Significance
Although plant metabolites are critical for both plant and human nutrition, the genetic control of the plant metabolome remains largely unknown. Here, a genetic analysis of the rice metabolome resulted in the identification of hundreds of metabolic quantitative trait loci with both high resolution and large effects. We observed distinct and overlapping accumulation patterns of metabolites and complex genetic regulation of metabolism in different tissues. Twenty-four candidate genes, mainly those underlying phenolics, were assigned and the corresponding pathways were reconstructed in rice. This study demonstrated a powerful tool and provided a vast amount of high-quality data for understanding the genetic and biochemical basis of the metabolome. It is also valuable for crop genetic improvement through metabolomics-assisted breeding.
Keywords: Oryza sativa, recombinant inbred line, metabolic profiling, gene function
Abstract
Plant metabolites are crucial for both plant life and human nutrition. Despite recent advance in metabolomics, the genetic control of plant metabolome remains largely unknown. Here, we performed a genetic analysis of the rice metabolome that provided over 2,800 highly resolved metabolic quantitative trait loci for 900 metabolites. Distinct and overlapping accumulation patterns of metabolites were observed and complex genetic regulation of metabolism was revealed in two different tissues. We associated 24 candidate genes to various metabolic quantitative trait loci by data mining, including ones regulating important morphological traits and biological processes. The corresponding pathways were reconstructed by updating in vivo functions of previously identified and newly assigned genes. This study demonstrated a powerful tool and provided a vast amount of high-quality data for understanding the plasticity of plant metabolome, which may help bridge the gap between the genome and phenome.
Plants are highly enriched in specific metabolites with extensive quantitative and qualitative variation both among and within different plant species (1, 2). Understanding the genes involved in metabolism and dissection of the metabolic pathway are essential to improve plant adaptation to environmental stresses, to improve food quality, and to increase crop yield. Recent advance in metabolomics together with transcriptomics and gene/metabolite coexpression networks analyses has proven to be powerful in functional gene elucidation, although it is specified for transcriptionally regulated genes with limited throughput so far.
A large number of metabolic quantitative trait loci (mQTLs) have been identified based on linkage maps using low-density markers, such as restriction fragment length polymorphism and simple sequence repeat markers (3–5). However, the underlying genes remain elusive because of the relatively low resolution of the genetic maps associated with these markers. In addition to the species-specific accumulation of the metabolites in plant, tissue-specific accumulation of metabolites, especially secondary metabolites, are of special importance for the survival and adaptation of plant species. Although the genetics of tissue-specific regulation gene expression across the tissues as revealed by expression quantitative trait locus analysis is one of the major topics in animal and plant, the genetics of tissue-specific metabolome is somewhat overlooked (5, 6). Despite the advance in metabolomics (2), the genetic control of the plant metabolome is still largely unknown.
Moreover, the elucidation of pathways for the biosyntheses of important metabolites such as amino acids, lysophosphatidylcholines (LPCs), and flavonoids, and the variation and the genetics control of them are vital for both basic research and breeding application. For example, flavonoids are one of the main groups of secondary metabolites that play important roles in a number of biological processes such as resistance to biotic/abiotic stresses (7) and confer health-promoting effects against chronic diseases such as cardiovascular diseases and certain cancers (8). The accumulation and regulation of O-glycosyl flavonols have been well studied in dicotyledonous plants (9). However, monocots such as rice and maize accumulate C-glycosylated flavones as the major flavonoids (10), whereas the regulation and modification of flavonoids in these crops are poorly understood (3, 11). Elucidation of the genetic bases of these pathways in rice would provide foundations for genetic improvement of important traits such as stress resistance and nutritional quality in one of the world’s most important crops.
Here, we report a genetic analysis of rice metabolome combining metabolic profiling (12) with an ultrahigh-density genetic map (13) using a recombinant inbred line (RIL) population. We demonstrated that the high-resolution mapping of the large number of mQTLs may greatly accelerate gene identification and pathway elucidation for metabolites, which will enhance our understanding of the genetic and biochemical basis of the metabolome and also be valuable for crop genetic improvement through metabolomics-assisted breeding.
Results
Widely Targeted Metabolic Profiling Analysis Using Liquid Chromatography–Electrospray Ionization–MS/MS.
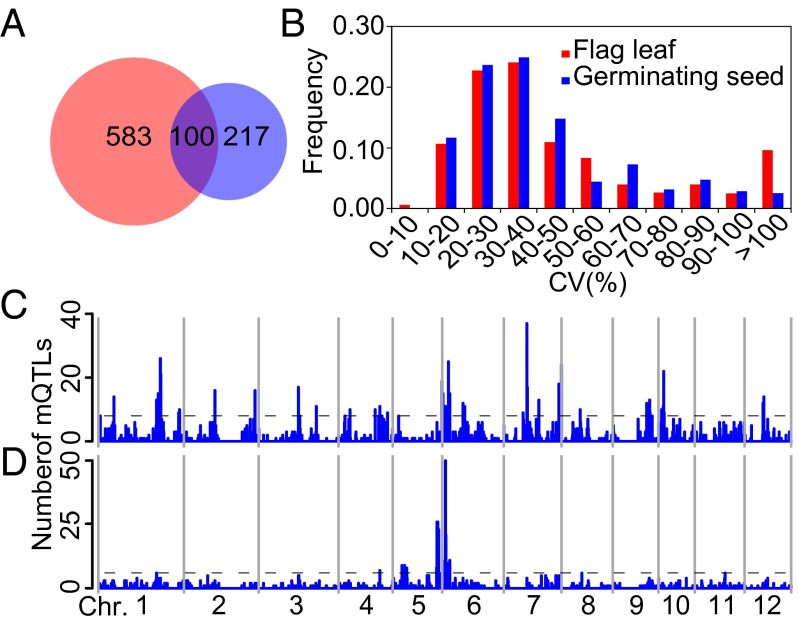
Samples of the flag leaf at the heading date (termed flag leaf hereafter) and seed at 72 h after germination (termed germinating seed hereafter) from 210 RILs derived from a cross between two elite indica rice varieties, Zhenshan 97 (ZS97) and Minghui 63 (MH63) (13) were collected, and an MS2 spectral tag (MS2T) library was constructed as described previously (12, 14) (Dataset S1). A data matrix was generated, consisting a total of 1,000 metabolites including 683 in flag leaf and 317 germinating seed, with 100 metabolites were detected in both tissues (Fig. 1A and Dataset S2). Among the 900 unique metabolites, 50 were structurally identified by direct comparison of their chromatographic and fragmental behaviors to those of the commercial standards, including amino acids, flavonoids, LPCs, and fatty acids, etc., and 303 metabolites were annotated (Dataset S2) using strategies described previously (12). Totally, 60 of the 100 metabolites detected in both tissues were identified or annotated (Fig. 1A and Dataset S2). High-throughput quantification of metabolites was then carried out by scheduled multiple-reaction monitoring (15). The content of metabolite varied substantially among the RILs with an average genetic coefficient of variation (CV) of 51.2% and 43.7% in flag leaf and germinating seed, respectively (Fig. 1B and Dataset S2).
