Significance
Drug resistance is an increasing problem in clinical settings with some bacterial pathogens now resistant to virtually all available drugs. Chlorhexidine is a commonly used antiseptic and disinfectant in hospital environments, and there is increasing resistance to chlorhexidine seen in some pathogenic bacteria, such as Acinetobacter baumannii. This paper examines the global gene expression of A. baumannii in response to chlorhexidine exposure and identifies a gene that we demonstrate to mediate chlorhexidine resistance. Biochemical investigation reveals that this gene encodes a previously uncharacterized type of drug efflux pump that actively transports chlorhexidine out of the cell.
Keywords: drug resistance, membrane transport, opportunistic pathogen
Abstract
Chlorhexidine is widely used as an antiseptic or disinfectant in both hospital and community settings. A number of bacterial species display resistance to this membrane-active biocide. We examined the transcriptomic response of a representative nosocomial human pathogen, Acinetobacter baumannii, to chlorhexidine to identify the primary chlorhexidine resistance elements. The most highly up-regulated genes encoded components of a major multidrug efflux system, AdeAB. The next most highly overexpressed gene under chlorhexidine stress was annotated as encoding a hypothetical protein, named here as AceI. Orthologs of the aceI gene are conserved within the genomes of a broad range of proteobacterial species. Expression of aceI or its orthologs from several other γ- or β-proteobacterial species in Escherichia coli resulted in significant increases in resistance to chlorhexidine. Additionally, disruption of the aceI ortholog in Acinetobacter baylyi rendered it more susceptible to chlorhexidine. The AceI protein was localized to the membrane after overexpression in E. coli. This protein was purified, and binding assays demonstrated direct and specific interactions between AceI and chlorhexidine. Transport assays using [14C]-chlorhexidine determined that AceI was able to mediate the energy-dependent efflux of chlorhexidine. An E15Q AceI mutant with a mutation in a conserved acidic residue, although unable to mediate chlorhexidine resistance and transport, was still able to bind chlorhexidine. Taken together, these data are consistent with AceI being an active chlorhexidine efflux protein and the founding member of a family of bacterial drug efflux transporters.
Drug resistance determinants in bacterial pathogens are generally held under some level of regulatory control, presumably expressed only when required to avoid superfluous expenditure of cellular resources. Therefore, changes in gene expression can be exploited to identify the core drug resistance factors of an organism. To this end, we applied genome-wide transcriptomic analyses to identify the key chlorhexidine resistance determinants in the representative Gram-negative nosocomial pathogen Acinetobacter baumannii.
Chlorhexidine is a bisbiguanide antimicrobial agent that is extensively used in a range of antiseptic products ranging from skin washes, soaps, mouthwash, disinfectants, and preservatives (1). Chlorhexidine is effective against both Gram-positive and Gram-negative bacteria, with the primary modes of action involving cytoplasmic membrane damage, as well as intracellular protein precipitation upon prolonged exposure (2). Nonetheless, a number of microbial pathogens, particularly those associated with hospital-acquired infections, display high-level resistance to chlorhexidine. For example, A. baumannii isolates from some health care settings can survive chlorhexidine concentrations of at least 1% (3). Given the importance of chlorhexidine in controlling the spread of nosocomial pathogens, it is important to understand the potential resistance mechanisms used by these organisms.
Bacterial chlorhexidine resistance has been shown or suggested to occur either through alterations of the outer membrane that affect membrane permeability (4) or active efflux mediated by integral membrane transport proteins (5). The majority of chlorhexidine efflux systems described to date are multidrug efflux systems that confer resistance to a broad range of structurally diverse antimicrobial compounds, e.g., in A. baumannii the resistance/nodulation/division (RND) superfamily efflux systems AdeABC (Acinetobacter drug efflux) and AdeIJK (6), the major facilitator superfamily (MFS) AedF(Acinetobacter exporter of the DHA2 family) transporter (7), and the small multidrug resistance (SMR) family exporter AdeS (8) have been shown to confer chlorhexidine resistance. This study aimed to identify the core chlorhexidine resistance mechanisms operating in Gram-negative bacteria using a whole-genome transcriptomic analysis of A. baumannii as a representative Gram-negative pathogen and to functionally characterize proteins involved in the resistance response.
Results and Discussion
The A. baumannii Transcriptional Response to Chlorhexidine Was Highly Focused.
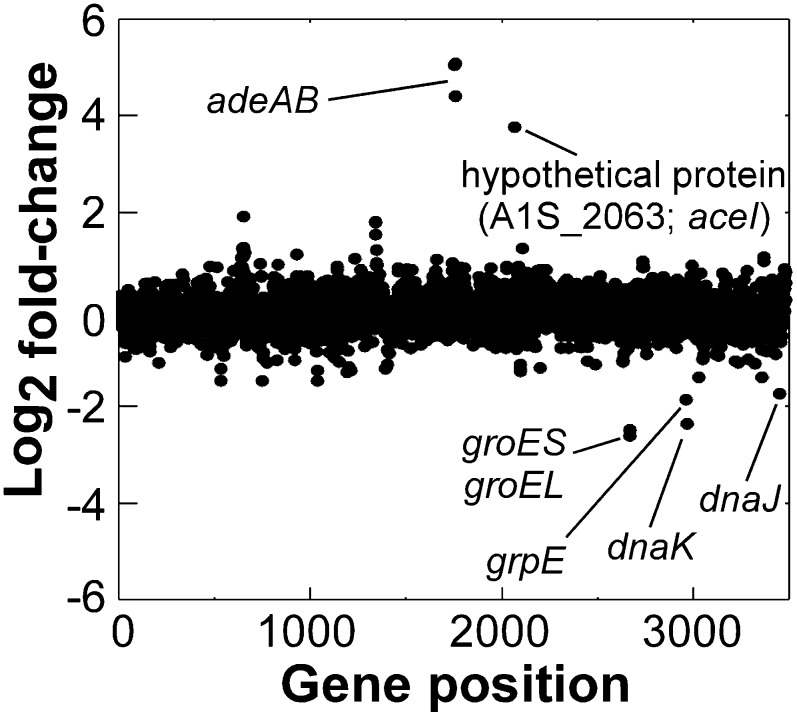
A whole-genome microarray was used to compare RNA transcript levels between cultures receiving a subinhibitory shock treatment with chlorhexidine (4 µg/mL) and cultures receiving no shock treatment. Surprisingly, after this treatment, only 22 and 35 genes were up-regulated and down-regulated by more than twofold, respectively, suggesting that the A. baumannii response to chlorhexidine is highly focused (Fig. 1 and Tables S1 and S2). The microarray expression results were validated by quantitative reverse-transcriptase PCR using a subset of differentially expressed genes (Fig. S1).
