in the cycle for LacY involves deprotonation of Glu325, which is thought to cause closure of the cytoplasmic cavity (61). As a result, the central cavity becomes closed on both sides. Although this conformation has not been observed crystallographically with LacY, such a conformation was observed with PepTSo (30), and a homology model for the LacY apo-intermediate is also available (41). The apo-intermediate can relax to the outside-open conformation, and the symporter reprotonates from the outside. An open-outward crystallographic model of FucP (21) and theoretical models for LacY (38) have been presented. Binding of substrate to the protonated open-outward conformer causes the protein to close around the substrate to form the occluded state. This conformation has been described in XylE (20) and PiPT (26). Interestingly, in both structures, the substrate-liganding side chains are located in the N- and C-terminal halves of the symporters. The occluded conformation can either reopen to the outside or open to the inside with release of substrate. In LacY, this global conformational change is postulated to be initiated by a localized scissors-like movement between helices V and I that is induced by galactoside binding (24). High-resolution X-ray structures of the inward-facing conformation of PepTGk (19), with bound inhibitor (pdbID: 4IKZ), show helix V with Asn166 displaced from its corresponding position in XylE (Asn415 on helix XI). It is likely that Asn166 ligates substrate in the fully occluded state and is no longer in contact with alafosfalin in the inward-facing conformation of PepTGk (Fig. 6A). Asn166 is presumably important for substrate binding in PepTGk (19), like Arg144 in LacY (43). Therefore, movement of helix V may initiate release of substrate on the cytoplasmic side. As discussed above with respect to LacY, helices V and I cross in the approximate middle of the membrane where Cys154 (helix V) and Gly24 (helix I) are in close proximity (8, 44). The C154G mutant binds ligand with high affinity but catalyzes almost no transport (44, 62). The X-ray structures of PepT (18, 19, 30) demonstrate that helix V crosses helix I in the approximate middle of the membrane in such a manner that Ala171 (helix V in PepTGk) lies close to Gly42 (helix I in PepTGk), thereby emphasizing the homology to this position in LacY. In the inward-facing conformation of PepTGk without bound substrate (pdbID: 4IKV) (Fig. 6B), further distortion of the substrate-binding site is observed. An induced-fit mechanism has already been suggested for LacY (9). Galactoside is thought to induce the formation of its binding site, primarily through side-chain movements without a global conformation change. Such a scenario is consistent with crystallographic structures observed with PepTGk (19).
It is obvious that the formation of the open-outward cavity to initiate another round of transport requires a conformation in which substrate is released without rebinding. The structure of the open-inward conformation of PepTSt (pdbID: 4APS) (31) provides structural insight in this regard. Here, the substrate-capping Tyr residue is rotated back toward the center of the cavity, blocking access of substrate. Further stabilization of this conformation is represented in the crystal structure of PepTSo (30) where the Tyr residue is stabilized in a conformation blocking access to substrate by H-bonding to an Arg32 (Fig. 6C).
Taken together, the striking similarities within the MFS clearly suggest that a common mechanistic pattern may be used for catalysis of symport by additional transporters in this superfamily.
Materials and Methods
Structure and Sequence Alignments.
The structural symmetry motifs according to Radestock and Forrest (38) were generated from the crystallographic coordinates of LacY (A, Thr7-Asn102; B, Leu104-Phe187; C, Lys220-Ser309; D, Ala311-Leu400) (10), FucP (A, Arg22-Met115; B, Asn116-Thr229; C, Arg258-Ala345; D, Gly347-Phe431) (21), XylE (A, Tyr5-Ile112; B, Tyr125-Pro221; C, Gly276-Thr365; D, Gly369-Glu465) (21), PiPT (A, Pro30-His125; B, Trp126-Arg229; C, Thr306-Ile410; D, Gly411-Arg518) (26), and PepT (A, His21-Ile114; B, Gly117-Leu211; C,Ala286-Ser392; D, His398-Met492) (19), and the superimposition of the helix-triplets was carried out as described in Madej et al. (12). In brief, computer programs COOT v0.7 (63) and UCSF-Chimera (64) were used for the structure-guided sequence alignment. No positional restraints were applied to functionally significant residues in the sequence or structure alignments. The superposition was visually inspected for the conservation of LacY functional markers. As functional markers, 22 polar residues in LacY were used where mutations to Cys cause greater than 50% (SI Appendix, Eq. S2: sAA25–50%) or 75% (SI Appendix, Eq. S2: sAA0–25%) inhibition of the transport rate with Cys-less LacY (7, 12).