Fig. 1.
The number of detected metabolites, distribution of the values of genetic coefficient of variation (CV), and metabolic quantitative trait loci (mQTLs) for metabolic traits. The number of metabolites detected (A) and distribution of genetic CVs of metabolites (B) in the RIL population. Red, flag leaf. Blue, germinating seed. Distribution of mQTLs in the rice genome in flag leaf (C) and germinating seed (D). The horizontal dashed line indicates the threshold for mQTL hot spots, represented by the maximum number of mQTLs expected to fall into any interval by chance alone with a genome-wide P = 0.01.
The distributions of broad-sense heritability (H2) across all metabolites revealed a H2 of over 0.6 for more than 47% and 58% of the metabolites in flag leaf and germinating seed, respectively (Fig. S1A), indicating a significant genetic contribution in determining the content of these metabolites in both tissues. Correlation-based network analysis of the 100 codetected metabolites indicated that the metabolic network of germinating seed is more coordinately regulated than that of the flag leaf (Fig. S2A). Further analysis of annotated metabolites showed that the content of most flavonoids exhibited even higher heritability with a mean H2 > 0.7 for both tissues (Fig. S1A and Dataset S2). The levels of most flavonoids were controlled coordinately within each tissue, while distinct regulations were observed between the two tissues (Fig. S2B). However, the existence of two seed flavonoids in the cluster of leaf flavonoids, and vice versa, suggests that there is some overlapped regulation of this pathway between the tissues.
mQTL Mapping Using an Ultrahigh-Density SNP Map in Different Tissues.
mQTL mapping using an ultrahigh-density map consisted of 1,619 bins generated by population sequencing (13) resulted in 1,884 and 937 mQTLs in flag leaf and germinating seed [logarithm of odds (LOD) > 3.0], respectively. More than 90% of the metabolites had at least one mQTLs detected (Dataset S2). The number of mQTL for each metabolite varied from one to nine in flag leaf, with 120 metabolites having more than four mQTLs (Dataset S2). Although mQTL was not found for 15 (4.7%) metabolites in germinating seed, 48 metabolites had more than four mQTLs in this tissue (Dataset S2).
Of the 1,884 mQTLs detected in flag leaf, 1,682, 129, and 73 mQTLs accounted for <20%, 20–50%, and over 50% of the variation of the corresponding metabolites, respectively (Fig. S1B and Dataset S2). In germinating seed, 831 mQTLs accounted for <20% of the variation in metabolites, whereas 22 mQTLs had effects of over 50% (Fig. S1C and Dataset S2). In total, 202 and 106 mQTLs with effects of more than 20% were obtained in flag leaf and germinating seed, respectively (Dataset S2).
Genome-wide analysis of mQTLs revealed a significant deviation from random distribution across the 12 chromosomes of both in flag leaf (χ2 = 322.92, P < 2.2e−16) and in germinating seed (χ2 = 605.38, P < 2.2e−16). The occurrence of mQTL enriched regions indicates that major genes controlling levels of large set of metabolites may exist within them. We identified 44 and 16 potential mQTL “hot spots” in flag leaf and germinating seed, respectively. mQTL hot spots in flag leaf were mainly located on chromosomes 1, 6, 7, and 10, but on chromosomes 5 and 6 in germinating seed (Fig. 1 C and D and Table S1). When mQTLs of individual metabolite were compared, we detected 463 distinct loci among the total of 509 loci detected for the 100 codetected metabolites in the two tissues (Dataset S2), suggesting that the majority of QTLs are under different genetic control. For example, in germinating seed, two major QTLs for the level of l-glutamic acid were mapped to the 27.2- to 27.8-Mb region on chromosome 5 and the 1.6- to 1.8-Mb region on chromosome 6 with LOD of 23.6 and 24.2, respectively. However, QTLs for the same metabolite were mapped to 4.0–4.8 Mb on chromosome 1 and 5.5–9.9 Mb on chromosome 6 in flag leaf with much smaller effects (Dataset S2).