Fig. 1.
Global transcriptional response of A. baumannii ATCC 17978 to chlorhexidine shock. Each dot represents a single ORF within the genome numbered according to locus tag along the x axis, and its fold-change (Log2) in expression in response to treatment with 4 µg/mL chlorhexidine for 30 min. Genes or gene clusters of particular interest are labeled.
The most prominently down-regulated genes under chlorhexidine stress were those encoding heat shock proteins (Fig. 1 and Table S1), which are likely to function as chaperones. Similarly, a number of these genes were down-regulated in response to chlorhexidine exposure in Pseudomonas aeruginosa (9) (Table S1).
Genes encoding the AdeAB efflux system were the most highly up-regulated under chlorhexidine stress; each was overexpressed by 20- to 35-fold (Fig. 1 and Table S2). The AdeABC efflux system has been shown to mediate resistance to chlorhexidine in clinical A. baumannii isolates (6). Studies conducted in P. aeruginosa and Burkholderia cenocepacia have shown that overexpression of homologous RND efflux systems is a major part of the transcriptional response to chlorhexidine stress (9, 10), indicating that these proteins are primary chlorhexidine resistance factors across a number of Gram-negative genera. Other known chlorhexidine efflux system genes found in A. baumannii were not induced by exposure to chlorhexidine, e.g., aedF encoding a multidrug efflux system, as reported (7).
A Small Hypothetical Membrane Protein Was Overexpressed in Response to Chlorhexidine.
With the exception of the adeAB efflux transporter genes, only one gene was overexpressed by more than 10-fold in response to chlorhexidine exposure in A. baumannii, A1S_2063, which was annotated as encoding a hypothetical protein. Orthologs of A1S_2063 in P. aeruginosa and B. cenocepacia were also among the most overexpressed genes in response to chlorhexidine challenge (9, 10). Therefore, these genes are a highly conserved component of a chlorhexidine-responsive regulatory circuit in several proteobacterial genera. Sequence analyses (11) suggested that the A1S_2063 gene product contains four predicted transmembrane α-helices (Fig. S2) and is likely localized in the cytoplasmic membrane. Pfam database searches showed that the protein contains two PF05232 (Bacterial Transmembrane Pair family; BTP) domains, which are conserved in a number of hypothetical proteins encoded primarily by proteobacteria (12) (Fig. 2). To date, no functional roles for any proteins from the BTP family have been determined.
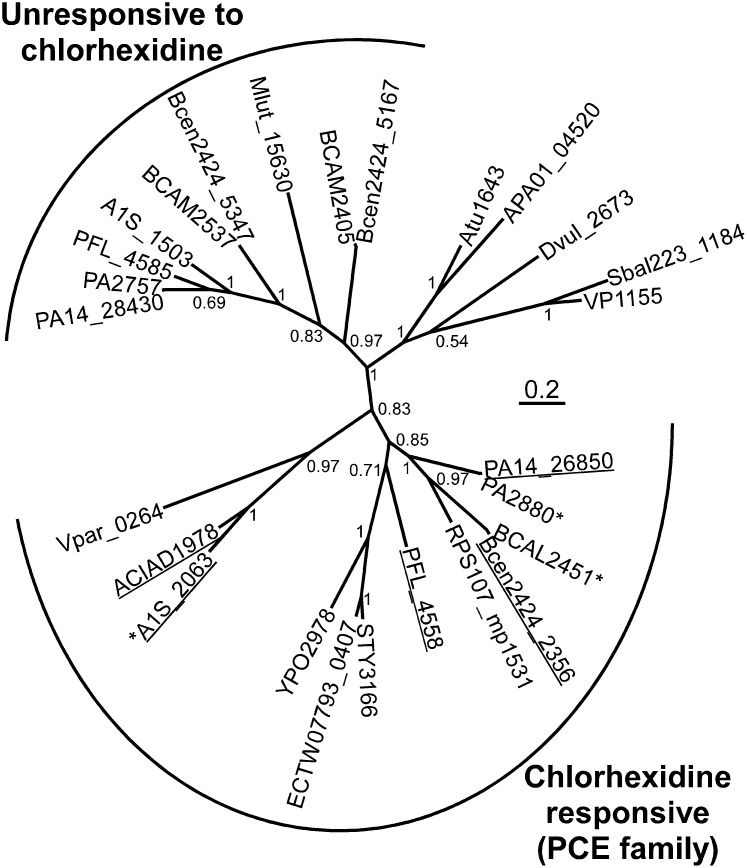
Fig. 2.
Phylogenetic tree generated by using MrBayes 3.2.1 (33) from a ClustalX2 alignment of representative BTP proteins. Sequences were obtained from the National Center for Biotechnology Information database. The interior node values are the clade credibility values (posterior probabilities) generated by MrBayes. Genes found to mediate chlorhexidine resistance in this study by heterologous expression in E. coli (Table 1) or using gene deletion strains (Fig. S4) are underlined. Genes found to be transcriptionally responsive to chlorhexidine exposure in this study, or elsewhere (9, 10) are marked with an asterisk (*).
