Summary
Hydrogen, the most abundant and lightest element in the universe, has much potential as a future energy source. Hydrogenases catalyse one of the simplest chemical reactions, 2H+ + 2e‐ ↔ H2, yet their structure is very complex. Biologically, hydrogen can be produced via photosynthetic or fermentative routes. This review provides an overview of microbial production of hydrogen by fermentation (currently the more favourable route) and focuses on biochemical pathways, theoretical hydrogen yields and hydrogenase structure. In addition, several examples of metabolic engineering to enhance fermentative hydrogen production are presented along with some examples of expression of heterologous hydrogenases for enhanced hydrogen production.
Fermentation versus photosynthesis for hydrogen
Hydrogen, the smallest biological substrate, has great potential as an alternative to limited fossil fuel resources (Das and Veziroglu, 2001). In addition to its higher energy content than fossil fuels (Chen et al., 2006), it is renewable if it is derived from renewable feedstocks (Hawkes et al., 2007), and the product of hydrogen oxidation is water; hence, the impact of hydrogen on the environment is not relevant (Das and Veziroglu, 2001).
Production of hydrogen by microorganisms is at ambient temperature and pressure; hence, it requires less energy compared with conventional thermal systems (steam methane reforming, 850°C, 25 bar) (Yi and Harrison, 2005) and compared with electrolytic processes. Microorganisms produce hydrogen via two main pathways: photosynthesis and fermentation. Photosynthesis is a light‐dependent process, including direct biophotolysis, indirect biophotolysis and photo‐fermentation, whereas, anaerobic fermentation, also known as dark fermentation, is a light‐independent process (Benemann, 1996; Hallenbeck and Benemann, 2002). Photosynthetic hydrogen production is performed by photosynthetic microorganisms, such as algae (Benemann, 2000), photosynthetic bacteria (Matsunaga et al., 2000) and cyanobacteria (Dutta et al., 2005). Fermentative hydrogen production is conducted by fermentative microorganisms, such as strict anaerobes [Clostridium strains (Levin et al., 2006), thermophiles (van Niel et al., 2002), rumen bacteria (Nandi and Sengupta, 1998) and methanogens (Valentine et al., 2000)], facultative anaerobes [Enterobacter strains (Kumar and Das, 2000; Shin et al., 2007), Escherichia coli(Yoshida et al., 2007) and Citrobacter species (Vatsala, 1992)], or mixed cultures (Lay, 2000).
Compared with photosynthetic processes, fermentative hydrogen production generally yields two orders of magnitude higher rates, does not rely on the availability of light, utilizes a variety of carbon sources such as organic compounds, low‐cost wastes, or insoluble cellulosic and cellobiose substrates, requires less energy, and is technically much simpler and more stable (Nandi and Sengupta, 1998; Levin et al., 2004; 2006; Chen et al., 2006; Kapdan and Kargi, 2006; Ust’ak et al., 2007). Although hydrogen production yields are usually higher with photosynthetic processes, oxygen is evolved during photosynthesis which inhibits the hydrogenase enzyme which is responsible for H2 production (Nath and Das, 2004). In addition, fermentative microorganisms have rapid growth and are not affected by oxygen as much as the main process is anaerobic (any residual oxygen is rapidly consumed at the onset). Therefore, fermentative hydrogen production is more advantageous than the photosynthetic hydrogen production and appears to have more potential for practical applications (Das and Veziroglu, 2001).
Formate hydrogen lyase system
The formate hydrogen lyase (FHL) system is a multienzyme complex responsible for molecular hydrogen production from formate (Peck and Gest, 1957). Formate hydrogen lyase activity has been found in various bacteria such as Salmonella typhimurium(Chippaux et al., 1977; Barrett et al., 1984), Klebsiella pneumoniae(Steuber et al., 1999), Rhodospirillum rubrum(Schön and Voelskow, 1976); (Voelskow and Schön, 1980), Methanobacterium formicicum(Baron and Ferry, 1989), E. coli, and many other coli‐aerogenes bacteria (Peck and Gest, 1957); however, the FHL complex of E. coli is the most studied and was discovered in 1931 (Stephenson and Stickland, 1931). The FHL system of E. coli is briefly reviewed here and more detailed information can be found in other reviews (Sawers, 1994; Sawers, 2005).
The E. coli genome (Hayashi et al., 2006) encodes four nickel‐iron hydrogenases: hydrogenase‐1 (Hyd‐1) (Menon et al., 1991), hydrogenase‐2 (Hyd‐2) (Menon et al., 1994), hydrogenase‐3 (Hyd‐3) (Böhm et al., 1990) and hydrogenase‐4 (Hyd‐4) (Andrews et al., 1997). From these four hydrogenases, Hyd‐3 is a part of the active anaerobic FHL complex and is encoded by the hycoperon (Fig. 1, Table 1) (Böhm et al., 1990; Sauter et al., 1992). Hyd‐1 and Hyd‐2 are known as uptake hydrogenases which catalyse hydrogen oxidation and are encoded by the hya(Menon et al., 1991) and hyb operons (Menon et al., 1994) respectively (Fig. 1, Table 1). Hyd‐3 was first thought to only have hydrogen‐producing activity (Bagramyan et al., 2002); however, Maeda and colleagues (2007a) recently showed that Hyd‐3 has hydrogen uptake activity as well. Hence, Hyd‐3 is a reversible hydrogenase, both producing and utilizing hydrogen although the synthesis reaction predominates. Hyd‐4, encoded by the hyf operon (Fig. 1, Table 1), has high homology with the hyc operon and was first proposed to possess a second FHL complex (Andrews et al., 1997; Bagramyan et al., 2002). However, Self and colleagues (2004) later showed that the Hyd‐4 did not replace the Hyd‐3 in hydrogen production and the hyf operon is not expressed in E. coli, but can be activated in the presence of effector‐independent FhlA (transcriptional activator of the FHL complex) mutant proteins (Self and Shanmugam, 2000; Self et al., 2001) or HyfR, which is an FhlA homologue for Hyd‐4 (Skibinski et al., 2002).
Figure 1.
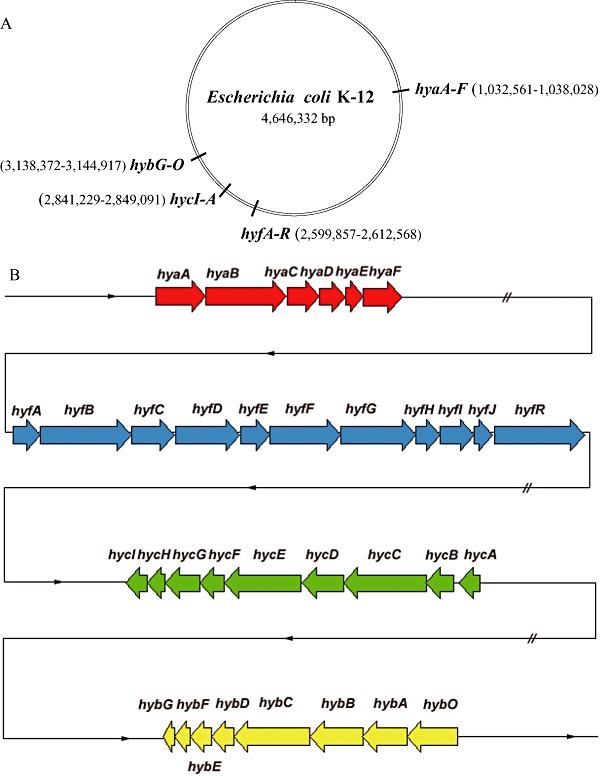
A. Location of the four structural hydrogenase operons on the E. coli K12 chromosome (AC000091) (Hayashi et al., 2006). The values in brackets signify the locations of the respective genes on the genome map. B. Organization of the genes of each hydrogenase operon in E. coli.Arrows indicate the direction of transcription. For details for each gene see Table 1 (Hayashi et al., 2006).
Table 1.
Genes of four hydrogenase operons (hya, hyb, hyc, hyf) in E. coli.
| Gene | Size, bp | Description |
|---|---|---|
| hyaA | 372 | Hydrogenase 1, small subunit |
| hyaB | 597 | Hydrogenase 1, large subunit |
| hyaC | 235 | Hydrogenase 1, b‐type cytochrome subunit |
| hyaD | 195 | Protein involved in processing of HyaA and HyaB proteins |
| hyaE | 132 | Protein involved in processing of HyaA and HyaB proteins |
| hyaF | 285 | Protein involved in nickel incorporation into hydrogenase‐1 proteins |
| hyfA | 205 | Hydrogenase 4, 4Fe‐4S subunit |
| hyfB | 672 | Hydrogenase 4, membrane subunit |
| hyfC | 315 | Hydrogenase 4, membrane subunit |
| hyfD | 479 | Hydrogenase 4, membrane subunit |
| hyfE | 216 | Hydrogenase 4, membrane subunit |
| hyfF | 526 | Hydrogenase 4, membrane subunit |
| hyfG | 555 | Hydrogenase 4, subunit |
| hyfH | 181 | Hydrogenase 4, Fe‐S subunit |
| hyfI | 252 | Hydrogenase 4, Fe‐S subunit |
| hyfJ | 137 | Predicted processing element hydrogenase |
| hyfR | 670 | DNA‐binding transcriptional activator, formate sensing |
| hycI | 156 | Protease involved in processing C‐terminal end of HycE |
| hycH | 136 | Protein required for maturation of hydrogenase 3 |
| hycG | 255 | Hydrogenase 3, small subunit |
| hycF | 180 | Formate hydrogenlyase complex Fe‐S protein |
| hycE | 569 | Hydrogenase 3, large subunit |
| hycD | 307 | Hydrogenase 3, membrane subunit |
| hycC | 608 | Hydrogenase 3, membrane subunit |
| hycB | 203 | Hydrogenase 3, Fe‐S subunit |
| hycA | 153 | Regulator of the transcriptional regulator FhlA |
| hybG | 82 | Hydrogenase 2 accessory protein |
| hybF | 113 | Protein involved with the maturation of hydrogenases 1 and 2 |
| hybE | 162 | Hydrogenase 2‐specific chaperone |
| hybD | 164 | Predicted maturation element for hydrogenase 2 |
| hybC | 567 | Hydrogenase 2, large subunit |
| hybB | 392 | Predicted hydrogenase 2 cytochrome b‐type component |
| hybA | 328 | Hydrogenase 2 4Fe‐4S ferredoxin‐type component |
| hybO | 372 | Hydrogenase 2, small subunit |
The FHL complex of E. coli is responsible for the conversion of formate to CO2 and H2under anaerobic conditions and in the absence of electron acceptors such as oxygen and nitrate (Axley et al., 1990). The active complex consists of seven proteins, six from the hyc operon (HycBCDEFG) (Böhm et al., 1990; Sauter et al., 1992) and formate dehydrogenase H (Fdh‐H encoded by fdhF) (Axley et al., 1990). In addition, the FHL system is controlled by a transcriptional activator FhlA (Schlensog and Böck, 1990; Hopper et al., 1994), which is required for the transcription of fdhF and the hyc operon, and a negative transcriptional regulator, HycA (Sauter et al., 1992). Transcription of the FHL complex is dependent on the presence of formate, acidic pH and the σ54 factor (Hopper et al., 1994; Self and Shanmugam, 2000). In the presence of formate, FhlA is activated and induces the transcription of the FHL complex by σ54‐RNA polymerase after binding to the upstream‐activating sequences. Molybdate also has a role in transcriptional control as a required second effector (Self and Shanmugam, 2000).
HycE (65 kDa) is the large subunit of Hyd‐3 and contains the nickel‐iron active site, whereas HycG (28 kDa) is the small subunit of Hyd‐3 (Sauter et al., 1992; Sawers, 1994). HycB (22 kDa) and HycF (20 kDa) are iron‐sulfur proteins of Hyd‐3 which function as intermediate electron carrier proteins within the FHL complex (Table 1). HycC (64 kDa) and HycD (33 kDa) are membrane proteins which probably function as an electron transfer protein (Sauter et al., 1992; Sawers, 1994).