Despite the overall tissue-specific regulation of metabolism, we found that 23 loci for 19 metabolites (15 known and 4 unknown) were detected simultaneously in both tissues (Dataset S2), suggesting some overlapped genetic control of metabolism between tissues. For instance, a QTL for the level of chrysoeriol O-rutinoside was mapped to the same 0.5-Mb region on chromosome 1 in both tissues, and a QTL for the content of Helonioside B that mapped to 0.3-Mb region on chromosome 11 in flag leaf was also detected in germinating seed. A major QTL for the level of pyridoxine O-hexoside was observed in flag leaf within a 0.8-Mb region on chromosome 7. The same QTL was detected for this metabolite in germinating seed in addition to another major tissue-specific QTL in this tissue (Dataset S2). The above data suggested that, although some mQTLs could be detected simultaneously in both tissues, most of them functioned in a tissue-specific manner.
Complex Genetic Control of Metabolism Revealed by mQTL Analysis.
Different types of genetic control of metabolism were revealed by our mQTL analysis. The levels of some metabolites were controlled by only one major mQTL, which can explain more than 70% of the variation, such as m0202-L, m0458-L, m0707-L, m0853-L, m0876-L (Fig. S3A and Dataset S2).
However, the levels of most metabolites are determined by multiple loci. To test the possible interactions between mQTLs, we calculated the pairwise epistatic interactions between the mQTL hotspots against the average accumulation of known metabolites within the RILs (16, 17). All 946 possible pairwise epistatic interactions between the 44 mQTL hot spots were tested in flag leaf, and a total of 5,063 significant interactions (P < 0.01) was detected for 343 known metabolites, varying from 1 to 67 epistatic interactions for a single metabolite (Dataset S3). Similarly, analysis of pairwise epistatic interactions between the 16 mQTL hot spots for 151 known metabolites in germinating seed revealed from 1 to 29 significant (P < 0.01) epistatic interactions for 61 metabolites (Dataset S3). For example, qm0681-1 (bin147) on chromosome 1 and qm0681-2 (bin288) on chromosome 2, which are the two major mQTLs for m0681-L (annotated as chrysoeriol O-malonylglucoside), had significant interaction (P < 0.001) in determining the content of m0681-L (Fig. S3B and Dataset S3), in which the effect of MH63 allele at qm0681-1 on increasing the content of m0681-L was dependent on the ZS97 allele at qm0681-2 locus.
Confirmation of the mQTLs by Introgression Lines in Flag Leaf.
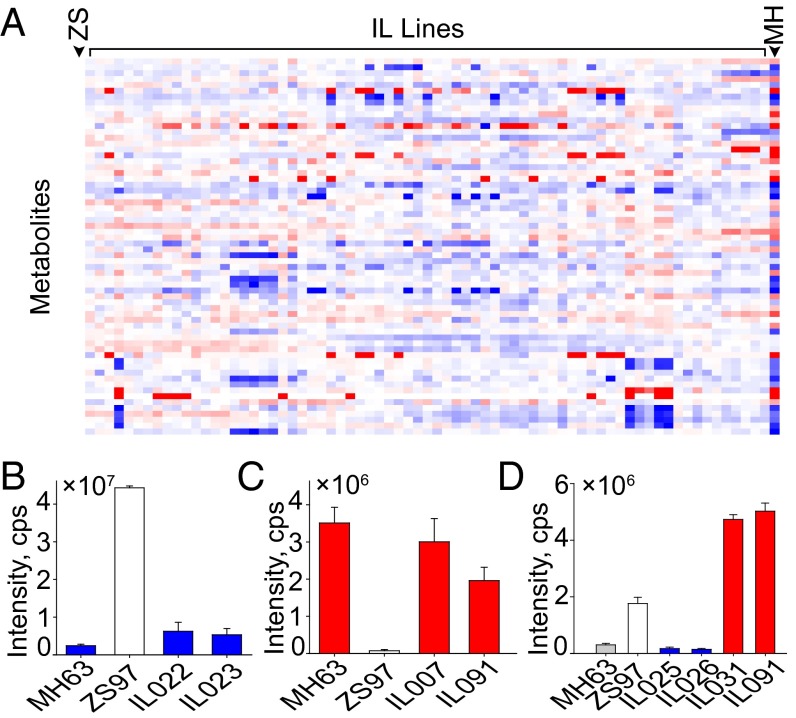
To evaluate the quality of our mQTL analyses, metabolic profiling of 64 selected metabolites was carried out using an introgression line (IL) population in which marker-defined genomic regions of ZS97 were replaced with homologous intervals of MH63 (Dataset S4). Fifty of the 64 mQTLs detected in RILs were confirmed in ILs that showed the expected variation for both the direction and amplitude of the variation, each of which was confirmed in at least two independent ILs (Fig. 2A). For instance, mQTL for m0723-L (annotated as tricin O-malonylhexoside) with a support interval from 16.6 to 18.1 Mb on chromosome 2 was validated by showing that the two ILs that both contain the 16.6- to 18.1-Mb segment of MH63 introduced into ZS97 background (IL022 and IL023) accumulated the MH63 level of m0723-L (Fig. 2B and Dataset S4). Similarly, the mQTL for m0873-L (annotated as C-pentosyl-apigenin O-caffeoylhexoside) was confirmed by examining the genotypes and comparing the content of the m0873-L of the two ILs (IL007 and IL091) relative to the parental lines (Fig. 2C).
Fig. 2.
Validation of the metabolic quantitative trait loci (mQTLs) results with introgression lines (ILs). (A) Overlay heat map of the metabolite profiles of the ILs in comparison with the parental control (ZS, ZS97; MH, MH63). Blue and red indicate that the metabolite contents are decreased or increased, respectively, after the introgression of MH63 segments. The content of m0723-L (tricin O-malonylhexoside) (B), m0873-L (C-pentosyl-apigenin O-caffeoylhexoside) (C), and m0681-L (chrysoeriol O-malonylhexoside) (D) in both parents and the corresponding ILs.