Several bacterial strains encode more than one BTP family protein. Indeed, A. baumannii ATCC 17978 itself encodes two of these proteins, although expression of the second, A1S_1503, was not affected by chlorhexidine challenge. P. aeruginosa and B. cenocepacia also encode 2–3 BTP family proteins. Of the genes encoding these proteins, those most closely related to A1S_2063, within the “chlorhexidine responsive clade” (Fig. 2), are transcriptionally responsive to chlorhexidine, whereas those related to A1S_1503, within the “unresponsive to chlorhexidine clade” (Fig. 2), are not transcriptionally responsive to chlorhexidine (9, 10).
The A1S_2063 Product and Orthologous BTP Family Proteins Confer Resistance to Chlorhexidine.
Despite their conservation across proteobacterial genera, BTP family proteins are absent from sequenced Escherichia strains, except for Escherichia coli TW07793 and KTE84, which carry a homolog of A1S_2063 that is flanked by transposase and phage genes, suggesting that it has been laterally acquired in these strains (Fig. 2). Initial characterization of A1S_2063 in chlorhexidine tolerance involved its expression as a His-tag (RGSH6) fusion in E. coli BL21(DE3) using plasmid pTTQ18 (13, 14). Western blot analyses determined that the A1S_2063 protein was localized to the membrane fraction in E. coli (Fig. S3 A and B). Minimum inhibitory concentration (MIC) analyses determined that E. coli BL21(DE3) was significantly more resistant to chlorhexidine when expressing A1S_2063, than when carrying an empty vector (Table 1), indicating that this gene is a previously uncharacterized chlorhexidine resistance determinant. Importantly, the A1S_2063 protein conferred chlorhexidine resistance in a suite of E. coli deletion strains with mutations to each of the 20 known E. coli drug efflux systems (15), demonstrating that it is not reliant on any of these proteins for function (Table S3).
Table 1.
Chlorhexidine resistance levels of cells overexpressing BTP family proteins
| Strain | Chlorhexidine MIC*, µg/mL |
| Parental | 0.16 |
| A1S_2063 (aceI) | 1.25 |
| ACIAD1978 | 1.25 |
| PA14_26850 | 0.63 |
| PFL_4558 | 1.25 |
| Bcen2424_2356 | 1.25 |
| A1S_1503 | 0.16 |
| PFL_4585 | 0.16 |
| E15Q A1S_2063 (E15Q aceI) | 0.16 |
| A1S_2057 (aedF)† | 2.5 |
The results shown were generated using the broth dilution method and are from a minimum of seven replicates including at least two biological replicates.
Results are shown for E. coli strain BL21(DE3) carrying empty pTTQ18 (parental) or pTTQ18 with the gene indicated and have been rounded to two decimal places. These studies were also conducted in E. coli AG100A, which contains a mutation in the gene encoding a major E. coli multidrug efflux protein AcrB (25). The fold changes in chlorhexidine resistance between cells expressing the genes listed and the parental strain were identical between BL21(DE3) and AG100A.
aedF encodes a previously characterized multidrug efflux protein (7) used as a control in this work.
To determine whether BTP family proteins from other proteobacterial strains could similarly mediate chlorhexidine resistance, several representatives were also expressed in E. coli (Fig. S3C). Genes orthologous to A1S_2063 from Acinetobacter baylyi ADP1 (ACIAD1978), P. aeruginosa PA14 (PA14_26850), Pseudomonas protegens Pf-5 (PFL_4558), and B. cenocepacia HI2424 (Bcen2424_2356) were also able to mediate chlorhexidine resistance in E. coli (Table 1 and Fig. 2). We were unable to construct a gene deletion mutant of A1S_2063 in A. baumannii, therefore, we generated a gene knockout mutant of the closely related ACIAD1978 gene in A. baylyi ADP1. As stated above, this gene confers chlorhexidine resistance in E. coli (Table 1). Furthermore, ACIAD1978 is 67% identical in amino acid sequence to the A1S_2063 protein in A. baumannii (Fig. 2) and its expression is inducible by chlorhexidine in a dose-dependent manner. All these observations show that ACAID1978 from A. baylyi is an excellent surrogate for investigation of this group of BTP family proteins. A. baylyi ADP1 ΔACIAD1978 showed increased chlorhexidine susceptibility compared with its isogenic parent (Fig. S4). The A. baumannii BTP family gene that was not transcriptionally responsive to chlorhexidine, A1S_1503, and its P. protegens Pf-5 ortholog, PFL_4585 (Fig. 2), did not mediate chlorhexidine resistance when expressed in E. coli (Table 1).
To explore additional resistance phenotypes that may be mediated by A1S_2063, the E. coli strain overexpressing this gene was tested for resistance to a range of antimicrobial compounds of different structures and valencies, including alexidine, benzalkonium, chloramphenicol, 4′,6-diamidino-2-phenylindole, ethidium, SDS, and tetracycline (Table S4). The A1S_2063 overexpression strain was no more resistant to these compounds than the parental strain. To examine sensitivity of the A1S_2063 overexpression strain to a broader panel of antimicrobials, as well as pH and osmolyte stresses, the Biolog OmniLog Phenotype MicroArray (PM) system was used with plates PM9-20 (16). Of the 240 different antimicrobial compounds included in the Biolog PM11-20 series, A1S_2063 was only able to mediate resistance to chlorhexidine (Fig. S5). Additionally, A1S_2063 did not improve the fitness of E. coli in any of the pH or osmolyte tests included in the Biolog PM9-10 plates.
An Intramembranous Glutamic Acid Residue Plays an Essential Role in AceI Transport Function.
The BTP family proteins examined in this study each contain a glutamic acid residue centrally within the first α-helical transmembrane segment (Fig. S6). To examine the functional importance of this conserved residue in A1S_2063, it was neutralized by site-directed mutagenesis, generating a glutamine encoding derivative, E15Q. Although the E15Q AceI mutant was expressed at a level similar to that of the wild-type protein in E. coli cells (Fig. S3D), it conferred no chlorhexidine resistance (Table 1). This mutant protein served as an important control in subsequent studies (see below).