Discrimination of Defective Alignments.
For discrimination of defective alignments, a similarity score, Q, was defined. A reduction matrix (SI Appendix, Table S1) of similar side chains was defined based on clustering of reduction groups in similar folds, omitting interlacing for different levels of reduction (65). This reduction matrix was modified with respect to the clustering for packing values of helix–helix contacts in the membrane, their natural occurrence in α-helical membrane proteins (66), and hydrophobicity (67) (SI Appendix, Fig. S4 and Table S1). The main difference is that His was grouped with positively charged residues Arg and Lys instead of with Asn in the first reduction level. Additionally, groups introducing helix irregularities and small side chains (Gly, Pro) are grouped with Cys at the last reduction level based on similar helix packing and hydrophobicity to Ser and Thr. Due to high abundance in helical membrane proteins, the hydrophobic side chains I, F, V, and L were ignored. The following value scoring of sequence conservation was used: conserved residue = 20, first reduction level = 10, second reduction level = 7, and third reduction level = 4. The sum of the scoring values (red Value) was divided by the total number of analyzed positions multiplied by the highest score (v = 20) for a conserved residue (SI Appendix, Eq. S1). For 100% conservation, Q should equal 1. The quantification of functionally overlapping positions was performed by weighting the scoring of functionally critical positions (AA0–25%, activity inhibited by at least 75% upon mutation to Cys) and functionally relevant positions (AA25–50%, activity inhibited by at least 50% upon mutation to Cys) using the reduction matrix (SI Appendix, Table S1) and an arbitrary weighting factor of 5 for functionally critical positions (SI Appendix, Eq. S2). For 100% conserved transporters with 100% conserved functional positions, this value, QAct, should equal 1. The best scoring structure alignments were inspected visually for conservation of the geometry in the functional site, i.e., if functional groups face each other in the alignment.
Supplementary Material
Acknowledgments
We are deeply indebted to Arthur Karlin for editorial contributions. This work was supported by National Institutes of Health Grants DK51131, DK069463, and GM073210, as well as National Science Foundation Grant MCB-1129551 (to H.R.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319754110/-/DCSupplemental.
References
- 1.Saier MH., Jr Families of transmembrane sugar transport proteins. Mol Microbiol. 2000;35(4):699–710. doi: 10.1046/j.1365-2958.2000.01759.x. [DOI] [PubMed] [Google Scholar]
- 2.Saier MH, Jr, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1(2):257–279. [PubMed] [Google Scholar]
- 3.Reddy VS, Shlykov MA, Castillo R, Sun EI, Saier MH., Jr The major facilitator superfamily (MFS) revisited. FEBS J. 2012;279(11):2022–2035. doi: 10.1111/j.1742-4658.2012.08588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell P. Translocations through natural membranes. Adv Enzymol Relat Areas Mol Biol. 1967;29:33–87. doi: 10.1002/9780470122747.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Guan L, Kaback HR. Lessons from lactose permease. Annu Rev Biophys Biomol Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smirnova I, Kasho V, Kaback HR. Lactose permease and the alternating access mechanism. Biochemistry. 2011;50(45):9684–9693. doi: 10.1021/bi2014294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: A novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12(13):1281–1299. doi: 10.1096/fasebj.12.13.1281. [DOI] [PubMed] [Google Scholar]