Moreover, mQTLs for metabolites controlled by more than one locus were also confirmed. For instance, the MH63 alleles of qm0681-1 and qm0681-2 contribute positively and negatively to m0681-L accumulation, respectively (Fig S3B and Dataset S2). The effect of each locus was confirmed by the fact that ILs (IL031 and IL091) with both the positive alleles had higher m0681-L level than that in either of the parents, whereas the opposite was the case for the ILs (IL025 and IL026) containing both of the negative alleles (Fig. 2D).
Identification of Candidate Genes for mQTLs.
The high resolution and large effects of our mQTLs facilitated the assignment of candidate genes for mQTLs. In most cases, the chemical structure of the metabolites, the existing knowledge of the pathway architecture, together with the annotated genome sequence allowed the tentative assignment of a protein or protein cluster as regulating the metabolic traits. Candidate gene mining of mQTLs for flavonoids allowed the tentative assignment of function to 24 candidate genes (Table 1 and Dataset S5) including 15 reported genes (10, 11, 18–25), with the majority of them characterized only in vitro.
Table 1.
The list of previously unidentified candidate genes for metabolic quantitative trait loci (mQTLs)
| Metabolite* | Chr | LOD† | Rsq‡ | Interval§ | Gene |
| Tri O-malhex | 2 | 146 | 0.95 | 16.5–18.1 | Os02g28340 |
| Tri O-hex-O-hex | 2 | 6 | 0.10 | 34.2–34.7 | Os02g56010 |
| m0434-L | 3 | 117 | 0.91 | 14.5–14.6 | Os03g25500 |
| Sin O-hex | 5 | 25 | 0.36 | 27.5–27.8 | Os05g47950 |
| l-Glutamic acid | 6 | 25 | 0.29 | 1.6–1.8 | Os06g03990 |
| Pyr O-hex | 7 | 19 | 0.17 | 0.0–0.2 | Os07g01020 |
| Tri O-hex der | 9 | 100 | 0.86 | 18.6–18.7 | Os09g30980 |
| Pyr O-hex | 10 | 24 | 0.30 | 0.0–0.8 | Os10g01080 |
| Api 7-O-rut | 11 | 44 | 0.29 | 14.2–15.4 | Os11g26950 |
The full names of metabolites’ abbreviation are given in SI Materials and Methods.
LOD, logarithm of odds.
Variation explained by the QTL.
1.5-LOD support interval of the QTL (in megabases).
An mQTL specific for three annotated flavonoid O-rutinosides (rather than flavonoid O-monoglycoside) was mapped to a 1.2-Mb region on chromosome 11 in flag leaf (Dataset S2), which suggested that a flavonoid O-glucoside: O-rhamnosyltransferase was involved. Os11g26950, one of the two annotated UDP-glucosyltransferases (UGTs) within the support interval, shows higher expression level and higher sequence identity to At5g54060 that encodes a flavonoid 3-O-glucoside: 2″-O-xylosyltransferase (26). Further phylogenetic analysis indicated that both Os11g26950 and At5g54060 clustered with UGTs that catalyze glycosyl transfer to a sugar moiety of flavonoid glycosides (Fig. S4A) (26). We therefore assign Os11g26950 as the candidate gene underlying this mQTL. Similarly, Os09g30980 (a putative UGT gene) was tentatively identified as the gene underlying the mQTL for m0837-L (putative tricin O-hexoside derivative) (Dataset S2) for its homology (36% identity at amino acid level) with At4g15280, which encodes a quercetin glucosyl transferase (27).
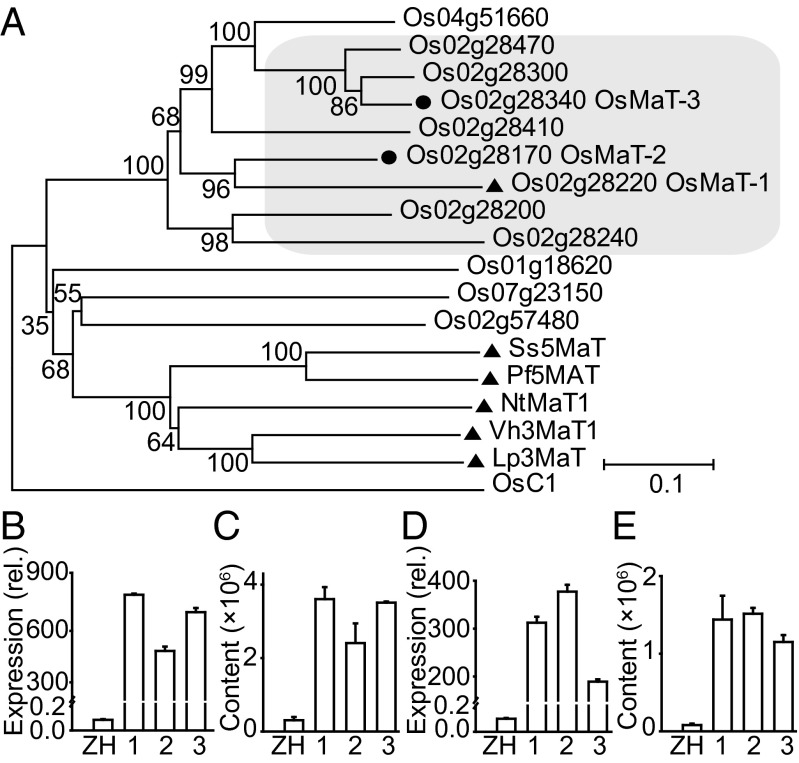
mQTLs for malonylated flavones were comapped to a 1.5-Mb region on chromosome 2 (Table 1 and Dataset S5). Candidate gene search within this region revealed a cluster of eight (putative) malonyltransferase genes (Fig. 3A) including OsMaT-2 that functioned as flavonol 3-O-glucoside malonyltransferase in vitro (20). OsMaT-2 and Os02g28340 (termed OsMaT-3 hereafter), which showed the highest expression levels among them and contained the conserved -YFGNC- motif typically detected in the anthocyanin/flavonoid BAHD acyltransferases (28), were assigned as the candidate genes, although effects of other genes could not be ruled out. Major mQTLs for a number of aromatically acylated flavonoids were comapped to a 0.5-Mb interval on chromosome 10 (Dataset S2), but not for their nonacylated precursors, suggesting the involvement of a gene encoding an acyltransferase. Only one acyltransferase gene Os10g11980 (OsAT1) was found within this region, encoding an acyltransferase that confers bacterial blight resistance in rice (22). Comparison of Os10g11980 sequences disclosed 1-aa difference of this gene between ZS97 (Ser) and MH63 (Ala), suggesting Os10g11980 to be the likely candidate gene underlying the mQTL for these acylated flavonoids.