Fdh‐H, encoded by the fdhF gene, is a 79 kDa cytoplasmic protein and contains selenocysteine, molybdenum and a [4Fe‐4S] cluster at its active site (Axley et al., 1990; Boyington et al., 1997). Recently a new reaction mechanism was proposed for Fdh‐H after re‐analysing the crystal structure data and finding that a loop region close to the molybdenum active site was mistraced (PDB:2IV2) (Raaijmakers and Romão, 2006). It was shown that selenocysteine‐140 is not a ligand for molybdenum hence is no longer bound to the metal after reduction of the enzyme with formate. The E. coli genome also encodes two other formate dehydrogenases, Fdh‐O and Fdh‐N, which can oxidize formate but are not a part of the FHL complex (Sawers, 2005). Fdh‐N is induced in the presence of nitrate under anaerobic conditions and encoded by the fdnGHI operon, whereas Fdh‐O is induced under aerobic as well as nitrate‐respiring conditions and encoded by the fdoGHI operon. The crystal structure of the Fdh‐N, a 600 kDa membrane protein, is also known (PDB:1KQF) (Jormakka et al., 2002).
Theoretical yield of hydrogen from glucose
The hydrogen yield is defined as the moles of H2 produced per mole of substrate. Carbohydrates, mainly glucose, are the preferred substrates for fermentative hydrogen production. Starch, cellulose, as well as organic wastes can also be used as substrates. Many microorganisms such as Enterobactersp., Clostridiumsp. and E. coliare capable of producing hydrogen. Depending on the pathways used by the microorganisms and end‐products, hydrogen yields may be variable (Levin et al., 2004; Nath and Das, 2004).
The fermentative route of hydrogen production starts with the conversion of glucose to pyruvate and NADH through glycolysis in both strict and facultative anaerobic bacteria (Hallenbeck, 2005) (Figs 2 and 3). In facultative anaerobes, such as E. coli(Fig. 2), pyruvate is then converted to acetyl‐CoA and formate, which is catalysed by pyruvate formate lyase (PFL). Hydrogen is produced from formate by the FHL complex. Because a maximum of two molecules of formate are produced from two pyruvate molecules, facultative anaerobic bacteria have a theoretical maximum yield of 2 mol of H2 per mole of glucose. There are several factors that influence the yield, such as whether some of the pyruvate is converted to lactate; these competing paths for obtaining reducing power from pyruvate lower the yield (Hallenbeck, 2005).
Figure 2.
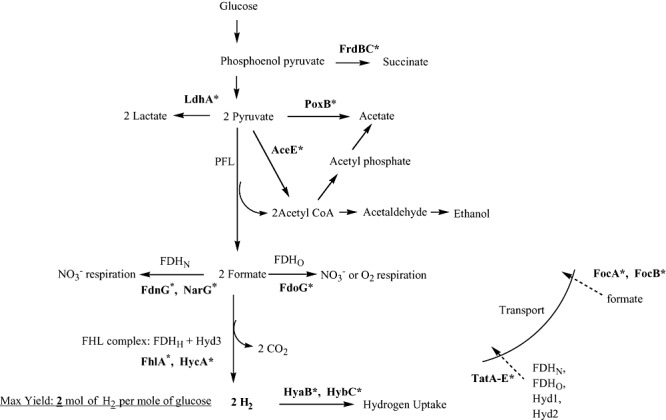
Fermentative hydrogen production from glucose by E. coli, a well‐studied facultative anaerobic bacterium. Hydrogen is produced through the action of the FHL complex. The maximum theoretical hydrogen yield is 2 mol of H2 per mole of glucose. The glucose metabolic pathway yields succinate, lactate, acetate, ethanol and formate, as fermentation end‐products. The proteins shown in bold with an asterisk have been studied through metabolic engineering in order to enhance the biohydrogen production. PFL, pyruvate formate lyase; FDH, formate dehydrogenase; FHL, formate hydrogen lyase; Hyd, hydrogenase; CoA, coenzyme A.
Figure 3.
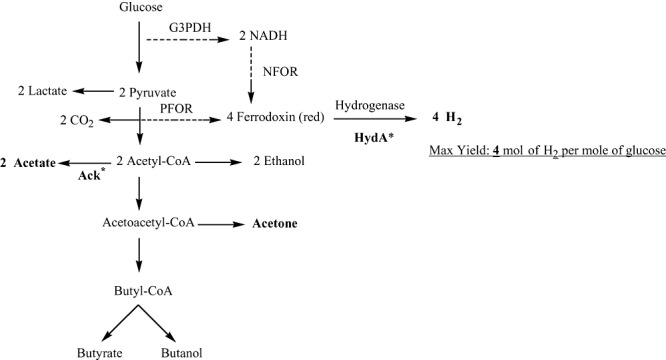
Fermentative hydrogen production from glucose by C. acetobutylicum, a strict anaerobic bacterium. Hydrogen can be produced through the action of PFOR and NFOR. The maximum theoretical hydrogen yield is 4 mol of H2 per mole of glucose, with acetate or acetone as the fermentation end‐product. The glucose metabolic pathway results in lactate, acetate, ethanol, acetone, butanol and butyrate as fermentation end‐products. The proteins shown in bold with an asterisk have been studied in Clostridium species through metabolic engineering in order to enhance biohydrogen production. G3PDH, glyceraldehyde‐3‐phosphate dehydrogenase; PFOR, pyruvate ferredoxin oxidoreductase; NFOR, NADH:ferredoxin oxidoreductase; NADH, nicotineamide‐adenine dinucleotide; red, reduced.
In strict anaerobes, such as Clostridiumsp. (Fig. 3), pyruvate is converted to acetyl‐CoA and CO2 through pyruvate ferredoxin oxidoreductase (PFOR), and this oxidation requires reduction of ferredoxin (Fd) (Hallenbeck, 2005; Kraemer and Bagley, 2007). Hydrogen is produced from the reduced Fd by the action of hydrogenase. This results with a maximum yield of 2 mol of H2 per mole of glucose. Two additional moles of hydrogen can be produced from the NADH produced during glycolysis. In this step, NADH is oxidized by Fd reduction by NADH:ferredoxin oxidoreductase (NFOR). Again, hydrogen can be produced from the reduced Fd by hydrogenase. Overall, strict anaerobic bacteria have a theoretical maximum yield of 4 mol of H2 per mole of glucose, which is the greatest theoretical yield possible. However, in practice, yields are lower, as the NADH oxidation by NFOR is inhibited under standard conditions and only proceeds at very low partial pressures of hydrogen (< 60 Pa) (Angenent et al., 2004; Hallenbeck, 2005; Kraemer and Bagley, 2007). The relevant reactions are:
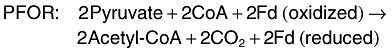 |
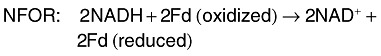 |
Products formed from acetyl‐CoA such as acetate, butyrate, butanol, acetone, lactate or ethanol in strict anaerobes determine the theoretical yield of hydrogen (Chin et al., 2003). One can obtain the greatest theoretical yield of hydrogen of 4 mol of H2 per mole of glucose when acetate or acetone is the fermentation end‐product:
When butyrate is the fermentation end‐product, the maximum theoretical yield of hydrogen is 2 mol of H2per mole of glucose:
When alcohols are the end‐products, lower yields of hydrogen are obtained as alcohols contain additional hydrogen atoms that have not been converted to hydrogen gas. Note that, the fermentation end‐products and the resultant hydrogen yields vary based on the environmental conditions even within the same bacterium (Hawkes et al., 2002).
A maximum yield of 4 mol of H2 per mole of glucose is still low for practical applications and a two‐stage process has been envisioned to obtain yields closer to the theoretical stoichiometric yield of 12 mol of H2 per mole of glucose (Nath et al., 2005; Hawkes et al., 2007). Combining dark and photo‐fermentation may be a possible way to achieve this goal; however, improvements in sunlight conversion efficiency (such as improvements in sunlight collection and light transfer systems) and photobioreactor development need to be performed to reduce the energy demand and to increase the productivity of the photo‐fermentation step (Hallenbeck and Benemann, 2002; Akkerman et al., 2003). In the integrated system, the first step, dark fermentation, produces fermentation end‐products, which can then be converted to hydrogen by photo‐fermentation. Theoretically, the maximum hydrogen yield of 12 mol of H2 per mole of glucose may be obtained when glucose is converted to acetate as the terminal product through dark fermentation, which can then be converted into H2 through photo‐fermentation (Nath et al., 2005):
There are some examples in the literature in which the integrated systems were shown to improve the hydrogen yields; however, the yields were still far lower than the theoretical yield of 12 mol of H2 per mole of glucose. Enterobacter cloacae strain DM11, a facultative anaerobe, produced 1.86 mol of H2from 1 mol of glucose through dark fermentation, and Rhodobacter sphaeroides strain O.U.001 produced 1.5–1.72 mol of H2from 1 mol of glucose through photo‐fermentation. The hydrogen yield was shown to be higher with the integrated system compare to single‐step fermentation (Nath et al., 2005).
Another example of an integrated system resulting in higher hydrogen yields was shown by Kim and colleagues (2006). In the dark fermentation step, Clostridium butyricum produced hydrogen with a yield of 2.58 mol of H2 per mole of glucose as well as intermediates such as formate, acetate, propionate and butyrate. These intermediates were then converted into 5.72 mol of H2 by R. sphaeroides KD131, resulting in a total yield of 8.3 mol of H2 from 1 mol of glucose.
The current status of mesophilic, continuous, dark, fermentative, hydrogen production using mixed microflora was reviewed by Hawkes and colleagues (2007). Possible second‐stage processes to follow the dark fermentation stage include photo‐fermentation, microbial fuel cells and anaerobic digestion; these second stages increase the hydrogen production yield, produce electricity or methane.
Theoretically, stoichiometric yields can be obtained under equilibrium conditions, meaning at very low partial pressures of hydrogen and very slow rates (Hallenbeck and Benemann, 2002). Under these conditions, which are not practical, Woodward and colleagues (2000) were able to achieve nearly complete conversion of glucose to H2 and obtained a hydrogen yield of 11.6 mol of H2 per mole of glucose 6‐phosphate using pentose phosphate cycle enzymes combined with the hydrogenase from Pyrococcus furiosusthat uses NADP+ as the electron carrier (Woodward et al., 2000).
Biochemical structure of hydrogenases
Hydrogenases, the enzymes responsible for the reversible conversion of molecular hydrogen into two protons and two electrons (H2 ↔ 2H+ + 2e‐), are complex metalloenzymes that can be classified into three groups based on the number and identity of the metals in their active sites: [NiFe]‐, [FeFe]‐ and [Fe]‐hydrogenases (Vignais et al., 2001; Vignais and Colbeau, 2004). They are phylogenetically not related but still share common properties at their active sites; for example, they all contain Fe and CO as a ligand to the Fe atom.
The more common [NiFe]‐hydrogenases consist of a small subunit of 30 kDa and a large subunit of 60 kDa which are tightly bound via a large, hydrophobic surface (Fig. 4) (Vignais et al., 2001; Frey, 2002; Vignais and Colbeau, 2004). The small subunit contains up to three iron‐sulfur clusters which mediate the electron transfer between the active site and electron acceptor (or donor) of hydrogenase, while the large subunit contains the nickel‐iron active site in which the hydrogen activation occurs. The dinuclear active centre generally consists of a (CysS)2Ni(µ‐‘O’)(µ‐CysS)2Fe(CN)2(CO) structure where µ is the prefix for a bridging ligand (Fig. 5). The nickel group is coordinated to the protein via four thiol groups from cysteine residues. The Ni and Fe atoms are connected by two of these cysteine residues in both the active and inactive form of the enzyme and by an oxygen atom only in the oxidized inactive form. In addition, one CO and two CN ligands are coordinated to the Fe atom. Hydrophobic channels connect the active centre with the protein surface and most probably mediate gas substrate and product transport. It has been suggested that a few oxygen‐resistant hydrogenases have more narrow channels that serve to block oxygen transport to the active site (Vignais et al., 2001; Frey, 2002; Volbeda and Fontecilla‐Camps, 2003; Vignais and Colbeau, 2004). The first crystal structure of a hydrogenase enzyme was the [NiFe]‐hydrogenase from the sulfate‐reducing bacterium Desulfovibrio gigas (PDB:1FRV, 2FRV) (Fig. 4) (Volbeda et al., 1995; 1996). Other known X‐ray crystal structures of [NiFe]‐hydrogenases were obtained from Desulfovibrio vulgaris Miyazaki F (PDB:1H2A, 1H2R) (Higuchi et al., 1997; 1999), D. fructosovorans (PDB:1FRF) (Rousset et al., 1998), D. desulfuricans ATCC 27774 (PDB:1E3D) (Matias et al., 1999) and Desulfomicrobium baculatum (PDB:1CC1) (Garcin et al., 1999).