The A1S_2063 Protein Functions as an Efflux System for Chlorhexidine.
There are a number of ways in which an integral inner-membrane protein may act to increase chlorhexidine resistance. For example, it may alter membrane permeability, either via lipid transport or enzymatic modification, it could act as a membrane-bound sensor that induces expression of chlorhexidine resistance mechanisms, or it could function as an active chlorhexidine efflux system. Of these possibilities, several lines of evidence suggested that the expressed gene product was a chlorhexidine efflux protein: (i) The protein is likely to function independently, i.e., it was active in heterologous hosts that do not encode a homologous protein and are therefore unlikely to encode any essential accessory proteins, which would not be expected if the protein were functioning as a membrane bound sensor, and (ii) of the 240 different antimicrobial compounds included in the Biolog PM series, the protein specifically conferred resistance to chlorhexidine, which would not be expected if the protein was acting to modify the membrane permeability.
Transport assays using [14C]-chlorhexidine were used to test directly the hypothesis that the A1S_2063 protein is a chlorhexidine efflux system. These assays were complicated by the amphiphilic nature and low solubility of chlorhexidine, which adsorbs nonspecifically to filter and biological membranes (17, 18). The undenatured AceI protein was reconstituted into proteoliposomes composed of E. coli total lipids for transport measurements. Because of nonspecific filter adsorption, we separated these proteoliposomes from nonassociated chlorhexidine by using miniultracentrifugation (400,000 × g/15 min). We carried out a number of protocols to measure transport, including driving transport with pH- and valinomycin/K+-generated electrochemical gradients, substrate-substrate counterflow exchange at modest substrate concentrations, and radiolabel overshoot counterflow experiments using different substrate concentrations (low outside and high inside). Unfortunately, a combination of the hydrophobic nature and low solubility of chlorhexidine resulted in our failure to obtain reliable transport data with the proteoliposome system.
Consequently, the assays were performed in whole E. coli cells, which could be rapidly isolated from supernatant by centrifugation; similar methods have been applied previously in chlorhexidine transport experiments (18). These assays used E. coli BL21(DE3) cells expressing the proteins: (i) A1S_2063; (ii) E15Q A1S_2063; (iii) the A. baumannii multidrug transport protein, AedF (7), which confers efflux-mediated chlorhexidine resistance, as a positive control; and (iv) no additional protein, as a negative control.
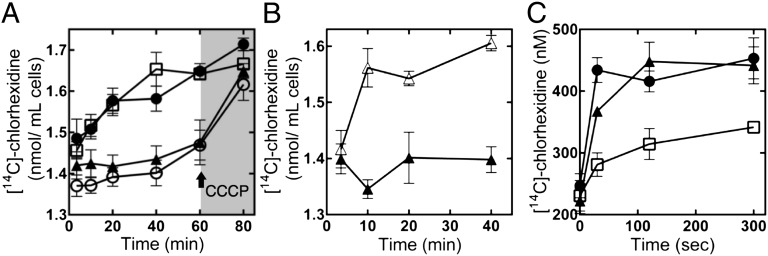
In accumulation assays, the addition of [14C]-chlorhexidine to all samples resulted in an immediate spike in the level of radioactivity associated with the cells, due to the propensity of chlorhexidine to associate rapidly and nonspecifically with bacterial cell membranes (18). Nonetheless, the amount of [14C]-chlorhexidine associated with the negative control cells increased steadily beyond this point over time, whereas the concentration associated with cells expressing AedF or the A1S_2063 protein remained relatively stable (Fig. 3A). To determine whether the lack of accumulation of [14C]-chlorhexidine in cells expressing these proteins was the result of an active, energy-requiring process, the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to deenergize the cells. Upon deenergization, [14C]-chlorhexidine was rapidly accumulated into both the A1S_2063 and AedF expressing cells to a level similar to that seen in the negative control cells (Fig. 3A). Addition of CCCP at the start of the assay also allowed the rapid accumulation of [14C]-chlorhexidine into A1S_2063 expressing cells, indicating that the maintenance of stable chlorhexidine levels observed in these cells is the result of an active energy requiring process, consistent with efflux (Fig. 3B). The concentration of CCCP used to deenergize the cells (∼4 µg/mL) did not alter the viability of the cells and was below the MIC for this compound in the strains tested (Table S4). In line with the lack of resistance, accumulation of [14C]-chlorhexidine into cells expressing the E15Q mutant A1S_2063 protein closely resembled that of the negative control cells (Fig. 3A).
Fig. 3.
Chlorhexidine transport mediated by A1S_2063 (AceI). (A) [14C]-chlorhexidine accumulation in E. coli BL21(DE3) cells carrying pTTQ18 (□; negative control), pTTQ18-aedF (○; positive control), pTTQ18-A1S_2063 (aceI) (▲), or pTTQ18-E15Q A1S_2063 (●). Cells were incubated in the presence of 2 µM [14C]-chlorhexidine and samples taken at the times indicated. After 60 min, cells were deenergized by using 10 µM CCCP at the point marked with an arrow. The rate of [14C]-chlorhexidine accumulation in the negative control or cells expressing the E15Q mutant protein was more rapid than in the positive control or cells expressing A1S_2063. Following deenergization with CCCP, all cell types contain similar [14C]-chlorhexidine levels. (B) [14C]-chlorhexidine accumulation in E. coli BL21(DE3) cells carrying pTTQ18-A1S_2063 (aceI) with 10 µM CCCP added after the 3.5-min time point (△), or no added CCCP (▲). (C) [14C]-chlorhexidine efflux from E. coli BL21(DE3) cells carrying pTTQ18 (□), pTTQ18-aedF (●), or pTTQ18-A1S_2063 (aceI) (▲). Cells were loaded with 4 µM [14C]-chlorhexidine in the presence of 10 µM CCCP and washed in assay buffer. Cells were r-energized and the assay initiated by using 0.4% glucose. Fractions of supernatant were collected over time to monitor efflux. Assays were performed at least four times, and the error bars show the SEM.