Fig. 3.
Functional identification of OsMaT-2 and OsMaT-3. (A) Phylogenetic analysis of 12 BAHD acyltransferases with OsC1 as an outgroup. The neighbor-joining tree was constructed using aligned full-length amino acid sequences. Bootstrap values from 1,000 replicates are indicated at each node. (Bar: 0.1-aa substitutions per site.) GenBank accession numbers are given in SI Materials and Methods. The mRNA level of OsMaT-2 (B) and the content of m0723-L (C) in OsMaT-2 overexpressors (1–3) (T1) and ZH11. The mRNA level of OsMaT-3 (D) and the content of m0723-L (E) in OsMaT-3 overexpressors (1–3) (T1) and ZH11. All data are given as mean ± SEM (n = 3).
mQTLs for both C- and O- glycosyl flavonoids with various structural classes were mapped to a 0.5-Mb region on chromosome 5, suggesting the involvement of a regulatory gene underlying these QTLs. Candidate gene search within this region revealed a gene OsMYB55 (Os05g48010) modulating amino acid metabolism that was involved in the tolerance to high temperature (29). Interestingly, Os05g48010 was coexpressed highly with several structural genes of the flavonoid pathway (Table S2). Sequence comparison of Os05g48010 indicates 1-aa difference between ZS97 (Leu) and MH63 (Ile). These data suggest that Os05g48010 could be the candidate gene underlying the mQTLs.
Candidate genes underlying mQTLs for metabolites from other pathways such as pyridoxine, amino acid, and LPCs were also tentatively assigned (Table 1 and Dataset S5). m0283 (annotated as pyridoxine O-glucoside) was mapped into a 0.2-Mb interval on chromosome 7 in both tissues investigated (Dataset S2). Os07g01020 (encoding a putative SOR/SNZ family protein) was tentatively identified as underlying this mQTL for its high homology (85% identity at amino acid level) with the pyridoxine synthase gene AtPDX1 (30). m0033-S (l-glutamic acid) was mapped to a 0.2-Mb interval on chromosome 6 in germinating seed. The high homology (51% identity at amino acid level) between Os06g03990, the only biochemical related gene in the interval, and the aminotransferase gene AtACS12 (31) suggests that Os06g03990 is likely the candidate gene underlying this QTL. m0297-S (annotated as sinapoyl O-hexoside) was mapped into a 0.3-Mb interval on chromosome 5 in germinating seed. Os05g47950, encoding a putative UGT, was assigned as candidate gene for these mQTLs for its homology (31% at amino acid level) with At3g21560 that catalyze the formation of sinapoyl glucose (32) and also because it was one of the two biochemically relevant genes within the support interval. QTLs for 10 putative LPCs colocalized to a 0.2-Mb region on chromosome 6 (Dataset S5). Candidate gene mining within the support interval revealed no gene involved in LPC metabolism/catabolism, but a gene (Os06g04200) that encodes starch synthase was detected (33) (Dataset S5). LPCs are the main phospholipids in rice kernels and their contents are an important determinant of starch quality. Previous results suggested that starch synthase IIIa (SSIIIa) and starch branching enzyme (BE) affect the metabolite composition including LPC levels in rice kernels (34). We therefore assign Os06g04200 as a candidate gene underlying the mQTLs regulating these LPCs.
The high resolution and large effects of our mQTLs also facilitate the assignment of candidate genes for QTLs of unknown metabolites. A QTL for m0434-L (m/z 427/397) detected in flag leaf was mapped to a region of 0.1 Mb on chromosome 3 and explained over 90% of the variation (Dataset S2). Examination of the expression profiles of genes within the region revealed a leaf-specific gene Os03g25500 (encoding putative cytochrome P450 72A1). Comparison of the sequences of Os03g25500 between ZS97 and MH63 revealed several SNPs, one of which (GAG for Glu in MH63 to TAG for stop codon in ZS97) caused a premature termination of Os03g25500 in ZS97 compared with MH63. We therefore assign Os03g25500 as the candidate gene underlying the mQTL for the level of m0434-L.
Functional Identification of Three Candidate Genes in Vivo.
Experimental validation of all candidate genes is beyond the scope of a single study, but we showed that confirmation is possible by demonstrating the contributions of candidate genes to the accumulation of the corresponding metabolites. To confirm the in vivo function of Os11g26950, a candidate gene for the level of m0760-L, this gene was introduced into ZH11 (a variety with low m0760-L content) and the results showed that the content of m0760-L was significantly increased in the transgenic plants than the control (Fig. S4 B and C), which is in accordance with the result of the QTL analysis. Similarly, when the two malonyltransferases were overexpressed in ZH11, the content of m0723-L was substantially increased in both the OsMaT-2 (Fig. 3 B and C) and OsMaT-3 (Fig. 3 D and E) overexpressors compared with the control, whereas malonylated flavonols were detectable in neither the control nor the transgenic plants, indicating that they both function as flavone malonyltransferase in vivo.