Figure 4.
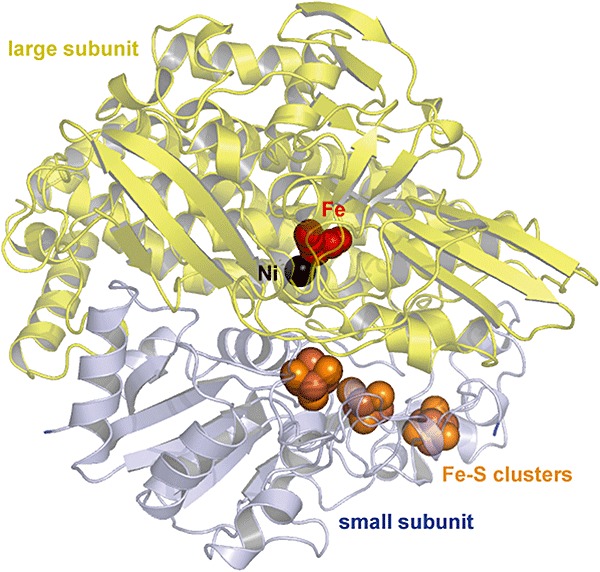
Three‐dimensional structure of [NiFe]‐hydrogenase from D. gigas(PDB:2FRV). The large subunit which contains the Ni‐Fe catalytic centre is shown in yellow. The small subunit which contains the Fe‐S clusters is shown in blue. Metals and sulfur atoms are depicted as spheres. Colour scheme: nickel is black, carbonmonoxide‐(dicyano) iron is red and Fe‐S cluster is orange.
Figure 5.
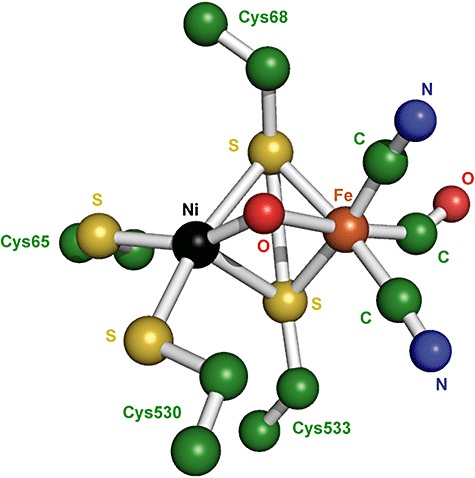
Structure of the oxidized [NiFe]‐hydrogenase active centre from D. gigas(PDB:2FRV). Colour scheme: nickel is black, iron is orange, oxygen is red, carbon is green and nitrogen is blue.
The large subunits of [NiFe]‐hydrogenases include hyaB for E. coli Hyd‐1 (Andrews et al., 1997), hybC for E. coli Hyd‐2, hycEfor E. coli Hyd‐3, hyfG for E. coli Hyd‐4, HoxH for Ralstonia eutropha(Burgdorf et al., 2002), HoxH for D. gigas(Volbeda et al., 1995) and HoxH for Synecocystis sp. PCC 6803 (Tamagnini et al., 2002). We have aligned these hydrogenases using Vector NTI software to determine their identity (Table 2). The bioinformatics analysis shows that the large subunits of these hydrogenases have very low homology. For example, the large subunit of E. coli Hyd‐3 has only 13.7% identity with HoxH of D. gigaswhose crystal structure is known (Fig. 4). Modelling by homology to predict the structure of E. coli Hyd‐3 using the structure of D. gigas is quite risky and may impede mutagenesis studies. This low homology illustrates the need for a crystal structure of E. coli Hyd‐3.
Table 2.
Identity between hydrogenase large subunits derived from Escherichia coli (E. coli), Ralstonia eutropha H16 (R. eutropha), Rhodoccus opacus MR11 (R. opacus), Synechocystis sp. PCC 6803 (Syn. PCC 6803) and Desulfovibrio gigas (D. gigas).
| E. coli Hyd‐1 | E. coli Hyd‐2 | E. coli Hyd‐3 | E. coli Hyd‐4 | R. eutropha HoxH | R. opacus HoxH | Syn. PCC 6803 HoxH | D. gigas large subunit | |
|---|---|---|---|---|---|---|---|---|
| E. coli Hyd‐1 | 40.7% | 13.9% | 11.9% | 18.8% | 18.3% | 16.1% | 39.9% | |
| E. coli Hyd‐2 | 40.7% | 13.8% | 11.0% | 17.3% | 18.1% | 19.0% | 41.6% | |
| E. coli Hyd‐3 | 13.9% | 13.8% | 69.8% | 13.1% | 12.2% | 12.4% | 13.7% | |
| E. coli Hyd‐4 | 11.9% | 11.0% | 69.8% | 12.7% | 12.0% | 13.0% | 12.4% | |
| R. eutropha HoxH | 18.8% | 17.3% | 13.1% | 12.7% | 85.2% | 42.7% | 19.9% | |
| R. opacus HoxH | 18.3% | 18.1% | 12.2% | 12.0% | 85.2% | 42.0% | 19.8% | |
| Syn. PCC 6803 HoxH | 16.1% | 19.0% | 12.4% | 13.0% | 42.7% | 42.0% | 17.8% | |
| D. gigas large subunit | 39.9% | 41.6% | 13.7% | 12.4% | 19.9% | 19.8% | 17.8% |
Hyd‐1, Hyd‐2, Hyd‐3, Hyd‐4 are HyaB, HybC, HycE and HyfG of E. coli hydrogenases 1, 2, 3 and 4 respectively. Protein sequences for the large subunit or HoxH of hydrogenase were aligned by using a VectroNTI alignment software.
The second class of [FeFe]‐hydrogenases (also known as Fe‐only hydrogenases) generally consist of a single catalytic subunit (Nicolet et al., 2000; Vignais et al., 2001; Frey, 2002; Vignais and Colbeau, 2004). The dinuclear active centre probably consists of a (H2O)(CO)(CN)Fe(µ‐CO)(µ‐NH(CH2S‐)2)Fe(CysS)(CN)(CO) overall structure. Both Fe atoms are connected by one CO and two S ligands of a dithiol‐bridging ligand [a di(thiomethyl)amine molecule (HN‐CH2S‐) or propane dithiol] (Zilberman et al., 2007). Each Fe atom has a CO and a CN ligand. In addition, a [4Fe‐4S] cluster is coordinated through the sulfur of a cysteine residue to one of the Fe atoms (Fig. 6) (Nicolet et al., 2000; Frey, 2002; Vignais and Colbeau, 2004; Albracht et al., 2006). The crystal structure of [FeFe]‐hydrogenases from Clostridium pasteurianum (PDB:1FEH) (Peters et al., 1998) and D. desulfuricans ATCC 7757 (PDB:1HFE) (Nicolet et al., 1999) revealed the similarities between [NiFe]‐ and [FeFe]‐hydrogenases such as the presence of iron‐sulfur clusters for electron transport, a dinuclear metal centre with CO and CN ligands, and hydrophobic channels connecting the molecular surface to the active site (Volbeda et al., 1995; 1996; Montet et al., 1997).
Figure 6.
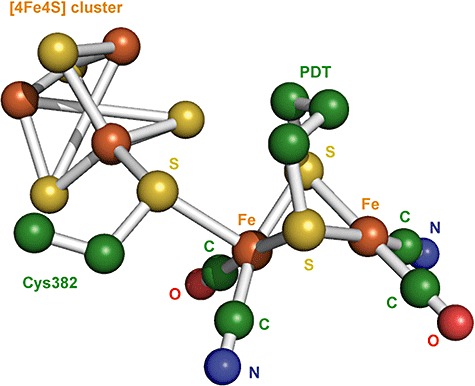
Structure of the [FeFe]‐hydrogenase active centre from Desulfovibrio desulfuricans ATCC 7757 (PDB:1HFE). The bridging CO that connects both Fe atoms and the water molecule that binds to the Fe atom can be viewed using the structure from C. pasteurianum (PDB:1FEH). Colour scheme is iron is orange, oxygen is red, carbon is green and nitrogen is blue. PDT, 1,3‐propanedithiol.
The third class of [Fe]‐hydrogenases (also known as iron‐sulfur cluster‐free hydrogenases or metal‐free hydrogenases) are found in some methanogenic archaea (Zirngibl et al., 1990). These enzymes are also referred to as H2‐forming methylene‐tetrahydromethanopterin dehydrogenase as the enzyme catalyses the reversible reduction of N5,N10‐methenyl‐tetrahydromethanopterin with H2 to N5,N10‐methylene‐tetrahydromethanopterin and a proton (Thauer et al., 1996). Different from [NiFe]‐ and [FeFe]‐hydrogenases, they do not catalyse the reversible conversion of molecular hydrogen into two protons and two electrons and contain neither Fe‐S clusters nor nickel (Zirngibl et al., 1992). They contain a single Fe atom bound with two CO, one S and one or two N/O ligands (Korbas et al., 2006). Recently, the crystal structure of the apoenzyme of the iron‐sulfur cluster‐free hydrogenase from Methanocaldococcus jannaschii (PDB:2B0J) (Pilak et al., 2006) was determined. The crystal structure of the mature iron‐sulfur cluster‐free hydrogenase and its catalytic mechanism are still unknown.
Maturation proteins
The biosynthesis of [NiFe]‐hydrogenases and [FeFe]‐hydrogenases are distinct from each other; however, they share some common properties such as requiring guanine nucleotide‐binding proteins (GTPases), with similar roles (Leach and Zamble, 2007). The biochemical pathways for the synthesis and incorporation of the NiFe(CO)(CN)2 metal centre in [NiFe]‐hydrogenases are briefly described here. By comparison, more progress needs to be made in understanding the biosynthesis of [FeFe]‐hydrogenases.
The maturation steps of [NiFe] biosynthesis include (i) the biosynthesis of CO and CN, the diatomic ligands of the active site, (ii) insertion of the one CO and two CN ligands to the Fe atom, (iii) insertion of the Fe(CN)2CO complex into the hydrogenase, (iv) insertion of the Ni atom into the active site of hydrogenase, and (v) correct folding before delivery to the membrane (Forzi and Sawers, 2007). More detailed information can be found on the maturation steps of [NiFe]‐hydrogenases in other review papers (Forzi and Sawers, 2007; Leach and Zamble, 2007).
There are many genes encoding accessory proteins that participate in the maturation of nickel‐containing hydrogenases. For example, at least seven proteins are required for the maturation of Hyd‐3 in E. coli, which is the most studied hydrogenase: six of the proteins are from the hyp (hydrogen pleiotropy) operon (hypABCDEF), and an endopeptidase is encoded by hycI(Blokesch et al., 2002). The function of these genes on the maturation process has been studied (Table 3); however, more research needs to be performed to understand the complete biochemical pathway of the active site synthesis.
Table 3.
Maturation proteins and their functions.
| Protein | Name | Function | PDB ID | Reference |
|---|---|---|---|---|
| HypA | Zn‐containing protein | Maturation of the large subunit of Hyd‐3 , nickel insertion along with SlyD, a peptidyl‐prolyl cis/trans‐isomerase that forms a complex with HypB | – | Hube et al. (2002); Atanassova and Zamble (2005); Zhang et al. (2005) |
| HypB | GTPase | Nickel liganding into hydrogenase large subunit | 2HF9 | Leach et al. (2005); Gasper et al. (2006) |
| HypC | Chaperone‐like protein | Maturation of hydrogenase 3 by catalysing Fe insertion | 2Z1C | Blokesch et al. (2001); Watanabe et al. (2007) |
| HypD | Fe/S protein | Possibly transferring the cyano group to the Fe atom or in the cyanation reaction | 2ZID | Blokesch and Böck (2006); Watanabe et al. (2007) |
| HypF | Carbamoyl phosphate phosphatase | Catalyses the synthesis of the CN– ligands of the active site iron of [NiFe]‐hydrogenases using carbamoylphosphate as a substrate along with HypE | 1GXTa | Rosano et al. (2002); Reissmann et al. (2003) |
| HypE | ATP‐dependent dehydratase | Hydrogenase maturation protein | 2Z1E, 2Z1F | Reissmann et al. (2003); Watanabe et al. (2007) |
| HybF | Zn‐containing protein | Maturation of Hyd‐1 and ‐2 (HypA homologous) | – | Hube et al. (2002) |
| HybG | Chaperone‐like protein | Maturation of Hyd‐1 and ‐2 (HypC homologous) | – | Blokesch et al. (2001) |
| HyaD | Endopeptidase | Maturation of Hyd‐1 | – | Forzi and Sawers (2007) |
| HybD | Endopeptidase | maturation of Hyd‐2 | 1CFZ | Fritsche et al. (1999); Forzi and Sawers (2007) |
| HycI | Endopeptidase | Recognizes the Ni bound state of Hyd3, maturation of the large subunit of Hyd‐3 | 2I8L | Forzi and Sawers (2007); Yang et al. (2007) |
| HydEHydG | S‐adenosylmethionine enzymes | Required for the biosynthesis of [FeFe]‐hydrogenases | – | King et al. (2006) |
| HydF | GTPase | Required for the biosynthesis of [FeFe]‐hydrogenases | – | King et al. (2006) |
The crystal structure of HypF N‐terminal acylphosphatase domain (residues 1–91).