- 8.Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301(5633):610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 9.Mirza O, Guan L, Verner G, Iwata S, Kaback HR. Structural evidence for induced fit and a mechanism for sugar/H+ symport in LacY. EMBO J. 2006;25(6):1177–1183. doi: 10.1038/sj.emboj.7601028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc Natl Acad Sci USA. 2007;104(39):15294–15298. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaptal V, et al. Crystal structure of lactose permease in complex with an affinity inactivator yields unique insight into sugar recognition. Proc Natl Acad Sci USA. 2011;108(23):9361–9366. doi: 10.1073/pnas.1105687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madej MG, Dang S, Yan N, Kaback HR. Evolutionary mix-and-match with MFS transporters. Proc Natl Acad Sci USA. 2013;110(15):5870–5874. doi: 10.1073/pnas.1303538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.le Coutre J, Kaback HR, Patel CK, Heginbotham L, Miller C. Fourier transform infrared spectroscopy reveals a rigid alpha-helical assembly for the tetrameric Streptomyces lividans K+ channel. Proc Natl Acad Sci USA. 1998;95(11):6114–6117. doi: 10.1073/pnas.95.11.6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patzlaff JS, Moeller JA, Barry BA, Brooker RJ. Fourier transform infrared analysis of purified lactose permease: A monodisperse lactose permease preparation is stably folded, alpha-helical, and highly accessible to deuterium exchange. Biochemistry. 1998;37(44):15363–15375. doi: 10.1021/bi981142x. [DOI] [PubMed] [Google Scholar]
- 15.Sayeed WM, Baenziger JE. Structural characterization of the osmosensor ProP. Biochim Biophys Acta. 2009;1788(5):1108–1115. doi: 10.1016/j.bbamem.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Kaback HR, Smirnova I, Kasho V, Nie Y, Zhou Y. The alternating access transport mechanism in LacY. J Membr Biol. 2011;239(1-2):85–93. doi: 10.1007/s00232-010-9327-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell P. A general theory of membrane transport from studies of bacteria. Nature. 1957;180(4577):134–136. doi: 10.1038/180134a0. [DOI] [PubMed] [Google Scholar]
- 18.Quistgaard EM, Löw C, Moberg P, Trésaugues L, Nordlund P. Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters. Nat Struct Mol Biol. 2013;20(6):766–768. doi: 10.1038/nsmb.2569. [DOI] [PubMed] [Google Scholar]
- 19.Doki S, et al. Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT. Proc Natl Acad Sci USA. 2013;110(28):11343–11348. doi: 10.1073/pnas.1301079110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L, et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1-4. Nature. 2012;490(7420):361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 21.Dang S, et al. Structure of a fucose transporter in an outward-open conformation. Nature. 2010;467(7316):734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 22.Sugihara J, Sun L, Yan N, Kaback HR. Dynamics of the L-fucose/H+ symporter revealed by fluorescence spectroscopy. Proc Natl Acad Sci USA. 2012;109(37):14847–14851. doi: 10.1073/pnas.1213445109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Madej MG, Kaback HR. Helix dynamics in LacY: Helices II and IV. J Mol Biol. 2010;396(3):617–626. doi: 10.1016/j.jmb.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Madej MG, Guan L, Nie Y, Kaback HR. An early event in the transport mechanism of LacY protein: Interaction between helices V and I. J Biol Chem. 2011;286(35):30415–30422. doi: 10.1074/jbc.M111.268433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang W, Guan L, Kaback HR. Helices VII and X in the lactose permease of Escherichia coli: Proximity and ligand-induced distance changes. J Mol Biol. 2002;315(1):53–62. doi: 10.1006/jmbi.2001.5206. [DOI] [PubMed] [Google Scholar]