Reconstruction of Metabolic Pathways from mQTL Results.
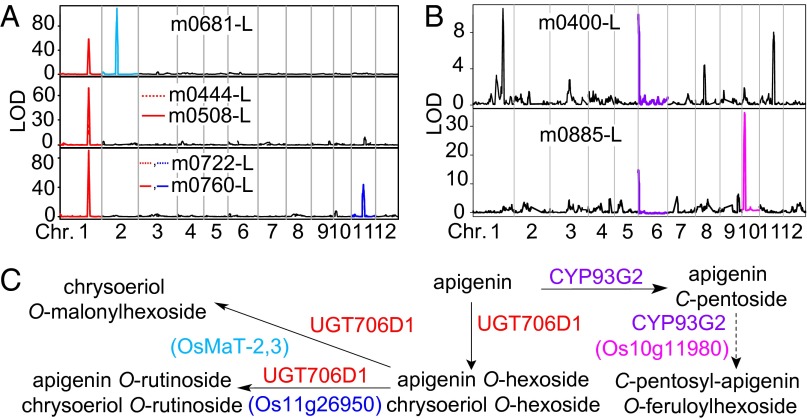
Combining prior knowledge of pathway architecture, the chemical structure of identified/annotated metabolites and the candidate genes revealed in the study, we updated and reconstructed pathways of the corresponding metabolites in rice (Figs. 4 and 5) using the genetic logistic approach as previously described (35, 36). m0508-L (chrysoeriol 7-O-hexoside) showed only one QTL at the UGT706D1 locus, whereas m0681-L (annotated as chrysoeriol O-malonylglucoside) mapped to UGT706D1 and OsMaT-2 loci, and m0760-L (annotated as chrysoeriol O-rutinoside) to UGT706D1 and Os11g26950 loci (Fig. 4A and Dataset S5). This enabled us to put both OsMaT-2 and Os11g26950 downstream of UGT706D1 with different branches (Fig. 4C). This deduction also applies to the biosynthesis and modification of C-glycosyl flavones (Fig. 4 B and C). Using this strategy, the pathways were updated and reconstructed (Fig. 5).
Fig. 4.
Typical pathway model for metabolic pathway reconstruction. (A) QTL mapping results of m0681-L (chrysoeriol O-malonylhexoside), m0444-L (apigenin O-hexoside), m0508-L (chrysoeriol O-hexoside), m0722-L (apigenin O-rutinoside), and m0760-L (chrysoeriol O-rutinoside). (B) QTL mapping results of m0400-L (apigenin C-pentoside) and m0885-L (C-pentosyl-apigenin O-feruloylhexoside). (C) The candidate genes of the aforementioned metabolites.
Fig. 5.
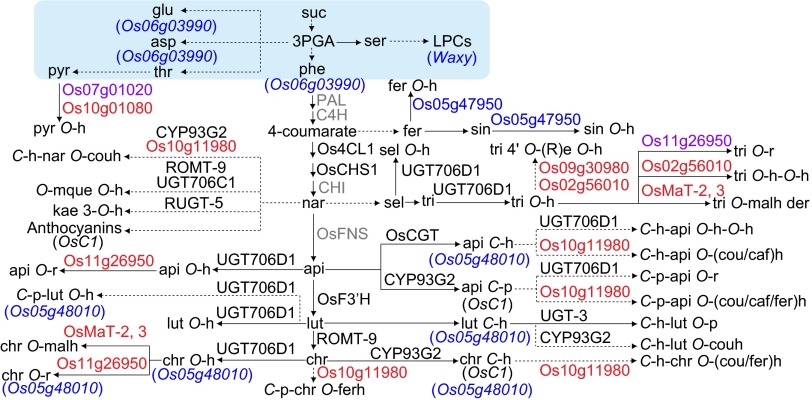
Reconstructed rice metabolic pathways based on the metabolomics and metabolic quantitative trait locus (mQTL) analysis. The candidate genes in red and blue indicate the newly mapped genes in flag leaf and germinating seed, respectively. The candidate genes in purple indicate the mapped genes in both tissues. Reported genes shown in black were mapped in this study, whereas those in gray were not. The box in light blue indicates primary metabolic pathways. The full names of metabolites’ abbreviation are given in SI Materials and Methods.
In the newly constructed pathways, most of the structural genes reported previously for flavonoids biosyntheses were mapped (Fig. 5). The broad substrate specificities of both CYP93G2, and UGT706D1 revealed in our study (Fig. 5 and Dataset S5) are consistent with previous studies (10, 37). OsMaT-2, a malonyltransferase that displayed higher activity toward glycosyl flavonols such as quercetin 3-O-glucoside in vitro (20), was reannotated with its in vivo function listed as specific to glycosyl flavones (Fig. 5 and Dataset S5). In addition, genes underlying the mQTLs for primary metabolites such as LPCs, amino acids, and pyridoxine with its derivative were also mapped (Fig. 5 and Dataset S5).
Tissue-specific regulations of most known metabolites (Fig. S2A) were reflected by the different genes assigned for mQTLs of the same metabolites between the two tissues tested (Fig. 5). However, Os11g26950 and Os07g01020 were assigned to be the candidate genes underlying the accumulation of flavones rutinoside and pyridoxine derivative, respectively, in both tissues (Fig. 5), indicating overlapping regulation of the pathway, to some extent, between the two tissues. Distinct regulation of the metabolism in the two tissues might arise partially from the tissue-specific expression of the responsible gene. For example, Os05g48010, which was tentatively assigned to be responsible for the accumulation of most of the flavonoids in germinating seed, was expressed in this tissue but not in flag leaf (Fig. 5).