Efflux from cells preloaded with [14C]-chlorhexidine in the presence of CCCP was also monitored after reenergization of the cells (Fig. 3C). All cell types displayed equal viability (approximately 99%) after loading, as determined by using a fluorescent vital stain. In the efflux assays, [14C]-chlorhexidine export from cells expressing AedF or A1S_2063 was significantly more rapid than from the negative control cells (Fig. 3C). These results are consistent with A1S_2063 encoding an active chlorhexidine efflux protein. Therefore, we have designated A1S_2063 as Acinetobacter chlorhexidine efflux protein I (AceI). AceI represents the founding member of a family of drug resistance efflux proteins that we have named the Proteobacterial Chlorhexidine Efflux (PCE) protein family (Fig. 2 and Fig. S6).
The Wild-Type and E15Q AceI Proteins Directly and Specifically Bind Chlorhexidine.
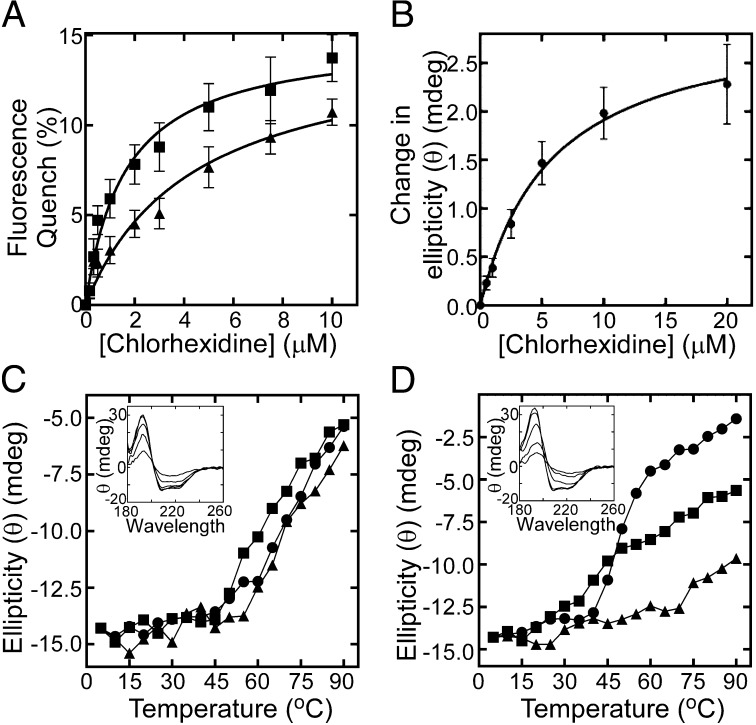
To observe direct interactions between the purified AceI protein and chlorhexidine, we used tryptophan (Trp) fluorescence quenching experiments. The cloned AceI protein contains three Trp residues, including two with predicted intramembranous localizations (Fig. S2). Significant Trp fluorescence quenching was observed upon chlorhexidine titration (Fig. 4A). Using 4.4 µM protein, we determined an apparent Kd value of 1.6 µM for the wild-type AceI–chlorhexidine interaction, indicative of high affinity binding (Fig. 4A). This direct interaction provides further support for an active chlorhexidine efflux mechanism operating in AceI. Assays of AceI Trp fluorescence conducted with 11 additional antimicrobial compounds did not induce significant fluorescence changes (Fig. S7), suggesting that these compounds are not substrates, in line with resistance data. Trp fluorescence quenching was also used to determine whether the transport negative E15Q mutant AceI protein maintained affinity for chlorhexidine. This protein showed an apparent Kd value of 4.4 µM, suggesting that the transport defect in this protein is unrelated to chlorhexidine binding (Fig. 4A).
Fig. 4.
AceI–chlorhexidine binding interactions. (A) Trp-fluorescence quenching of 4.4 µM wild-type AceI (■) and E15Q AceI (▲) protein upon chlorhexidine titration. Samples were excited at 295 nm, and the fluorescence emission was measured at 330 nm. The apparent Kd for the wild-type and E15Q AceI-chlorhexidine interactions determined via nonlinear regression were 1.6 µM and 4.4 µM, respectively. (B) Average change in ellipticity (θ) (mdeg) of 20 µM wild-type AceI protein across wavelengths 258–280 nm during titration of chlorhexidine. The apparent Kd determined for the AceI–chlorhexidine interaction in this experiment was 5.8 µM. Error bars show the SEM ellipticity change from different wavelengths. (C and D) Thermal stability of 33 µM wild-type (C) and E15Q (D) AceI protein in the presence and absence of chlorhexidine as determined by synchrotron radiation CD spectrophotometry. The ellipticity at 209 nm is shown for protein only (●), protein plus 100 µM chlorhexidine (■; 1:3 molar ratio) and protein plus 500 µM chlorhexidine (▲; 1:15 molar ratio) at increasing temperature. Insets show the characteristic α-helical protein far-UV spectrum of the respective proteins at increasing temperature in the absence of chlorhexidine.
Binding of chlorhexidine to purified AceI protein was also demonstrated by using near-UV synchrotron radiation circular dichroism (SRCD) spectroscopy. Using 20 µM protein, significant saturable increases in the CD signal were observed across the near-UV wavelengths with the average change across the whole region (258–280 nm), giving an apparent Kd value of 5.8 µM (Fig. 4B and Fig. S8).
The secondary structure and thermal stability of purified wild-type and E15Q AceI proteins were examined by using far-UV SRCD spectroscopy. These experiments demonstrated that both proteins were largely α-helical in line with their hydropathy profiles (Fig. 4 C and D and Fig. S2). The loss of α-helical structure at increasing temperature was used to measure the thermal stability of these proteins. The wild-type protein was stable at temperatures below 60 °C (Fig. 4C). However, the E15Q AceI mutant displayed a lower thermal stability, with denaturation apparent at 40 °C (Fig. 4D). Interestingly, chlorhexidine greatly increased the thermal stability of the E15Q protein in a dose-dependent manner (Fig. 4D). This observation provides additional evidence that the E15Q protein retains affinity for chlorhexidine.