The crystal structure or the NMR solution structure is given by the Protein Data Bank identification number (PDB ID).
The crystal structures of HypB from M. jannaschii(Gasper et al., 2006), HypC, HypD and HypE from Thermococcus kodakaraensis(Watanabe et al., 2007), HybD from E. coli (Fritsche et al., 1999), HypF N‐terminal acylphosphatase domain (residues 1–91) from E. coli(Rosano et al., 2002), and the NMR solution structure of HycI from E. coli (Yang et al., 2007) are known (Table 3) and provide insight into their functions in maturation process.
Maturation of [FeFe]‐hydrogenases may not require as many proteins as it only requires the biosynthesis and insertion of the catalytic iron‐sulfur cluster known as the H‐cluster. Initial studies on how [FeFe]‐hydrogenases are biosynthesized showed that the HydE, HydF and HydG proteins are required (Table 3) (King et al., 2006). McGlynn and colleagues (2007) showed that the inactive hydrogenase structural protein (HydA) of Clostridium saccharobutylicum can be rapidly activated by the maturation proteins of Clostridium acetobutylicum which are expressed in concert (HydEFG) in E. coli(McGlynn et al., 2007). The group proposed that the HydE, HydF and HydG proteins form an H‐cluster precursor, which is transferred to the inactive hydrogenase structural protein in order to active it.
Metabolic engineering of fermentative systems
There are several examples of metabolic engineering applications to increase the fermentative or photosynthetic hydrogen production. Some of the examples concerning enhanced photosynthetic hydrogen production can be found elsewhere (Miyake et al., 1999; Vignais et al., 2006; Rey et al., 2007). Examples discussed here concern improving fermentative hydrogen production pathways.
Recombinant strains can be created through genetic and metabolic engineering, which leads to higher levels of hydrogen production. The hydrogen yield is suboptimal in many organisms containing uptake hydrogenases, as some of the produced hydrogen is consumed. Therefore, knocking out the genes encoding uptake hydrogenases is one way to enhance hydrogen production. Other ways to increase hydrogen production include overexpression of hydrogen‐evolving hydrogenases, shutting down metabolic pathways that compete for hydrogen production, and overexpression of cellulases, hemicellulases and lignases that can maximize glucose availability (Levin et al., 2004; Nath and Das, 2004). Table 4 summarizes the H2 production rates and yields for some of the recombinant and wild‐type strains.
Table 4.
Productivities and yields of recombinant and wild‐type systems.
| Strain | Productivity, µmol H2 (mg protein)−1 h−1 | Yield | Comments | Reference |
|---|---|---|---|---|
| E. coli BW25113 hyaB hybC hycA fdoG fhlA+ | 113 | 1.2 mol of H2 per mole of formate | Low partial pressure batch reactor using low cell density with complex‐formate medium | Maeda et al. (2007b) |
| E. coli BW25113 hyaB hybC hycA fdoG ldhA frdC aceE | 32 | 1.3 mol of H2 per mole of glucose | Low partial pressure batch reactor with complex glucose | Maeda et al. (2007c) |
| E. coli TG1/pBS(Kan)Synhox | 10 | – | Hydrogen production from E. coli cells expressing cyanobacterium Synechocystis sp. PCC6803 hoxEFUYH | Maeda et al. (2007d) |
| E. coli SR15 (ΔldhAΔfrdBC) | 27 | 1.8 mol of H2 per mole of glucose | Reactor equipped with pH sensor and ports for NaOH feed, gas exhaustion, substrate feed and sampling | Yoshida et al. (2006) |
| E. coli DADE (ΔtatA–E) | 4.4 | – | Reaction at 100 mM glucose | Penfold et al. (2006) |
| E. coli HD701 (ΔhycA) | 5.7 | – | Reaction at 100 mM glucose | Penfold et al. (2003) |
| E. coli SR13 (ΔhycA fhlA+) | 254 | – | Reaction using high cell density and 25 mM formic acid | Yoshida et al. (2005) |
| E. coli SR14 (ΔhycA fhlA+ΔldhAΔfrdBC) | 288 | – | Reaction by using a metabolite excretion system to remove excess metabolites from the medium | Yoshida et al. (2007) |
| E. coli BL21 hydA+ | 5.6 | 3.1 mol of H2 per mole of glucose | Hydrogen production with E. coli BL21 expressing the [Fe]‐hydrogenase gene (hydA) from E. cloacae IIT‐BT‐08 | Chittibabu et al. (2006) |
| E. coli HD701 (+invertase) | 3.2 | – | Hydrogen production from sucrose with E. coli HD701 strain expressing an invertase activity | Penfold and Macaskie (2004) |
| C. paraputrificum M‐21 hydA+ | – | 2.4 mol of H2 per mole of GlcNAc | Cultured using GS medium with 1% GlcNAc as a carbon source | Morimoto et al. (2005) |
| C. tyrobutyricum (Δack) | – | 2.2 mol of H2 per mole of glucose | Fed‐batch fermentations of glucose by free cells | Liu et al. (2006) |
| E. aerogenes AY‐2 (double mutant) | 6.8 | 1.1 mol of H2 per mole of glucose | Reaction by self‐flocculated cells in a packed‐bed reactor in a minimal medium | Rachman et al. (1998) |
| E. cloacae IIT‐BT‐08 (double mutant) | – | 3.4 mol of H2 per mole of glucose | Mutant with lower alcohol dehydrogenase and butanediol dehydrogenase activity, reaction using immobilized bioreactor | Kumar et al. (2001) |
| E. cloacae IIT‐BT‐08 (wild‐type) | 59 | 6 mol of H2 per mole of sucrose 2.2 mol of H2 per mole of glucose | A Gram‐negative hydrogen producing facultative anaerobe, reaction using sucrose at 36°C | Kumar and Das (2000) |
| E. asburiae SNU‐1 (wild type) | 76 | 0.4 mol of H2 per mole of formate | A new fermentative hydrogen‐producing bacterium isolated from a domestic landfill | Shin et al. (2007) |
| Citrobacter sp. Y19 (wild type) | 65 | 2.5 mol of H2 per mole of glucose | CO‐dependent H2 production | Oh et al. (2003) |
| Klesiella oxytoca HP1 (wild type) | 30 | 3.2 mol of H2 per mole of sucrose | Hydrogen‐producing bacterial strain isolated from a hot spring | Minnan et al. (2005) |
| Rhodopseudomonas palustris JA1 (wild type) | 60 | – | A purple non‐sulfur bacterium | Archana et al. (2003) |
| R. palustris P4 (wild type) | 41 | – | A photosynthetic bacterium isolated from an anaerobic wastewater | Jung et al. (1999) |
| Caldicellulosiruptor saccharolyticus (wild type) | 21 | 6.6 mol of H2 per mole of sucrose | Hydrogen‐producing extreme thermophilic bacterium | van Niel et al. (2002) |
GlcNAc, N‐acetylglucosamine.
The metabolic pathway for the fermentation of glucose by E. coliis shown in Fig. 2. The fermentation products are hydrogen, acetate, ethanol, lactate, formate and some succinate (Clark, 1989). Although several microorganisms can produce hydrogen through dark fermentation, E. colihas many advantages, such as rapid growth and simple nutritional requirements. It is also easy to perform metabolic engineering to increase hydrogen production in E. coli, as E. coli is easily manipulated through genetic engineering. To enhance hydrogen production, recombinant strains of E. coli were developed having mutations in several genes, for example, in the large subunit of uptake Hyd‐1 and ‐2 (hyaBand hybCrespectively) (Maeda et al., 2007b,c), in lactate dehydrogenase (ldhA) (Bisaillon et al., 2006; Yoshida et al., 2006; Maeda et al., 2007c), in the FHL repressor (hycA) (Penfold et al., 2003; 2006; Yoshida et al., 2005; 2006; Maeda et al., 2007b,c), in the FHL activator (fhlA) (Yoshida et al., 2005; 2006; Bisaillon et al., 2006; Maeda et al., 2007b,c), in fumarate reductase (frdBC) (Yoshida et al., 2006; Maeda et al., 2007c), in the Tat system (tatA–E) (Penfold et al., 2003; 2006), in the alpha subunit of the formate dehydrogenase‐N and ‐O (fdnGand fdoGrespectively) (Maeda et al., 2007b,c), in the alpha subunit of nitrate reductase A (narG) (Maeda et al., 2007b,c), in pyruvate dehydrogenase (aceE) (Maeda et al., 2007c), in pyruvate oxidase (poxB) (Maeda et al., 2007c) and in proteins that transport formate (focA and focB) (Maeda et al., 2007b,c) (Fig. 2).
Yoshida and colleagues (2005) enhanced fermentative biohydrogen production from formic acid by overexpressing the FHL activator encoded by the fhlAand by inactivating the FHL repressor encoded by the hycA in E. coliK‐12 strain W3110. The strain in which fhlAwas overexpressed had 1.7‐fold higher hydrogen production, whereas the hycA‐inactivated strain had 1.2‐fold higher hydrogen compared with the wild‐type strain. The hydrogen production rate was 2.8‐fold higher with both fhlAoverexpressed and hycA inactivated (recombinant strain SR13). The fhlAoverexpression and hycA inactivation caused a 6.5‐fold increase in the formate dehydrogenase expression encoded by fdhFand a sevenfold increase in the Hyd‐3 large subunit expression encoded by hycE.Further enhancement in hydrogen production rate from formic acid was achieved using the mutant strain SR13 under the high cell density [254 µmol (mg protein)−1 h−1] (Table 4) (Yoshida et al., 2005).
In another study by Yoshida and colleagues (2006), enhanced hydrogen yields from glucose was demonstrated by blocking the competing lactate (via deleting ldhA) and succinate (via deleting frdBC) production pathways. The maximum hydrogen yield was found to be 1.08 mol of H2 per mole of glucose with the wild‐type E. coliK‐12 strain W3110, whereas the E. colistrain with fhlAoverexpressed, hycA inactivated, and ldhA and frdBC deleted (SR14) had a maximum hydrogen yield of 1.87 mol of H2 per mole of glucose. The maximum hydrogen yield with ldhA and frdBC deleted (SR15) was also around 1.82 mol of H2 per mole of glucose; hence, fhlAoverexpression and hycA inactivation did not cause a significant increase in hydrogen yield from glucose. Moreover, the hydrogen production rate from glucose was 1.4‐fold higher with mutant strain SR15 [27 µmol (mg protein)−1 h−1] (Table 4) compared with the wild‐type strain (Yoshida et al., 2006).
In a study by Penfold and colleagues (2003), the maximum hydrogen production rate with hycA inactivated in E. coli(strain HD701) was around twofold higher [5.7 µmol (mg protein)−1 h−1] (Table 4), compared with the wild‐type strain MC4100 at a glucose concentration of 100 mM. The difference in rates between wild‐type and the recombinant strain was better (up to 14‐fold) at lower glucose concentrations. Increases in hydrogen production rates were similar when industrial waste water with high sugar content was used instead of glucose as a substrate (Penfold et al., 2003). Introducing an invertase gene, which is responsible for converting sucrose to glucose and fructose, enabled strain HD701 to utilize sucrose and produce hydrogen with a rate of 3.2 µmol (mg protein)−1 h−1 (Table 4) (Penfold and Macaskie, 2004). Deletion of tatCto inactivate the twin arginine translocation (Tat) translocase in strain HD701 did not affect hydrogen production significantly probably because the saturated FHL system did not use the excess amount of formate (Penfold and Macaskie, 2004).