Discussion
The large number, high resolution, and large effects of the mQTL detected in our study were mainly due to the high coverage, sensitivity, and accuracy of the metabolomic method used, and the high density of SNP markers. The characteristics of our combined omics strategy have enabled us to identify candidate genes directly from the genetic mapping and to generate a large number of hypotheses for follow-up studies (Table 1 and Dataset S5). In addition to high-throughput annotation of unknown metabolites and candidate genes for various metabolites, including those associated with morphological traits, our study greatly updated the knowledge of pathways of both primary and secondary metabolites in rice in terms of biosynthesis, modification, and regulation. Transcription factor and enzymes with unknown functions have been identified/annotated, and functional annotation of genes previously identified in vitro has been confirmed/updated in vivo. The distinct and overlapping regulation of metabolic pathways was also revealed in the two different tissues (Fig. 5). Our study demonstrates the potential of a combined omics strategy in understanding the genetic basis of rice metabolome.
Research in humans has shown that revealing genetic influences on metabolic phenotypes is crucial to dissect complex disorders and the definition of the pathophysiological basis of disease susceptibility (38). The dissection and the reconstruction of metabolic pathways in rice using mQTL analysis provides approaches for engineering the compounds that have health-promoting effects for humans (8) or confer resistance to fungi pathogens (39). The discovery of Os07g01020 as a tentative determinator of pyridoxine O-glucoside, Os10g11980 (OsAT1) with a potential activity in aromatic acylated flavonoid biosynthesis that confers blight resistance, and Os06g04200, which encodes starch synthase with the proposed role in regulating LPCs (Table 1 and Dataset S5), not only provides potential targets for quality improvement but also demonstrates the potential in uncovering the mechanisms of complex agronomic traits.
Integrating metabolomics strategies with other high-throughput technologies will play important roles in genomics-assisted selection for crop improvement. Although the linkage mapping strategy described here proved to be a powerful tool in detecting large number of mQTLs with high resolution, further increase in the number of mQTLs might be limited by the availability of appropriate mapping populations, with further increase of resolution hindered by limited recombinant events, hence long linkage blocks (40). Advance in genome-wide association studies (GWAS) with high-throughput genotyping techniques has made it possible to land close to the gene of interests for agronomical important traits in rice (41) and more recently in maize (42). Appling our widely targeted metabolic profiling methodology into GWAS has the potential to further accelerate the pace of functional genomics in major crops such as rice and maize, thus providing a powerful tool for crop improvement.
Materials and Methods
Details on experimental materials and methods are presented in SI Materials and Methods. A summary is given below.
The mapping population consisted of 210 RILs derived from a cross between ZS97 and MH63. Metabolic profiling was carried out as previously described (12). An ultrahigh-density map was used for the mQTL mapping (13). Seventy-one ILs generated from the same parents as the RILs were used for validating the mQTL results. Primers used in this study are shown in Table S3.
Supplementary Material
Acknowledgments
This work was supported by Major State Basic Research Development Program of China (973 Program) Grant 2013CB127001, National High Technology R&D Program of China (863 Program) Grant 2012AA10A304, National Natural Science Foundation of China Grant 31070267, and the Program for New Century Excellent Talents in University of Ministry of Education in China (NCET-09-0401).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319681110/-/DCSupplemental.
References
- 1.Dixon RA, Strack D. Phytochemistry meets genome analysis, and beyond. Phytochemistry. 2003;62(6):815–816. doi: 10.1016/s0031-9422(02)00712-4. [DOI] [PubMed] [Google Scholar]
- 2.Saito K, Matsuda F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu Rev Plant Biol. 2010;61:463–489. doi: 10.1146/annurev.arplant.043008.092035. [DOI] [PubMed] [Google Scholar]
- 3.Matsuda F, et al. Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J. 2012;70(4):624–636. doi: 10.1111/j.1365-313X.2012.04903.x. [DOI] [PubMed] [Google Scholar]
- 4.Keurentjes JJB, et al. The genetics of plant metabolism. Nat Genet. 2006;38(7):842–849. doi: 10.1038/ng1815. [DOI] [PubMed] [Google Scholar]
- 5.Toubiana D, et al. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genet. 2012;8(3):e1002612. doi: 10.1371/journal.pgen.1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrari F, et al. Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 2006;142(4):1380–1396. doi: 10.1104/pp.106.088534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol. 2002;5(3):218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
- 8.Luo J, et al. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008;56(2):316–326. doi: 10.1111/j.1365-313X.2008.03597.x. [DOI] [PubMed] [Google Scholar]
- 9.Mehrtens F, Kranz H, Bednarek P, Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138(2):1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du Y, Chu H, Chu IK, Lo C. CYP93G2 is a flavanone 2-hydroxylase required for C-glycosylflavone biosynthesis in rice. Plant Physiol. 2010;154(1):324–333. doi: 10.1104/pp.110.161042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brazier-Hicks M, et al. The C-glycosylation of flavonoids in cereals. J Biol Chem. 2009;284(27):17926–17934. doi: 10.1074/jbc.M109.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen W, et al. A novel integrated method for large-scale detection, identification and quantification of widely-targeted metabolites: Application in study of rice metabolomics. Mol Plant. 2013 doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 13.Yu H, et al. Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS One. 2011;6(3):e17595. doi: 10.1371/journal.pone.0017595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernie AR, et al. Recommendations for reporting metabolite data. Plant Cell. 2011;23(7):2477–2482. doi: 10.1105/tpc.111.086272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dresen S, Ferreirós N, Gnann H, Zimmermann R, Weinmann W. Detection and identification of 700 drugs by multi-target screening with a 3200 Q TRAP LC-MS/MS system and library searching. Anal Bioanal Chem. 2010;396(7):2425–2434. doi: 10.1007/s00216-010-3485-2. [DOI] [PubMed] [Google Scholar]