Conclusions
The most highly up-regulated genes in A. baumannii ATCC 17978 in response to chlorhexidine exposure were those encoding the RND efflux system AdeAB. A gene encoding a previously uncharacterized membrane protein, AceI, was also highly expressed under chlorhexidine stress. The data presented indicate that this protein functions as an active chlorhexidine efflux system. There are two lines of evidence that AceI functions independently to mediate chlorhexidine resistance: (i) AceI conferred chlorhexidine resistance in E. coli, a heterologous host that does not encode any aceI homologs (Table 1); and (ii) AceI-mediated chlorhexidine resistance in E. coli is not reliant on any one of the 20 known drug exporters encoded this organism (Table S3). There are also two pieces of direct evidence for transport: (i) AceI prevents accumulation of chlorhexidine in an energy-dependent manner (Fig. 3 A and B); and (ii) AceI promotes active efflux of chlorhexidine (Fig. 3C). Finally, direct AceI-chlorhexidine interactions were demonstrated by using Trp-fluorescence quenching (Fig. 4A) and near-UVCD (Fig. 4B), and were inferred from the increased thermal stability of the E15Q derivative in the presence of chlorhexidine (Fig. 4D).
AceI is a prototype of the previously undescribed PCE family of efflux systems. The discovery of this family of drug efflux systems through transcriptomic analysis of a drug shock treatment demonstrates the significant value of genome-wide expression studies in identifying novel drug resistance factors in bacteria.
The E15Q AceI mutant was inactive in conferring chlorhexidine resistance or transport but still retained binding affinity for chlorhexidine, indicating that E15 could be involved in an aspect of transport unrelated to substrate binding, possibly in an ion coupling reaction. This suggestion is consistent with the fact that this residue is also conserved in BTP family proteins that do not confer chlorhexidine resistance (Fig. S6). The SMR family of multidrug efflux transporters that display a similar size and secondary structure with the PCE family transporters (19) also have an essential conserved glutamic acid located within the first transmembrane segment, although the two families share no detectable sequence homology (Fig. S6). In the SMR family member EmrE, this acidic residue is functionally irreplaceable because of its role in the binding of both protons and substrate at different stages of the translocation cycle (20).
The primary features of the A. baumannii chlorhexidine resistance response, i.e., up-regulation of genes encoding RND and PCE family efflux systems, are conserved in other γ-proteobacteria (9), as well as β-proteobacteria (10). Given the phylogenetic distance of these organisms, this similarity of regulatory responses to a compound that has only been synthesized since the early 20th century is intriguing. RND efflux systems are well recognized for their broad protective functions in Gram-negative bacteria and it is not unusual for these systems to constitute part of a general stress response and to mediate resistance to foreign compounds, particularly amphipathic antimicrobials such as chlorhexidine. However, the involvement of the PCE family proteins in seemingly specific resistance to chlorhexidine is unexpected. It is likely that the physiological function(s) of this transporter family were originally unrelated to chlorhexidine resistance and that these proteins provide a fortuitous intrinsic resistance capacity.
Materials and Methods
Bacterial Strains, Reagents, and Growth Media.
The bacterial strains used were A. baumannii ATCC 17978 (21), A. baylyi ADP1 (22), E. coli BL21(DE3) (23), AG100A (24), BL21KAMR (25), and mutant strains harboring disrupted drug efflux system genes (15) from the Keio collection (26). Bacterial strains were routinely cultured in Luria–Bertani broth unless otherwise stated. The plasmid used for protein expression was pTTQ18 (14) modified to include an RGSH6 tag (27). E. coli cells carrying pTTQ18-based plasmids were cultured in media containing 100µg/mL ampicillin. Ampicillin and isopropyl-β-d-galactopyranoside (IPTG) were obtained from Amresco, [14C]-chlorhexidine diacetate was obtained from American Radiolabeled Chemicals,and Mueller–Hinton (MH) medium was obtained from Oxoid. All other chemicals were obtained from Sigma.
Cell Treatments and RNA Isolation.
A. baumannii ATCC 17978 cells were grown in an INFORS HT shaking incubator at 37 °C with shaking (200 rpm) in 35-mL cultures in MH broth to OD600 = 0.75, at which time they were split into 15-mL cultures. One 15-mL sample was treated with 4 µg/mL chlorhexidine (0.5× MIC), whereas the other was not treated and used as a reference. Cultures were allowed to grow for an additional 30 min (to an average final OD600 ∼ 1.35 for untreated cells and 1.25 for chlorhexidine-treated samples), when cells were harvested by centrifugation and immediately suspended in TRIzol reagent (Invitrogen). Total RNA was extracted as described (28).
Microarray Processing.
A custom-designed 8× 15,000 spot Agilent microarray for A. baumannii ATCC 17978 described was used (28). cDNA synthesis, labeling, and hybridizations were conducted at the Ramaciotti Centre for Gene Function Analysis, Australia, as described (28). Statistical analyses were performed by using the SAM algorithms (29), and all results reported were found to be significant by using a false discovery rate of 5%. The microarray data were deposited into the GEO database under the accession no. GSE51525. The microarray results were validated by qRT-PCR on a subset of differentially regulated genes (Fig. S1) as described (28).
Gene Cloning, Mutagenesis, and Construction of Deletion Strains.
Genes were amplified by PCR with Pfx supermix (Invitrogen) and cloned into the pTTQ18-RGSH6 vector as reported (7). Site-directed mutagenesis was conducted by using the QuikChange method as described (30), but using KOD hot start polymerase (Novagen). The ACIAD1978 gene was deleted from the A. baylyi ADP1 genome by allelic exchange as described (31).
Antimicrobial Susceptibility Testing.