The Tat translocase of E. coli MC4100 transports two uptake hydrogenases, Hyd‐1 and ‐2, and the two formate dehydrogenases, FdH‐N and FdH‐O, to the cell membrane (Penfold et al., 2006). The Tat system consists of the TatABCE proteins. Deletion of tatC (strain FTD701) or tatA–E(strain DADE) resulted in a twofold enhancement in hydrogen production rate compared with wild‐type E. coliMC4100 [4.4 µmol (mg protein)−1 h−1 with strain DADE and 2.2 µmol (mg protein)−1 h−1 with wild type] (Table 4). Deletion of hycA (strain HD701) caused a 2.5‐fold increase in hydrogen production rate compared with wild‐type E. coliMC4100. Combining the tatC and hycAmutations did not improve hydrogen production further. Hence, inactivating the Tat system in E. coli is another way of producing a recombinant system with enhanced hydrogen productivity (Penfold et al., 2006).
In a study by Bisaillon and colleagues (2006), the effect of mutations in uptake hydrogenases (Hyd‐1, Hyd‐2) as well as in ldhAand fhlA of E. colistrain BW545 was investigated (Bisaillon et al., 2006). Escherichia coli strain JW135, which lacks uptake Hyd‐1 and Hyd‐2, showed a 37% increase in hydrogen production rate compared with wild‐type strain BW545. Mutations in ldhA and in fhlA caused an 18% and 11% increase in hydrogen production, respectively, compared with the wild‐type E. coli BW545, and a 47% increase was obtained with the double mutant (ldhA and fhlA). Hydrogen yield approached 2 mol of H2 per mole of glucose at low glucose concentrations with E. coli recombinant strain DJT135, which lacks Hyd‐1 and Hyd‐2, and has mutations in ldhA and fhlA(Bisaillon et al., 2006).
Maeda and colleagues (2007b,c) obtained the largest increase in hydrogen production to date by introducing various combinations of mutations in E. coli cells (Maeda et al., 2007b,c). To enhance hydrogen production from formate, the hyaB and hybCgenes were deleted to remove the hydrogen uptake activity by Hyd‐1 and ‐2; the hycA gene (FHL repressor) was also deleted and the fhlA gene (FHL activator) was overexpressed to change the regulation of the FHL complex. In addition, the alpha subunit of formate dehydrogenase‐N encoded by fdnG, the alpha subunit of formate dehydrogenase‐O encoded by fdoGand the alpha subunit of nitrate reductase A encoded by narGwere deleted to inactivate formate consumption other than FHL. focA and focB deletions were also made to prevent export of formate (Maeda et al., 2007b). The quintuple mutant strain of BW25113 with hyaB, hybC, hycA and fdoGdeleted and fhlAoverexpressed was shown to be the best mutant strain, and its hydrogen production rate from formate [113 ± 12 µmol (mg protein)−1 h−1] (Table 4) was 141‐fold higher compared with the wild‐type strain BW25113. The theoretical maximum hydrogen yield of 1 mol of H2 per mole of formate was also achieved with the quintuple mutant (Maeda et al., 2007b). This paper is also noteworthy as the authors made 28 isogenic deletion mutants using a novel method involving successive P1 transformations and the E. coli Keio collection (Baba et al., 2006).
To increase the hydrogen production from glucose, Maeda and colleagues (2007c) studied four more mutations in addition to the nine mutations discussed above (Maeda et al., 2007c). The fumarate reductase encoded by frdC was deleted to prevent phosphoenol pyruvate from forming succinate (this should increase pyruvate for hydrogen), and to prevent pyruvate from forming anything but formate, the lactate dehydrogenase encoded by ldhA, the pyruvate dehydrogenase encoded by aceE and the pyruvate oxidase encoded by the poxB were deleted. The E. coliseptuple mutant with hyaB, hybC, hycA, fdoG, frdC, ldhA and aceEdeletions in strain BW25113 had the highest hydrogen production rate from glucose [4.6‐fold higher compared with the wild‐type strain BW25113, 32 ± 6 µmol (mg protein)−1 h−1] (Table 4). This septuple mutant also had a hydrogen yield of 1.3 mol of H2 per mole of glucose compared with 0.65 mol of H2 per mole of glucose with the wild‐type BW25113 cells (Maeda et al., 2007c).
The strict anaerobe Clostridiumsp. usually produces compounds such as acetate (acetic acid), butyrate (butyric acid), lactate, acetone, butanol and ethanol (Chin et al., 2003) (Fig. 3). Mutants of Clostridium sp. were also developed using metabolic and/or genetic engineering to enhance hydrogen production. Overexpression of its own hydA gene encoding a [Fe]‐hydrogenase in Clostridium paraputrificum M‐21 resulted in a 1.7‐fold enhancement in hydrogen production from N‐acetylglucosamine (GlcNAc) (2.4 mol of H2 per mole of GlcNAc versus 1.4 mol of H2 per mole of GlcNAc) (Table 4) (Morimoto et al., 2005). The improved hydrogen yield in recombinant C. paraputrificum M‐21 was due to enhanced acetic acid production by overoxidation of NADH and drastic reduction of lactic acid production.
Another study showed that inactivation of ackencoding acetate kinase for acetate formation in Clostridium tyrobutyricumresulted in a 1.5‐fold enhancement in hydrogen production from glucose (2.2 mol of H2 per mole of glucose versus 1.4 mol of H2 per mole of glucose) (Table 4) (Liu et al., 2006). The mutant also had 1.4‐fold increase in hydrogenase activity.
One must note that, although the theoretical hydrogen yields are greater with strict anaerobes such as Clostridiumsp., facultative anaerobes such as E. coli or Enterobacter sp. are more favourable for hydrogen production, as they are less sensitive to oxygen, able to recover their activities if accidentally exposed to oxygen, and have faster growth and hydrogen production rates (Shin et al., 2007).
Similar metabolic engineering studies have been performed using Enterobacter species as well. Blocking alcohol and some of the organic acid formation pathways in E. cloacae IIT‐BT‐08 (Kumar and Das, 2000) through mutagenesis resulted in enhanced hydrogen production from glucose (Kumar et al., 2001). An E. cloacae double mutant gave a hydrogen yield of 3.4 mol of H2 per mole of glucose (Table 4), whereas the wild‐type strain gave a yield of 2.2 mol of H2 per mole of glucose. Improvement of hydrogen yields in the double mutant was due to the lower amounts of ethanol and butanediol production. Similarly, due to the lower amounts of ethanol and butanediol production with Enterobacter aerogenesdouble mutant, the hydrogen yield was enhanced by twofold (Rachman et al., 1997). Enterobacter aerogenes double mutant strain AY‐2 had a hydrogen yield of 1.2 mol of H2 per mole of glucose, whereas the wild‐type strain (HU‐101) had a yield of 0.56 mol of H2 per mole of glucose in a batch cultures. In addition, the hydrogen production rate using a packed‐bed reactor in a minimal medium was around twofold higher with mutant AY‐2 [6.8 µmol (mg protein)−1 h−1] (Table 4), compared with wild‐type HU‐101 (Rachman et al., 1998).
Recombinant hydrogenases
One of the earliest studies for creating strains with recombinant hydrogenases was performed by Karube and colleagues (1983). Cloning and expressing the hydrogenase gene from C. butyricum in E. colistrain HK16 (Hyd‐, lacking the native hydrogenase activity) in CB medium resulted in a 3‐ and 3.5‐fold enhancement in hydrogenase activity compared with wild‐type C. butyricum and E. coli C600 (Hyd+) respectively (Karube et al., 1983).
In another study, the hydrogen yield was enhanced when hydA, the [Fe]‐hydrogenase gene from E. cloacae IIT‐BT‐08 was overexpressed in non‐hydrogen‐producing E. coli BL‐21 as a glutathione–HydA fusion protein (Mishra et al., 2004; Chittibabu et al., 2006). The recombinant E. coli BL21 had a yield of 3.12 mol of H2 per mole of glucose, which was higher than the wild‐type strain E. cloacae IIT‐BT‐08. Moreover, the maximum hydrogen production rate was reported to be 5.6 µmol (mg protein)−1 h−1 from glucose with the recombinant strain using a continuous immobilized whole‐cell bioreactor in MYG medium (Table 4) (Chittibabu et al., 2006).
Recently, a cyanobacterial enzyme was successfully cloned and expressed in E. colifor the first time to improve hydrogen production (Maeda et al., 2007d). The hydrogen yield was enhanced up to 41‐fold when the reversible hydrogenase encoded by hoxEFUYH from the cyanobacterium Synechocystis sp. PCC 6803 was cloned in E. coli. DNA microarrays were used to show that expression of the native E. coli hydrogenases was not affected by the expression of the cyanobacterial HoxEFUYH, and a series of isogenic knockout mutants were used to discern that the enhanced hydrogen production was due to the inhibition of the native hydrogen uptake activity (by E. coliHyd‐1 and ‐2) by HoxEFUYH (Maeda et al., 2007d).
Reactor‐based methods to increase hydrogen production
There are also non‐metabolic engineering ways to increase fermentative hydrogen yields, such as heat treatment for mixed cultures to select for spore‐forming bacteria in mixed cultures, sparging and operational controls (Nath and Das, 2004; Kraemer and Bagley, 2007). For example, gas sparging is usually beneficial for hydrogen production (Kraemer and Bagley, 2007), and we have found that using a low partial pressure system increases productivity with E. coli (Maeda et al., 2007b). In addition, sufficient mixing is beneficial for mass transfer of hydrogen from the fermentation to the headspace (Kraemer and Bagley, 2007).
Compounds other than formate and glucose stimulate hydrogen production by bacteria. For example, thiosulfate elevated hydrogen production by twofold for immobilized E. coli cells by limiting the consumption of glucose by metabolic paths other than those that produce hydrogen; a similar effect was observed by adding succinate (Nandi et al., 2001). The concentrations of phosphate (when growth limiting) also affect hydrogen yield (Bisaillon et al., 2006). In addition, low concentrations of nitrogen (less than 1 mM) are significant for enhancing hydrogen yields (reached 2 mol of H2 per mole of glucose), and the hydrogen yield was dramatically decreased for high concentrations of ammonium (Bisaillon et al., 2006).
Yoshida and colleagues (2005) increased hydrogen production over two orders of magnitude to 254 µmol (mg protein)−1 h−1 by using high‐cell density and 25 mM formic acid (Table 4); they estimated that 1 kW of energy could be supplied with 2 l of bacteria producing hydrogen at this rate. Also, they improved hydrogen productivity from glucose by using a metabolite excretion system to remove excess metabolites from the medium; the maximum hydrogen production rate using E. coli SR14 (ΔhycAΔldhAΔfrdBC overexpressing fhlA) was 288 µmol (mg protein)−1 h−1 at a dilution rate of 2 h−1 compared with 74 µmol (mg protein)−1 h−1 in batch‐mode incubation (Table 4) (Yoshida et al., 2007). Hence, reactor design is important for hydrogen production.
Perspectives
Metabolic engineering is clearly an important approach to improve fermentative hydrogen production, but further research and development are needed for converting cheap feedstocks to hydrogen. For example, hydrogen productivities are now such that reactors of reasonable size are required based on glucose and formate (2–500 l); however, if 24 mol of hydrogen per hour is required for 1 kW of electricity from a fuel cell (roughly enough to power a home) (Levin et al., 2004), then the annual cost of electricity if formate is used for the fermentation is ∼$172 000 (assuming $12 per kilogram of formate and 1 mol of H2 per mole of formate) and ∼$6400 if glucose is used (assuming $0.22 per kilogram of glucose and 1.3 mol of H2 per mole of glucose). Because the theoretical yield has been achieved with formate (Maeda et al., 2007b) and further increases in productivity will reduce the size of the reactor but not the cost of the reactants, then the main way to reduce these costs are to increase the yield from glucose or convert less expensive feedstocks into hydrogen.
Due to the complexity of the hydrogenases and their maturation proteins and due to the fact that 14% of the E. coligenome remains completely uncharacterized (14% is unknown, 32% is predicted by computational analysis and 54% is experimentally determined) (Riley et al., 2006; Willenbrock et al., 2006), unknown pathways need to be explored (rather than just deleting known pathways). One possible way to achieve this is by applying random chemical mutagenesis followed by the application of DNA microarrays to determine which paths are affected and to identify bottlenecks.