- 16.Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell. 2008;20(5):1199–1216. doi: 10.1105/tpc.108.058131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou G, et al. Genetic composition of yield heterosis in an elite rice hybrid. Proc Natl Acad Sci USA. 2012;109(39):15847–15852. doi: 10.1073/pnas.1214141109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gui J, Shen J, Li L. Functional characterization of evolutionarily divergent 4-coumarate:coenzyme a ligases in rice. Plant Physiol. 2011;157(2):574–586. doi: 10.1104/pp.111.178301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim BG, Lee Y, Hur HG, Lim Y, Ahn JH. Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry. 2006;67(4):387–394. doi: 10.1016/j.phytochem.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 20.Kim DH, Kim SK, Kim J-H, Kim B-G, Ahn J-H. Molecular characterization of flavonoid malonyltransferase from Oryza sativa. Plant Physiol Biochem. 2009;47(11-12):991–997. doi: 10.1016/j.plaphy.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Shih CH, et al. Functional characterization of key structural genes in rice flavonoid biosynthesis. Planta. 2008;228(6):1043–1054. doi: 10.1007/s00425-008-0806-1. [DOI] [PubMed] [Google Scholar]
- 22.Mori M, et al. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol. 2007;63(6):847–860. doi: 10.1007/s11103-006-9130-y. [DOI] [PubMed] [Google Scholar]
- 23.Kim BG, et al. Flavonoid O-diglucosyltransferase from rice: Molecular cloning and characterization. J Plant Biol. 2009;52:41–48. [Google Scholar]
- 24.Ko JH, Kim BG, Hur HG, Lim Y, Ahn JH. Molecular cloning, expression and characterization of a glycosyltransferase from rice. Plant Cell Rep. 2006;25(7):741–746. doi: 10.1007/s00299-006-0119-4. [DOI] [PubMed] [Google Scholar]
- 25.Saitoh K, Onishi K, Mikami I, Thidar K, Sano Y. Allelic diversification at the C (OsC1) locus of wild and cultivated rice: Nucleotide changes associated with phenotypes. Genetics. 2004;168(2):997–1007. doi: 10.1534/genetics.103.018390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yonekura-Sakakibara K, et al. Two glycosyltransferases involved in anthocyanin modification delineated by transcriptome independent component analysis in Arabidopsis thaliana. Plant J. 2012;69(1):154–167. doi: 10.1111/j.1365-313X.2011.04779.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim EK, Ashford DA, Hou B, Jackson RG, Bowles DJ. Arabidopsis glycosyltransferases as biocatalysts in fermentation for regioselective synthesis of diverse quercetin glucosides. Biotechnol Bioeng. 2004;87(5):623–631. doi: 10.1002/bit.20154. [DOI] [PubMed] [Google Scholar]
- 28.D’Auria JC. Acyltransferases in plants: A good time to be BAHD. Curr Opin Plant Biol. 2006;9(3):331–340. doi: 10.1016/j.pbi.2006.03.016. [DOI] [PubMed] [Google Scholar]
- 29.El-Kereamy A, et al. The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One. 2012;7(12):e52030. doi: 10.1371/journal.pone.0052030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li JZ, Zhang ZW, Li YL, Wang QL, Zhou YG. QTL consistency and meta-analysis for grain yield components in three generations in maize. Theor Appl Genet. 2011;122(4):771–782. doi: 10.1007/s00122-010-1485-4. [DOI] [PubMed] [Google Scholar]
- 31.Yamagami T, et al. Biochemical diversity among the 1-amino-cyclopropane-1-carboxylate synthase isozymes encoded by the Arabidopsis gene family. J Biol Chem. 2003;278(49):49102–49112. doi: 10.1074/jbc.M308297200. [DOI] [PubMed] [Google Scholar]
- 32.Lim EK, et al. Identification of glucosyltransferase genes involved in sinapate metabolism and lignin synthesis in Arabidopsis. J Biol Chem. 2001;276(6):4344–4349. doi: 10.1074/jbc.M007263200. [DOI] [PubMed] [Google Scholar]
- 33.Tian Z, et al. Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc Natl Acad Sci USA. 2009;106(51):21760–21765. doi: 10.1073/pnas.0912396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusano M, et al. Deciphering starch quality of rice kernels using metabolite profiling and pedigree network analysis. Mol Plant. 2012;5(2):442–451. doi: 10.1093/mp/ssr101. [DOI] [PubMed] [Google Scholar]
- 35.Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics. 2001;159(1):359–370. doi: 10.1093/genetics/159.1.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kliebenstein D, Pedersen D, Barker B, Mitchell-Olds T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics. 2002;161(1):325–332. doi: 10.1093/genetics/161.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko JH, et al. Four glucosyltransferases from rice: cDNA cloning, expression, and characterization. J Plant Physiol. 2008;165(4):435–444. doi: 10.1016/j.jplph.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Suhre K, et al. CARDIoGRAM Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulpice R, et al. Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA. 2009;106(25):10348–10353. doi: 10.1073/pnas.0903478106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mauricio R. Mapping quantitative trait loci in plants: Uses and caveats for evolutionary biology. Nat Rev Genet. 2001;2(5):370–381. doi: 10.1038/35072085. [DOI] [PubMed] [Google Scholar]
- 41.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 42.Riedelsheimer C, et al. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA. 2012;109(23):8872–8877. doi: 10.1073/pnas.1120813109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.