- 26.Pedersen BP, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496(7446):533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301(5633):616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 28.Kasho VN, Smirnova IN, Kaback HR. Sequence alignment and homology threading reveals prokaryotic and eukaryotic proteins similar to lactose permease. J Mol Biol. 2006;358(4):1060–1070. doi: 10.1016/j.jmb.2006.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugihara J, Smirnova I, Kasho V, Kaback HR. Sugar recognition by CscB and LacY. Biochemistry. 2011;50(51):11009–11014. doi: 10.1021/bi201592y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newstead S, et al. Crystal structure of a prokaryotic homologue of the mammalian oligopeptide-proton symporters, PepT1 and PepT2. EMBO J. 2011;30(2):417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solcan N, et al. Alternating access mechanism in the POT family of oligopeptide transporters. EMBO J. 2012;31(16):3411–3421. doi: 10.1038/emboj.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan H, et al. Structure and mechanism of a nitrate transporter. Cell Rep. 2013;3(3):716–723. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497(7451):647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312(5774):741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guettou F, et al. Structural insights into substrate recognition in proton-dependent oligopeptide transporters. EMBO Rep. 2013;14(9):804–810. doi: 10.1038/embor.2013.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hvorup RN, Saier MH., Jr Sequence similarity between the channel-forming domains of voltage-gated ion channel proteins and the C-terminal domains of secondary carriers of the major facilitator superfamily. Microbiology. 2002;148(Pt 12):3760–3762. doi: 10.1099/00221287-148-12-3760. [DOI] [PubMed] [Google Scholar]
- 37.Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62(1):1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radestock S, Forrest LR. The alternating-access mechanism of MFS transporters arises from inverted-topology repeats. J Mol Biol. 2011;407(5):698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Nasie I, Steiner-Mordoch S, Gold A, Schuldiner S. Topologically random insertion of EmrE supports a pathway for evolution of inverted repeats in ion-coupled transporters. J Biol Chem. 2010;285(20):15234–15244. doi: 10.1074/jbc.M110.108746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guan L, Hu Y, Kaback HR. Aromatic stacking in the sugar binding site of the lactose permease. Biochemistry. 2003;42(6):1377–1382. doi: 10.1021/bi027152m. [DOI] [PubMed] [Google Scholar]
- 41.Madej MG, Soro SN, Kaback HR. Apo-intermediate in the transport cycle of lactose permease (LacY) Proc Natl Acad Sci USA. 2012;109(44):E2970–E2978. doi: 10.1073/pnas.1211183109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frillingos S, Gonzalez A, Kaback HR. Cysteine-scanning mutagenesis of helix IV and the adjoining loops in the lactose permease of Escherichia coli: Glu126 and Arg144 are essential. off. Biochemistry. 1997;36(47):14284–14290. doi: 10.1021/bi972314d. [DOI] [PubMed] [Google Scholar]
- 43.Sahin-Tóth M, et al. Characterization of Glu126 and Arg144, two residues that are indispensable for substrate binding in the lactose permease of Escherichia coli. Biochemistry. 1999;38(2):813–819. doi: 10.1021/bi982200h. [DOI] [PubMed] [Google Scholar]
- 44.Ermolova NV, Smirnova IN, Kasho VN, Kaback HR. Interhelical packing modulates conformational flexibility in the lactose permease of Escherichia coli. Biochemistry. 2005;44(21):7669–7677. doi: 10.1021/bi0502801. [DOI] [PubMed] [Google Scholar]
- 45.Carrasco N, et al. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989;28(6):2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- 46.Kaback HR, Sahin-Tóth M, Weinglass AB. The kamikaze approach to membrane transport. Nat Rev Mol Cell Biol. 2001;2(8):610–620. doi: 10.1038/35085077. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Celma JJ, Smirnova IN, Kaback HR, Fendler K. Electrophysiological characterization of LacY. Proc Natl Acad Sci USA. 2009;106(18):7373–7378. doi: 10.1073/pnas.0902471106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Celma JJ, Ploch J, Smirnova I, Kaback HR, Fendler K. Delineating electrogenic reactions during lactose/H+ symport. Biochemistry. 2010;49(29):6115–6121. doi: 10.1021/bi100492p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Padan E, Patel L, Kaback HR. Effect of diethylpyrocarbonate on lactose/proton symport in Escherichia coli membrane vesicles. Proc Natl Acad Sci USA. 1979;76(12):6221–6225. doi: 10.1073/pnas.76.12.6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Garcia ML, Patel L, Padan E, Kaback HR. Mechanism of lactose transport in Escherichia coli membrane vesicles: Evidence for the involvement of histidine residue(s) in the response of the lac carrier to the proton electrochemical gradient. Biochemistry. 1982;21(23):5800–5805. doi: 10.1021/bi00266a012. [DOI] [PubMed] [Google Scholar]