To determine which pathways need to be optimized for hydrogen production, linear programming could be conducted with a metabolic model. Metabolic flux analysis (MFA) is the technique by which flux distributions through metabolic pathways are either determined or predicted (Varma and Palsson, 1994; Stephanopoulos et al., 1998). Fluxes are calculated through the development of stoichiometric models of the metabolic reaction network. Flux balance analysis (FBA) is a variant of MFA, in which the goal is to find all the feasible flux distributions for an organism under prescribed conditions (Varma and Palsson, 1994; Schilling et al., 2000; Edwards et al., 2002; Reed and Palsson, 2003). The benefit of FBA is that it may be carried out at the genome scale with limited data and still provide insight into how the organism will behave (Edwards et al., 2002; Schilling et al., 2002; Price et al., 2004; Reed and Palsson, 2004).
More effort is also needed for creating recombinant strains expressing foreign hydrogenases. The cloned gene could be from an organism that produces an oxygen‐insensitive hydrogenase such as Rhodobacter capsulatus(Vignais et al., 1997), R. eutropha(Burgdorf et al., 2005) or Aquifex aeolicus(Guiral et al., 2006) or it can be from E. coli itself. In addition, protein‐engineering techniques, such as DNA shuffling (Canada et al., 2002), may be used to create novel, oxygen‐tolerant hydrogenases and hydrogenases with enhanced activity; however, an efficient high‐throughput screening method is required to effectively screen thousands of colonies expressing hydrogenase variants. Along these lines, Nagy and colleagues (2007) successfully applied gene shuffling for the first time to create recombinant [FeFe]‐hydrogenase libraries of C. acetobutylicumand C. saccharobutylicum; these enzymes have 67% protein identity. A single‐stranded DNA approach was used on the hyaAsequences although no screening method was reported. By choosing 62 random clones, one mutant was found with a 30% increase in activity. Because it was necessary to coexpress the accessory proteins HydEFG to achieve activity of the Clostridialproteins in this heterologous host, it may be easier to shuffle native E. coli hydrogenases and make use of its native maturation proteins. This approach has been used successfully by T. Maeda and T.K. Wood who have increased activity of Hyd‐3 of E. coli by over 30‐fold using a hydrogen‐sensitive membrane (Seibert et al., 1998) to detect hydrogen directly (unpublished). Another rapid screening method based on the spectrophotometric detection of the consumption of formate in E. colihas recently been developed (Maeda and Wood, submitted); this is an indirect method that relies on formate consumption being directly related to more active hydrogenases.
The latest project of J. Craig Venter Institute (Rockville, MD), which accumulates DNA from marine microorganisms, may also provide us with naturally engineered novel hydrogenases with interesting metabolic processes; the Venter team already has announced the discovery of a novel oxygen‐insensitive hydrogenase (pers. comm.). Discovering novel hydrogenases and metabolic pathways through genetic engineering, high‐throughput genomic sequencing, environmental genomics and/or metagenomic technologies (Handelsman, 2004; Riesenfeld et al., 2004; Binnewies et al., 2006) may also help to make biological hydrogen production more favourable, practical and commercially competitive.
Acknowledgments
This research was supported by DARPA (HR0011‐06‐1‐0001). We thank Dr Ranjan Srivastava for his ideas on genome‐scale modelling.
References
- Akkerman I., Janssen M., Rocha J.M.S., Reith J.H., Wijffels R.H. Photobiological hydrogen production: photochemical efficiency and bioreactor design. In: Reith J.H., Wijffels R.H., Barten H., editors. Dutch Biological Hydrogen Foundation; 2003. pp. 124–145. [Google Scholar]
- Albracht S.P.J., Roseboom W., Hatchikian E.C. The active site of the [FeFe]‐hydrogenase from Desulfovibrio desulfuricans. I. Light sensitivity and magnetic hyperfine interactions as observed by electron paramagnetic resonance. J Biol Inorg Chem. 2006;11:88–101. doi: 10.1007/s00775-005-0039-8. [DOI] [PubMed] [Google Scholar]
- Andrews S.C., Berks B.C., McClay J., Ambler A., Quail M.A., Golby P., Guest J.R. A 12‐cistron Escherichia coli operon (hyf) encoding a putative proton‐translocating formate hydrogenlyase system. Microbiology. 1997;143:3633–3647. doi: 10.1099/00221287-143-11-3633. [DOI] [PubMed] [Google Scholar]
- Angenent L.T., Karim K., Al‐Dahhan M.H., Wrenn B.A., Domiguez‐Espinosa R. Production of bioenergy and biochemicals from industrial and agricultural wastewater. Trends Biotechnol. 2004;22:477–485. doi: 10.1016/j.tibtech.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Archana A., Sasikala C., Ramana C.V. Augmentation of H2 photoproduction in Rhodopseudomonas palustris by N‐heterocyclic aromatic compounds. Biotechnol Lett. 2003;25:79–82. doi: 10.1023/a:1021717424268. [DOI] [PubMed] [Google Scholar]
- Atanassova A., Zamble D.B. Escherichia coli HypA is a zinc metalloprotein with a weak affinity for nickel. J Bacteriol. 2005;187:4689–4697. doi: 10.1128/JB.187.14.4689-4697.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axley M.J., Grahame D.A., Stadtman T.C. Escherichia coli formate‐hydrogen lyase. Purification and properties of the selenium‐dependent formate dehydrogenase component. J Biol Chem. 1990;265:18213–18218. [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M. Construction of Escherichia coli K‐12 in‐frame, single‐gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006–0008. doi: 10.1038/msb4100050. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagramyan K., Mnatsakanyan N., Poladian A., Vassilian A., Trchounian A. The roles of hydrogenases 3 and 4, and the F0F1‐ATPase, in H2 production by Escherichia coli at alkaline and acidic pH. FEBS Lett. 2002;516:172–178. doi: 10.1016/s0014-5793(02)02555-3. [DOI] [PubMed] [Google Scholar]
- Baron S.F., Ferry J.G. Reconstitution and properties of a coenzyme F420‐mediated formate hydrogenlyase system in Methanobacterium formicicum. J Bacteriol. 1989;171:3854–3859. doi: 10.1128/jb.171.7.3854-3859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E.L., Kwan H.S., Macy J. Anaerobiosis, formate, nitrate, and pyrA are involved in the regulation of formate hydrogenlyase in Salmonella typhimurium. J Bacteriol. 1984;158:972–977. doi: 10.1128/jb.158.3.972-977.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benemann J. Hydrogen biotechnology: progress and prospects. Nat Biotechnol. 1996;14:1101–1103. doi: 10.1038/nbt0996-1101. [DOI] [PubMed] [Google Scholar]
- Benemann J.R. Hydrogen production by microalgae. J Appl Phycol. 2000;12:291–300. [Google Scholar]
- Binnewies T.T., Motro Y., Hallin P.F., Lund O., Dunn D., La T. Ten years of bacterial genome sequencing: comparative‐genomics‐based discoveries. Funct Integr Genomics. 2006;6:165–185. doi: 10.1007/s10142-006-0027-2. et al. [DOI] [PubMed] [Google Scholar]
- Bisaillon A., Turcot J., Hallenbeck P.C. The effect of nutrient limitation on hydrogen production by batch cultures of Escherichia coli. Int J Hydrogen Energy. 2006;31:1504–1508. [Google Scholar]
- Blokesch M., Böck A. Properties of the [NiFe]‐hydrogenase maturation protein HypD. FEBS Lett. 2006;580:4065–4068. doi: 10.1016/j.febslet.2006.06.045. [DOI] [PubMed] [Google Scholar]
- Blokesch M., Magalon A., Böck A. Interplay between the specific chaperone‐like proteins HybG and HypC in maturation of hydrogenases 1, 2, and 3 from Escherichia coli. J Bacteriol. 2001;183:2817–2822. doi: 10.1128/JB.183.9.2817-2822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blokesch M., Paschos A., Theodoratou E., Bauer A., Hube M., Huth S., Böck A. Metal insertion into NiFe‐hydrogenases. Biochem Soc Trans. 2002;30:674–680. doi: 10.1042/bst0300674. [DOI] [PubMed] [Google Scholar]
- Boyington J.C., Gladyshev V.N., Khangulov S.V., Stadtman T.C., Sun P.D. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science. 1997;275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- Burgdorf T., De Lacey A.L., Friedrich B. Functional analysis by site‐directed mutagenesis of the NAD‐reducing hydrogenase from Ralstonia eutropha. J Bacteriol. 2002;184:6280–6288. doi: 10.1128/JB.184.22.6280-6288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgdorf T., Lenz O., Buhrke T., Van Der Linden E., Jones A.K., Albracht S.P.J., Friedrich B. NiFe]‐hydrogenases of Ralstonia eutropha H16: modular enzymes for oxygen‐tolerant biological hydrogen oxidation. J Mol Microbiol Biotechnol. 2005;10:181–196. doi: 10.1159/000091564. [DOI] [PubMed] [Google Scholar]
- Böhm R., Sauter M., Böck A. Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol Microbiol. 1990;4:231–243. doi: 10.1111/j.1365-2958.1990.tb00590.x. [DOI] [PubMed] [Google Scholar]
- Canada K.A., Iwashita S., Shim H., Wood T.K. Directed evolution of toluene ortho‐monooxygenase for enhanced 1‐naphthol synthesis and chlorinated ethene degradation. J Bacteriol. 2002;184:344–349. doi: 10.1128/JB.184.2.344-349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Sun Y., Xiu Z., Li X., Zhang D. Stoichiometric analysis of biological hydrogen production by fermentative bacteria. Int J Hydrogen Energy. 2006;31:539–549. [Google Scholar]
- Chin H.‐L., Chen Z.‐S., Chou C.P. Fedbatch operation using Clostridium acetobutylicum suspension culture as biocatalyst for enhancing hydrogen production. Biotechnol Prog. 2003;19:383–388. doi: 10.1021/bp0200604. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Pascal M.‐C., Casse F. Formate hydrogenlyase system in Salmonella typhimurium LT2. Eur J Biochem. 1977;72:149–155. doi: 10.1111/j.1432-1033.1977.tb11234.x. [DOI] [PubMed] [Google Scholar]
- Chittibabu G., Nath K., Das D. Feasibility studies on the fermentative hydrogen production by recombinant Escherichia coli BL‐21. Process Biochem. 2006;41:682–688. [Google Scholar]
- Clark D.P. The fermentation pathways of Escherichia coli. FEMS Microbiol Rev. 1989;63:223–234. doi: 10.1016/0168-6445(89)90033-8. [DOI] [PubMed] [Google Scholar]
- Das D., Veziroglu T.N. Hydrogen production by biological processes: a survey of literature. Int J Hydrogen Energy. 2001;26:13–28. [Google Scholar]
- Dutta D., De D., Chaudhuri S., Bhattacharya S.K. Hydrogen production by Cyanobacteria. Microb Cell Fact. 2005;4:36. doi: 10.1186/1475-2859-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J.S., Covert M., Palsson B. Metabolic modelling of microbes: the flux‐balance approach. Environ Microbiol. 2002;4:133–140. doi: 10.1046/j.1462-2920.2002.00282.x. [DOI] [PubMed] [Google Scholar]
- Forzi L., Sawers R.G. Maturation of [NiFe]‐hydrogenases in Escherichia coli. Biometals. 2007;20:565–578. doi: 10.1007/s10534-006-9048-5. [DOI] [PubMed] [Google Scholar]
- Frey M. Hydrogenases: hydrogen‐activating enzymes. Chembiochem. 2002;3:153–160. doi: 10.1002/1439-7633(20020301)3:2/3<153::AID-CBIC153>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Fritsche E., Paschos A., Beisel H.‐G., Böck A., Huber R. Crystal structure of the hydrogenase maturating endopeptidase HybD from Escherichia coli. J Mol Biol. 1999; 288:989–998. doi: 10.1006/jmbi.1999.2719. [DOI] [PubMed] [Google Scholar]