MIC analyses were conducted in MH media using the broth dilution method essentially as described (32). MICs of E. coli strains carrying pTTQ18-based plasmids included 0.05 mM IPTG. Cell growth was determined spectrophotometrically.
PM were used for high-throughput screening of resistance phenotypes conferred by A1S_2063 (aceI). E. coli BL21(DE3) cells carrying either pTTQ18 or pTTQ18-A1S_2063 (aceI) were inoculated into plates PM09-PM20 according to the manufacturer’s instructions, except for the addition of 0.05 mM IPTG.
[14C]-Chlorhexidine Accumulation Assays.
E. coli BL21(DE3) cells carrying pTTQ18, pTTQ18-aedF, pTTQ18-A1S_2063 (aceI), or pTTQ18-E15Q A1S_2063 (E15Q aceI) were grown and induced as described for Western blot analyses. The cells were washed three times in 20 mM Mops at pH 6.6, 140 mM NaCl, and 10 mM KCl, resuspended to a final density of OD600 = 1.0 in this buffer, and equilibrated to 37 °C. Glycerol (20 mM) was added to provide an energy source, and the assay was initiated by the addition of 2 µM [14C]-chlorhexidine. Samples were taken at 3.5, 10, 20, 40, and 60 min, and cells pelleted by centrifugation (15,000 × g for 20 s). After 60 min, the cells were deenergized by the addition of 10µM CCCP and the assay was allowed to continue for a further 20 min, when a final sample was collected. Radioactivity associated with the cells was determined by liquid scintillation analysis. The assays were repeated at least four times by using independent biological samples.
[14C]-Chlorhexidine Efflux Assays.
E. coli BL21(DE3) cells carrying pTTQ18, pTTQ18-aedF or pTTQ18-A1S_2063 (aceI) were grown and induced as described for Western blot analyses. Cells were washed three times in 20 mM Mops at pH 7.0, 140 mM NaCl, and 10 mM KCl, and the cell density was adjusted to OD600 = 1.0 in this buffer. CCCP was added to a final concentration of 10µM and the cells were loaded with 4 µM [14C]-chlorhexidine for 1.5 h at 37 °C. Cells were washed three times and resuspended in one volume of the above buffer. A time 0 sample was collected and transport was initiated by the addition of 0.4% (wt/vol) glucose. Samples (250 µL) were taken at 30, 120, and 300 s, pelleted by brief centrifugation and 200 µL of supernatant collected for liquid scintillation analysis. The results are from at least four independent biological assays. To confirm the loading protocol did not affect cell viability, loading was conducted by using unlabeled chlorhexidine, and cells were stained with Live/Dead BacLight bacterial viability probe (Molecular Probes) and visualized by fluorescence microscopy according to the manufacturer's instructions.
Fluorescence Measurements.
Steady-state spectrophotofluorimetry was undertaken on the purified proteins (SI Materials and Methods) by using the Photon Technology International spectrofluorimeter. Purified A1S_2063 (AceI) protein or E15Q A1S_2063 (E15Q AceI) (4.4 μM in 10mM Tris⋅HCl at pH 7.5, 0.05% (wt/vol) n-dodecyl-β-d-maltoside (DDM), and 2.5% vol/vol glycerol) were analyzed at 20 °C. Protein samples were excited at 295nm and fluorescence emission was analyzed at 330 nm. Microliter additions of each compound dissolved in water or 100% ethanol for valinomycin and cytochalasin B were added from 0 to 10 μM and accounted for <3% of the final volume. Samples were mixed for 1.5 min after each addition before the fluorescence emission was monitored. Nonlinear regression analysis was completed by using GraphPad Prism 6.
Circular Dichroism Measurements.
UV SRCD measurements were performed on the purified proteins (SI Materials and Methods) by using a nitrogen-flushed instrument on Beamline B23 at the Diamond Light Source. Samples used in far-UV SRCD contained 33µM purified DDM-solubilized A1S_2063 (AceI) protein or E15Q A1S_2063 (E15Q AceI) in NaPi buffer (10 mM, pH 7.5) with 0.05% (wt/vol) DDM in a cell with 0.02 cm pathlength (nominal volume 30 µL). Measurements were made with the sample held at a range of successive temperatures from 5 to 90 °C in steps of 5 °C over the wavelength range 180–260 nm by using an integration time of 1 s, increment of 0.5 nm, and slit widths of 0.5 mm. Chlorhexidine was included at concentrations of 100 or 500 µM in the samples as appropriate. Near-UV SRCD measurements were recorded using 20µM purified DDM-solubilized A1S_2063 (AceI) protein with increasing concentrations of chlorhexidine. Data are presented in units of ellipticity (θ), and all measurements had PMT values that were less than 600 V.
Supplementary Material
Acknowledgments
We thank David Sharples, University of Leeds, for protein expression; Dr. Louise Brown, Macquarie University, for advice on reconstitution; Liping Li and Hasinika Ariyaratne, Macquarie University, for testing gene expression in ADP1; and Diamond Light Source Ltd for instrument time on CD Beamline B23 with assistance from Giuliano Siligardi and Rohanah Hussain. This work was supported by National Health and Medical Research Council (Australia) Project Grant 535053 (to I.T.P. and M.H.B.) and Australian Research Council Discovery Grant DP110102680 (to I.T.P. and K.A.H.). Collaboration between the Australian and UK laboratories is supported by an EU International Research Staff Exchange Scheme BacMT 247634 (to Prof. Anne-Brit Kolstø, University of Oslo, and P.J.F.H.). Additional travel support was provided by the Australian Academy of Science (I.T.P.). Work in the P.J.F.H. laboratory was supported by EU European Drug Initiative for Channels and Transporters Grant 201924.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.I.Z. is a guest editor invited by the Editorial Board.
Data deposition: Transcriptomic data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE51525).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317052110/-/DCSupplemental.