- 51.Padan E, Sarkar HK, Viitanen PV, Poonian MS, Kaback HR. Site-specific mutagenesis of histidine residues in the lac permease of Escherichia coli. Proc Natl Acad Sci USA. 1985;82(20):6765–6768. doi: 10.1073/pnas.82.20.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Püttner IB, Sarkar HK, Poonian MS, Kaback HR. lac permease of Escherichia coli: Histidine-205 and histidine-322 play different roles in lactose/H+ symport. Biochemistry. 1986;25(16):4483–4485. doi: 10.1021/bi00364a003. [DOI] [PubMed] [Google Scholar]
- 53.Püttner IB, Kaback HR. lac permease of Escherichia coli containing a single histidine residue is fully functional. Proc Natl Acad Sci USA. 1988;85(5):1467–1471. doi: 10.1073/pnas.85.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smirnova IN, Kasho VN, Sugihara J, Choe JY, Kaback HR. Residues in the H+ translocation site define the pKa for sugar binding to LacY. Biochemistry. 2009;48(37):8852–8860. doi: 10.1021/bi9011918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou Y, Jiang X, Kaback HR. Role of the irreplaceable residues in the LacY alternating access mechanism. Proc Natl Acad Sci USA. 2012;109(31):12438–12442. doi: 10.1073/pnas.1210684109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahin-Toth M, Kaback HR. Arg-302 facilitates deprotonation of Glu-325 in the transport mechanism of the lactose permease from Escherichiacoli. Proc Natl Acad Sci USA. 2001;98(11):6068–6073. doi: 10.1073/pnas.111139698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vázquez-Ibar JL, Guan L, Svrakic M, Kaback HR. Exploiting luminescence spectroscopy to elucidate the interaction between sugar and a tryptophan residue in the lactose permease of Escherichia coli. Proc Natl Acad Sci USA. 2003;100(22):12706–12711. doi: 10.1073/pnas.1835645100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klingenberg M. Ligand-protein interaction in biomembrane carriers: The induced transition fit of transport catalysis. Biochemistry. 2005;44(24):8563–8570. doi: 10.1021/bi050543r. [DOI] [PubMed] [Google Scholar]
- 59.Kaczorowski GJ, Kaback HR. Mechanism of lactose translocation in membrane vesicles from Escherichia coli. 1. Effect of pH on efflux, exchange, and counterflow. Biochemistry. 1979;18(17):3691–3697. doi: 10.1021/bi00584a009. [DOI] [PubMed] [Google Scholar]
- 60.Smirnova I, Kasho V, Sugihara J, Vázquez-Ibar JL, Kaback HR. Role of protons in sugar binding to LacY. Proc Natl Acad Sci USA. 2012;109(42):16835–16840. doi: 10.1073/pnas.1214890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersson M, et al. Proton-coupled dynamics in lactose permease. Structure. 2012;20(11):1893–1904. doi: 10.1016/j.str.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Menick DR, Sarkar HK, Poonian MS, Kaback HR. cys154 Is important for lac permease activity in Escherichia coli. Biochem Biophys Res Commun. 1985;132(1):162–170. doi: 10.1016/0006-291x(85)91002-2. [DOI] [PubMed] [Google Scholar]
- 63.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Li T, Fan K, Wang J, Wang W. Reduction of protein sequence complexity by residue grouping. Protein Eng. 2003;16(5):323–330. doi: 10.1093/protein/gzg044. [DOI] [PubMed] [Google Scholar]
- 66.Eilers M, Shekar SC, Shieh T, Smith SO, Fleming PJ. Internal packing of helical membrane proteins. Proc Natl Acad Sci USA. 2000;97(11):5796–5801. doi: 10.1073/pnas.97.11.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.White SH, Wimley WC. Membrane protein folding and stability: physical principles. Annu Rev Biophys Biomol Struct. 1999;28(4):319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.