- Garcin E., Vernede X., Hatchikian E.C., Volbeda A., Frey M., Fontecilla‐Camps J.C. The crystal structure of a reduced [NiFeSe] hydrogenase provides an image of the activated catalytic center. Structure. 1999;7:557–566. doi: 10.1016/s0969-2126(99)80072-0. [DOI] [PubMed] [Google Scholar]
- Gasper R., Scrima A., Wittinghofer A. Structural insights into HypB, a GTP‐binding protein that regulates metal binding. J Biol Chem. 2006;281:27492–27502. doi: 10.1074/jbc.M600809200. [DOI] [PubMed] [Google Scholar]
- Guiral M., Tron P., Belle V., Aubert C., Léger C., Guigliarelli B., Giudici‐Orticoni M.‐T. Hyperthermostable and oxygen resistant hydrogenases from a hyperthermophilic bacterium Aquifex aeolicus: physicochemical properties. Int J Hydrogen Energy. 2006;31:1424–1431. [Google Scholar]
- Hallenbeck P.C. Fundamentals of the fermentative production of hydrogen. Water Sci Technol. 2005;52:21–29. [PubMed] [Google Scholar]
- Hallenbeck P.C., Benemann J.R. Biological hydrogen production; fundamentals and limiting processes. Int J Hydrogen Energy. 2002;27:1185–1193. [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes F.R., Dinsdale R., Hawkes D.L., Hussy I. Sustainable fermentative hydrogen production: challenges for process optimisation. Int J Hydrogen Energy. 2002;27:1339–1347. [Google Scholar]
- Hawkes F.R., Hussy I., Kyazze G., Dinsdale R., Hawkes D.L. Continuous dark fermentative hydrogen production by mesophilic microflora: principles and progress. Int J Hydrogen Energy. 2007;32:172–184. [Google Scholar]
- Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S. Highly accurate genome sequences of Escherichia coli K‐12 strains MG1655 and W3110. Mol Syst Biol. 2006:2006–0007. doi: 10.1038/msb4100049. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Yagi T., Yasuoka N. Unusual ligand structure in Ni‐Fe active center and an additional Mg site in hydrogenase revealed by high resolution X‐ray structure analysis. Structure. 1997;5:1671–1680. doi: 10.1016/s0969-2126(97)00313-4. [DOI] [PubMed] [Google Scholar]
- Higuchi Y., Ogata H., Miki K., Yasuoka N., Yagi T. Removal of the bridging ligand atom at the Ni‐Fe active site of [NiFe] hydrogenase upon reduction with H2, as revealed by X‐ray structure analysis at 1.4 Å resolution. Structure. 1999;7:549–556. doi: 10.1016/s0969-2126(99)80071-9. [DOI] [PubMed] [Google Scholar]
- Hopper S., Babst M., Schlensog V., Fischer H.‐M., Hennecke H., Böck A. Regulated expression in vitro of genes coding for formate hydrogenlyase components of Escherichia coli. J Biol Chem. 1994;269:19597–19604. [PubMed] [Google Scholar]
- Hube M., Blokesch M., Böck A. Network of hydrogenase maturation in Escherichia coli: role of accessory proteins HypA and HybF. J Bacteriol. 2002;184:3879–3885. doi: 10.1128/JB.184.14.3879-3885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jormakka M., Törnroth S., Byrne B., Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase‐N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- Jung G.Y., Jung H.O., Kim J.R., Ahn Y., Park S. Isolation and characterization of Rhodopseudomonas palustris P4 which utilizes CO with the production of H2. Biotechnol Lett. 1999;21:525–529. [Google Scholar]
- Kapdan I.K., Kargi F. Bio‐hydrogen production from waste materials. Enzyme Microb Tech. 2006;38:569–582. [Google Scholar]
- Karube I., Urano N., Yamada T., Hirochika H., Sakaguchi K. Cloning and expression of the hydrogenase gene from Clostridium butyricum in Escherichia coli. FEBS Lett. 1983;158:119–122. doi: 10.1016/0014-5793(83)80689-9. [DOI] [PubMed] [Google Scholar]
- Kim M.‐S., Baek J.‐S., Yun Y.‐S., Sim S.J., Park S., Kim S.‐C. Hydrogen production from Chlamydomonas reinhardtii biomass using a two‐step conversion process: anaerobic conversion and photosynthetic fermentation. Int J Hydrogen Energy. 2006;31:812–816. [Google Scholar]
- King P.W., Posewitz M.C., Ghirardi M.L., Seibert M. Functional studies of [FeFe] hydrogenase maturation in an Escherichia coli biosynthetic system. J Bacteriol. 2006;188:2163–2172. doi: 10.1128/JB.188.6.2163-2172.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbas M., Vogt S., Meyer‐Klaucke W., Bill E., Lyon E.J., Thauer R.K., Shima S. The iron‐sulfur cluster‐free hydrogenase (Hmd) is a metalloenzyme with a novel iron binding motif. J Biol Chem. 2006;281:30804–30813. doi: 10.1074/jbc.M605306200. [DOI] [PubMed] [Google Scholar]
- Kraemer J.T., Bagley D.M. Improving the yield from fermentative hydrogen production. Biotechnol Lett. 2007;29:685–695. doi: 10.1007/s10529-006-9299-9. [DOI] [PubMed] [Google Scholar]
- Kumar N., Das D. Enhancement of hydrogen production by Enterobacter cloacae IIT‐BT 08. Process Biochem. 2000;35:589–593. [Google Scholar]
- Kumar N., Ghosh A., Das D. Redirection of biochemical pathways for the enhancement of H2 production by Enterobacter cloacae. Biotechnol Lett. 2001;23:537–541. [Google Scholar]
- Lay J.‐J. Modeling and optimization of anaerobic digested sludge converting starch to hydrogen. Biotechnol Bioeng. 2000;68:270–278. doi: 10.1002/(sici)1097-0290(20000505)68:3<269::aid-bit5>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Leach M.R., Zamble D.B. Metallocenter assembly of the hydrogenase enzymes. Curr Opin Chem Biol. 2007;11:159–165. doi: 10.1016/j.cbpa.2007.01.011. [DOI] [PubMed] [Google Scholar]
- Leach M.R., Sandal S., Sun H., Zamble D.B. Metal binding activity of the Escherichia coli hydrogenase maturation factor HypB. Biochemistry. 2005;44:12229–12238. doi: 10.1021/bi050993j. [DOI] [PubMed] [Google Scholar]
- Levin D.B., Pitt L., Love M. Biohydrogen production: prospects and limitations to practical application. Int J Hydrogen Energy. 2004;29:173–185. [Google Scholar]
- Levin D.B., Islam R., Cicek N., Sparling R. Hydrogen production by Clostridium thermocellum 27405 from cellulosic biomass substrates. Int J Hydrogen Energy. 2006;31:1496–1503. [Google Scholar]
- Liu X., Zhu Y., Yang S.‐T. Construction and characterization of ack deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid and hydrogen production. Biotechnol Prog. 2006;22:1265–1275. doi: 10.1021/bp060082g. [DOI] [PubMed] [Google Scholar]
- McGlynn S.E., Ruebush S.S., Naumov A., Nagy L.E., Dubini A., King P.W. In vitro activation of [FeFe] hydrogenase: new insights into hydrogenase maturation. J Biol Inorg Chem. 2007;12:443–447. doi: 10.1007/s00775-007-0224-z. et al. [DOI] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Escherichia coli hydrogenase 3 is a reversible enzyme possessing hydrogen uptake and synthesis activities. Appl Microbiol Biotechnol. 2007a;76:1035–1042. doi: 10.1007/s00253-007-1086-6. [DOI] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Metabolic engineering to enhance bacterial hydrogen production. Microb Biotechnol. 2007b doi: 10.1111/j.1751-7915.2007.00003.x. , and [WWW document]. URL http://www.blackwell‐synergy.com/toc/mbt/0/0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Sanchez‐Torres V., Wood T.K. Enhanced hydrogen production from glucose by a metabolically‐engineered Escherichia coli. Appl Environ Microbiol. 2007c doi: 10.1007/s00253-007-1217-0. [DOI] [PubMed] [Google Scholar]
- Maeda T., Vardar G., Self W.T., Wood T.K. Inhibition of hydrogen uptake in Escherichia coli by expressing the hydrogenase from the cyanobacterium Synechocystis sp. PCC 6803. BMC Biotechnol. 2007d;7:25. doi: 10.1186/1472-6750-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias P.M., Saraiva L.M., Soares C.M., Coelho A.V., LeGall J., Carrondo M.A. Nine‐haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774: primary sequence determination, crystallographic refinement at 1.8 and modelling studies of its interaction with the tetrahaem cytochrome c3. J Biol Inorg Chem. 1999;4:478–494. doi: 10.1007/s007750050334. [DOI] [PubMed] [Google Scholar]
- Matsunaga T., Hatano T., Yamada A., Matsumoto M. Microaerobic hydrogen production by photosynthetic bacteria in a double‐phase photobioreactor. Biotechnol Bioeng. 2000;68:647–651. doi: 10.1002/(sici)1097-0290(20000620)68:6<647::aid-bit7>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Menon N.K., Robbins J., Wendt J.C., Shanmugam K.T., Przybyla A.E. Mutational analysis and characterization of the Escherichia coli hya operon, which encodes [NiFe] hydrogenase 1. J Bacteriol. 1991;173:4851–4861. doi: 10.1128/jb.173.15.4851-4861.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon N.K., Chatelus C.Y., Dervartanian M., Wendt J.C., Shanmugam K.T., Peck H.D., Jr, Przybyla A.E. Cloning, sequencing, and mutational analysis of the hyb operon encoding Escherichia coli hydrogenase 2. J Bacteriol. 1994;176:4416–4423. doi: 10.1128/jb.176.14.4416-4423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnan L., Jinli H., Xiaobin W., Huijuan X., Jinzao C., Chuannan L. Isolation and characterization of a high H2‐producing strain Klebsiella oxytoca HP1 from a hot spring. Res Microbiol. 2005;156:76–81. doi: 10.1016/j.resmic.2004.08.004. et al. [DOI] [PubMed] [Google Scholar]
- Mishra J., Khurana S., Kumar N., Ghosh A.K., Das D. Molecular cloning, characterization, and overexpression of a novel [Fe]‐hydrogenase isolated from a high rate of hydrogen producing Enterobacter cloacae IIT‐BT 08. Biochem Biophys Res Commun. 2004;324:679–685. doi: 10.1016/j.bbrc.2004.09.108. [DOI] [PubMed] [Google Scholar]
- Miyake J., Miyake M., Asada Y. Biotechnological hydrogen production: research for efficient light conversion. J Biotechnol. 1999;70:89–101. [Google Scholar]
- Montet Y., Amara P., Volbeda A., Vernede X., Hatchikian E.C., Field M.J. Gas access to the active site of Ni‐Fe hydrogenases probed by X‐ray crystallography and molecular dynamics. Nat Struct Biol. 1997;4:523–526. doi: 10.1038/nsb0797-523. et al. [DOI] [PubMed] [Google Scholar]
- Morimoto K., Kimura T., Sakka K., Ohmiya K. Overexpression of a hydrogenase gene in Clostridium paraputrificum to enhance hydrogen gas production. FEMS Microbiol Lett. 2005;246:229–234. doi: 10.1016/j.femsle.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Nagy L.E., Meuser J.E., Plummer S., Seibert M., Ghirardi M.L., King P.W. Application of gene‐shuffling for the rapid generation of novel [FeFe]‐hydrogenase libraries. Biotechnol Lett. 2007;29:421–430. doi: 10.1007/s10529-006-9254-9. et al. [DOI] [PubMed] [Google Scholar]
- Nandi R., Sengupta S. Microbial production of hydrogen: an overview. Crit Rev Microbiol. 1998;24:61–84. doi: 10.1080/10408419891294181. [DOI] [PubMed] [Google Scholar]
- Nandi R., Dey S., Sengupta S. Thiosulphate improves yield of hydrogen production from glucose by the immobilized formate hydrogenlyase system of Escherichia coli. Biotechnol Bioeng. 2001;75:492–494. doi: 10.1002/bit.10091. [DOI] [PubMed] [Google Scholar]
- Nath K., Das D. Improvement of fermentative hydrogen production: various approaches. Appl Microbiol Biotechnol. 2004;65:520–529. doi: 10.1007/s00253-004-1644-0. [DOI] [PubMed] [Google Scholar]
- Nath K., Kumar A., Das D. Hydrogen production by Rhodobacter sphaeroides strain O.U.001 using spent media of Enterobacter cloacae strain DM11. Appl Microbiol Biotechnol. 2005;68:533–541. doi: 10.1007/s00253-005-1887-4. [DOI] [PubMed] [Google Scholar]