References
- 1.McDonnell G, Russell AD. Antiseptics and disinfectants: Activity, action, and resistance. Clin Microbiol Rev. 1999;12(1):147–179. doi: 10.1128/cmr.12.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell AD. Chlorhexidine: Antibacterial action and bacterial resistance. Infection. 1986;14(5):212–215. doi: 10.1007/BF01644264. [DOI] [PubMed] [Google Scholar]
- 3.Brooks SE, Walczak MA, Hameed R, Coonan P. Chlorhexidine resistance in antibiotic-resistant bacteria isolated from the surfaces of dispensers of soap containing chlorhexidine. Infect Control Hosp Epidemiol. 2002;23(11):692–695. doi: 10.1086/501996. [DOI] [PubMed] [Google Scholar]
- 4.Tattawasart U, Hann AC, Maillard JY, Furr JR, Russell AD. Cytological changes in chlorhexidine-resistant isolates of Pseudomonas stutzeri. J Antimicrob Chemother. 2000;45(2):145–152. doi: 10.1093/jac/45.2.145. [DOI] [PubMed] [Google Scholar]
- 5.Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56(1):20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- 6.Rajamohan G, Srinivasan VB, Gebreyes WA. Novel role of Acinetobacter baumannii RND efflux transporters in mediating decreased susceptibility to biocides. J Antimicrob Chemother. 2010;65(2):228–232. doi: 10.1093/jac/dkp427. [DOI] [PubMed] [Google Scholar]
- 7.Hassan KA, et al. Roles of DHA2 family transporters in drug resistance and iron homeostasis in Acinetobacter spp. J Mol Microbiol Biotechnol. 2011;20(2):116–124. doi: 10.1159/000325367. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasan VB, Rajamohan G, Gebreyes WA. Role of AbeS, a novel efflux pump of the SMR family of transporters, in resistance to antimicrobial agents in Acinetobacter baumannii. Antimicrob Agents Chemother. 2009;53(12):5312–5316. doi: 10.1128/AAC.00748-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nde CW, Jang HJ, Toghrol F, Bentley WE. Global transcriptomic response of Pseudomonas aeruginosa to chlorhexidine diacetate. Environ Sci Technol. 2009;43(21):8406–8415. doi: 10.1021/es9015475. [DOI] [PubMed] [Google Scholar]
- 10.Coenye T, et al. Molecular mechanisms of chlorhexidine tolerance in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother. 2011;55(5):1912–1919. doi: 10.1128/AAC.01571-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 12.Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40(Database issue):D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ward A, et al. The amplified expression, identification, purification, assay, and properties of hexahistidine-tagged bacterial membrane transport proteins. In: Baldwin SA, editor. Membrane Transport: A Practical Approach. Oxford: Oxford Univ Press; 2000. pp. 141–166. [Google Scholar]
- 14.Stark MJ. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene. 1987;51(2-3):255–267. doi: 10.1016/0378-1119(87)90314-3. [DOI] [PubMed] [Google Scholar]
- 15.Nishino K, Yamaguchi A. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J Bacteriol. 2001;183(20):5803–5812. doi: 10.1128/JB.183.20.5803-5812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bochner BR. New technologies to assess genotype-phenotype relationships. Nat Rev Genet. 2003;4(4):309–314. doi: 10.1038/nrg1046. [DOI] [PubMed] [Google Scholar]
- 17.Denton GW. Chlorhexidine. In: Block SS, editor. Disinfection, Sterilization, and Preservation. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2000. p. 321. [Google Scholar]
- 18.Fitzgerald KA, Davies A, Russell AD. Uptake of 14C-chlorhexidine diacetate to Escherichia coli and Pseudomonas aeruginosa and its release by azolectin. FEMS Microbiol Lett. 1989;51(3):327–332. doi: 10.1016/0378-1097(89)90419-9. [DOI] [PubMed] [Google Scholar]
- 19.Paulsen IT, et al. The SMR family: A novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol Microbiol. 1996;19(6):1167–1175. doi: 10.1111/j.1365-2958.1996.tb02462.x. [DOI] [PubMed] [Google Scholar]
- 20.Yerushalmi H, Schuldiner S. A common binding site for substrates and protons in EmrE, an ion-coupled multidrug transporter. FEBS Lett. 2000;476(1-2):93–97. doi: 10.1016/s0014-5793(00)01677-x. [DOI] [PubMed] [Google Scholar]
- 21.Smith MG, et al. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007;21(5):601–614. doi: 10.1101/gad.1510307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barbe V, et al. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 2004;32(19):5766–5779. doi: 10.1093/nar/gkh910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Studier FW, Moffatt BA. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 24.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178(1):306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HS, Nagore D, Nikaido H. Multidrug efflux pump MdtBC of Escherichia coli is active only as a B2C heterotrimer. J Bacteriol. 2010;192(5):1377–1386. doi: 10.1128/JB.01448-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baba T, et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed]
- 27.Hassan KA, et al. Optimized production and analysis of the staphylococcal multidrug efflux protein QacA. Protein Expr Purif. 2009;64(2):118–124. doi: 10.1016/j.pep.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Eijkelkamp BA, Hassan KA, Paulsen IT, Brown MH. Investigation of the human pathogen Acinetobacter baumannii under iron limiting conditions. BMC Genomics. 2011;12:126. doi: 10.1186/1471-2164-12-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hassan KA, Souhani T, Skurray RA, Brown MH. Analysis of tryptophan residues in the staphylococcal multidrug transporter QacA reveals long-distance functional associations of residues on opposite sides of the membrane. J Bacteriol. 2008;190(7):2441–2449. doi: 10.1128/JB.01864-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brzoska AJ, Hassan KA, de Leon EJ, Paulsen IT, Lewis PJ. Single-step selection of drug resistant Acinetobacter baylyi ADP1 mutants reveals a functional redundancy in the recruitment of multidrug efflux systems. PLoS ONE. 2013;8(2):e56090. doi: 10.1371/journal.pone.0056090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiegand I, Hilpert K, Hancock RE. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.