- Nicolet Y., Piras C., Legrand P., Hatchikian C.E., Fontecilla‐Camps J.C. Desulfovibrio desulfuricans iron hydrogenase: the structure shows unusual coordination to an active site Fe binuclear center. Structure. 1999;7:13–23. doi: 10.1016/s0969-2126(99)80005-7. [DOI] [PubMed] [Google Scholar]
- Nicolet Y., Lemon B.J., Fontecilla‐Camps J.C., Peters J.W. A novel FeS cluster in Fe‐only hydrogenases. Trends Biochem Sci. 2000;25:138–143. doi: 10.1016/s0968-0004(99)01536-4. [DOI] [PubMed] [Google Scholar]
- Van Niel E.W.J., Budde M.A.W., De Haas G.G., Van Der Wal F.J., Claassen P.A.M., Stams A.J.M. Distinctive properties of high hydrogen producing extreme thermophiles, Caldicellulosiruptor saccharolyticus and Thermotoga elfii. Int J Hydrogen Energy. 2002;27:1391–1398. [Google Scholar]
- Oh Y.‐K., Seol E.‐H., Kim J.R., Park S. Fermentative biohydrogen production by a new chemoheterotrophic bacterium Citrobacter sp. Y19. Int J Hydrogen Energy. 2003;28:1353–1359. [Google Scholar]
- Peck H.D., Jr, Gest H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli‐aerogenes bacteria. J Bacteriol. 1957;73:706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold D.W., Macaskie L.E. Production of H2 from sucrose by Escherichia coli strains carrying the pUR400 plasmid, which encodes invertase activity. Biotechnol Lett. 2004;26:1879–1883. doi: 10.1007/s10529-004-6035-1. [DOI] [PubMed] [Google Scholar]
- Penfold D.W., Forster C.F., Macaskie L.E. Increased hydrogen production by Escherichia coli strain HD701 in comparison with the wild‐type parent strain MC4100. Enzyme Microb Tech. 2003;33:185–189. [Google Scholar]
- Penfold D.W., Sargent F., Macaskie L.E. Inactivation of the Escherichia coli K‐12 twin‐arginine translocation system promotes increased hydrogen production. FEMS Microbiol Lett. 2006;262:135–137. doi: 10.1111/j.1574-6968.2006.00333.x. [DOI] [PubMed] [Google Scholar]
- Peters J.W., Lanzilotta W.N., Lemon B.J., Seefeldt L.C. X‐ray crystal structure of the Fe‐only hydrogenase (CpI) from Clostridium pasteurianum to 1.8 Angstrom resolution. Science. 1998;282:1853–1858. doi: 10.1126/science.282.5395.1853. [DOI] [PubMed] [Google Scholar]
- Pilak O., Mamat B., Vogt S., Hagemeier C.H., Thauer R.K., Shima S. The crystal structure of the apoenzyme of the iron‐sulfur cluster‐free hydrogenase. J Mol Biol. 2006;358:798–809. doi: 10.1016/j.jmb.2006.02.035. et al. [DOI] [PubMed] [Google Scholar]
- Price N.D., Reed J.L., Palsson B.O. Genome‐scale models of microbial cells: evaluating the consequences of constraints. Nat Rev. Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- Raaijmakers H.C.A., Romão M.J. Formate‐reduced E. coli formate dehydrogenase H: the reinterpretation of the crystal structure suggests a new reaction mechanism. J Biol Inorg Chem. 2006;11:849–854. doi: 10.1007/s00775-006-0129-2. [DOI] [PubMed] [Google Scholar]
- Rachman M.A., Furutani Y., Nakashimada Y., Kakizono T., Nishio N. Enhanced hydrogen producton in altered mixed acid fermentation of glucose by Enterobacter aerogenes. J Ferm Bioeng. 1997;83:358–363. [Google Scholar]
- Rachman M.A., Nakashimada Y., Kakizono T., Nishio N. Hydrogen production with high yield and high evolution rate by self‐flocculated cells of Enterobacter aerogenes in a packed‐bed reactor. Appl Microbiol Biotechnol. 1998;49:450–454. [Google Scholar]
- Reed J.L., Palsson B.O. Thirteen years of building constraint‐based in silico models of Escherichia coli. J Bacteriol. 2003;185:2692–2699. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.L., Palsson B.O. Genome‐scale in silico models of E. coli have multiple equivalent phenotypic states: assessment of correlated reaction subsets that comprise network states. Genome Res. 2004;14:1797–1805. doi: 10.1101/gr.2546004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reissmann S., Hochleitner E., Wang H., Paschos A., Lottspeich F., Glass R.S., Böck A. Taming of a poison: biosynthesis of the NiFe‐hydrogenase cyanide ligands. Science. 2003;299:1067–1070. doi: 10.1126/science.1080972. [DOI] [PubMed] [Google Scholar]
- Rey F.E., Heiniger E.K., Harwood C.S. Redirection of metabolism for biological hydrogen production. Appl Environ Microbiol. 2007;73:1665–1671. doi: 10.1128/AEM.02565-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenfeld C.S., Schloss P.D., Handelsman J. Metagenomics: genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]
- Riley M., Abe T., Arnaud M.B., Berlyn M.K., Blattner F.R., Chaudhuri R.R. Escherichia coli K‐12: a cooperatively developed annotation snapshot‐2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosano C., Zuccotti S., Bucciantini M., Stefani M., Ramponi G., Bolognesi M. Crystal structure and anion binding in the prokaryotic hydrogenase maturation factor HypF acylphosphatase‐like domain. J Mol Biol. 2002;321:785–796. doi: 10.1016/s0022-2836(02)00713-1. [DOI] [PubMed] [Google Scholar]
- Rousset M., Montet Y., Guigliarelli B., Forget N., Asso M., Bertrand P. 3Fe‐4S] to [4Fe‐4S] cluster conversion in Desulfovibrio fructosovorans [NiFe] hydrogenase by site‐directed mutagenesis. Proc Natl Acad Sci USA. 1998;95:11625–11630. doi: 10.1073/pnas.95.20.11625. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter M., Böhm R., Böck A. Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol Microbiol. 1992;6:1523–1532. doi: 10.1111/j.1365-2958.1992.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Sawers G. The hydrogenases and formate dehydrogenases of Escherichia coli. Antonie Van Leeuwenhoek. 1994;66:57–88. doi: 10.1007/BF00871633. [DOI] [PubMed] [Google Scholar]
- Sawers R.G. Formate and its role in hydrogen production in Escherichia coli. Biochem Soc Trans. 2005;33:42–46. doi: 10.1042/BST0330042. [DOI] [PubMed] [Google Scholar]
- Schilling C.H., Edwards J.S., Letscher D., Palsson B.O. Combining pathway analysis with flux balance analysis for the comprehensive study of metabolic systems. Biotechnol Bioeng. 2000;71:286–306. [PubMed] [Google Scholar]
- Schilling C.H., Covert M.W., Famili I., Church G.M., Edwards J.S., Palsson B.O. Genome‐scale metabolic model of Helicobacter pylori 26695. J Bacteriol. 2002;184:4582–4593. doi: 10.1128/JB.184.16.4582-4593.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlensog V., Böck A. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol. 1990;4:1319–1327. doi: 10.1111/j.1365-2958.1990.tb00711.x. [DOI] [PubMed] [Google Scholar]
- Schön G., Voelskow H. Pyruvate fermentation in Rhodospirillum rubrum and after transfer from aerobic to anaerobic conditions in the dark. Arch Microbiol. 1976;107:87–92. doi: 10.1007/BF00427872. [DOI] [PubMed] [Google Scholar]
- Seibert M., Flynn T., Benson D., Tracy E., Gharardi M. Development of selection and screening procedures for rapid identification of H2‐producing algal mutants with increased O2 tolerance. In: Zaborsky O.R., editor. Plenum Publishing corporation; 1998. pp. 227–234. [Google Scholar]
- Self W.T., Shanmugam K.T. Isolation and characterization of mutated FhlA proteins which activate transcription of the hyc operon (formate hydrogenlyase) of Escherichia coli in the absence of molybdate. FEMS Microbiol Lett. 2000;184:47–52. doi: 10.1111/j.1574-6968.2000.tb08988.x. [DOI] [PubMed] [Google Scholar]
- Self W.T., Hasona A., Shanmugam K.T. N‐terminal truncations in the FhlA protein result in formate‐ and MoeA‐independent expression of the hyc (formate hydrogenlyase) operon of Escherichia coli. Microbiology. 2001;147:3093–3104. doi: 10.1099/00221287-147-11-3093. [DOI] [PubMed] [Google Scholar]
- Self W.T., Hasona A., Shanmugam K.T. Expression and regulation of a silent operon, hyf, coding for hydrogenase 4 isoenzyme in Escherichia coli. J Bacteriol. 2004;186:580–587. doi: 10.1128/JB.186.2.580-587.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J.‐H., Yoon J.H., Ahn E.K., Kim M.‐S., Sim S.J., Park T.H. Fermentative hydrogen production by the newly isolated Enterobacter asburiae SNU‐1. Int J Hydrogen Energy. 2007;32:192–199. [Google Scholar]
- Skibinski D.A.G., Golby P., Chang Y.‐S., Sargent F., Hoffman R., Harper R. Regulation of the hydrogenase‐4 operon of Escherichia coli by the σ54‐dependent transcriptional activators FhlA and HyfR. J Bacteriol. 2002;184:6642–6653. doi: 10.1128/JB.184.23.6642-6653.2002. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephanopoulos G.N., Aristidou A.A., Nielsen J. Academic Press; 1998. [Google Scholar]
- Stephenson M., Stickland L.H. Hydrogenase: a bacterial enzyme activating molecular hydrogen. Biochem J. 1931;25:205–214. doi: 10.1042/bj0250205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuber J., Krebs W., Bott M., Dimroth P. A membrane‐bound NAD(P)‐reducing hydrogenase provides reduced pyridine nucleotides during citrate fermentation by Klebsiella pneumoniae. J Bacteriol. 1999;181:241–245. doi: 10.1128/jb.181.1.241-245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamagnini P., Axelsson R., Lindberg P., Oxelfelt F., Wünschiers R., Lindblad P. Hydrogenases and hydrogen metabolism of cyanobacteria. Microbiol Mol Biol Rev. 2002;66:1–20. doi: 10.1128/MMBR.66.1.1-20.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer R.K., Klein A.R., Hartmann G.C. Reactions with molecular hydrogen in microorganisms: evidence for a purely organic hydrogenation catalyst. Chem Rev. 1996;96:3031–3042. doi: 10.1021/cr9500601. [DOI] [PubMed] [Google Scholar]
- Ust’ak S., Havrland B., Muñoz J.O.J., Fernández E.C., Lachman J. Experimental verification of various methods for biological hydrogen production. Int J Hydrogen Energy. 2007;32:1736–1741. [Google Scholar]
- Valentine D.L., Blanton D.C., Reeburgh W.S. Hydrogen production by methanogens under low‐hydrogen conditions. Arch Microbiol. 2000;174:415–421. doi: 10.1007/s002030000224. [DOI] [PubMed] [Google Scholar]
- Varma A., Palsson B.O. Metabolic flux balancing – basic concepts scientific and practical use. Bio-Technol. 1994;12:994–998. [Google Scholar]
- Vatsala T.M. Hydrogen production from (cane‐molasses) stillage by Citrobacter freundii and its use in improving methanogenesis. Int J Hydrogen Energy. 1992;17:923–927. [Google Scholar]
- Vignais P.M., Colbeau A. Molecular biology of microbial hydrogenases. Curr Issues Mol Biol. 2004;6:159–188. [PubMed] [Google Scholar]
- Vignais P.M., Dimon B., Zorin N.A., Colbeau A., Elsen S. HupUV proteins of Rhodobacter capsulatus can bind H2: evidence from the H–D exchange reaction. J Bacteriol. 1997;179:290–292. doi: 10.1128/jb.179.1.290-292.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignais P.M., Billoud B., Meyer J. Classification and phylogeny of hydrogenases. FEMS Microbiol Rev. 2001;25:455–501. doi: 10.1111/j.1574-6976.2001.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Vignais P.M., Magnin J.‐P., Willison J.C. Increasing biohydrogen production by metabolic engineering. Int J Hydrogen Energy. 2006;31:1478–1483. [Google Scholar]
- Voelskow H., Schön G. H2 production of Rhodospirillum rubrum during adaptation to anaerobic dark conditions. Arch Microbiol. 1980;125:245–249. doi: 10.1007/BF00964263. [DOI] [PubMed] [Google Scholar]
- Volbeda A., Fontecilla‐Camps J.C. The active site and catalytic mechanism of NiFe hydrogenases. Dalton Trans. 2003;21:4030–4038. [Google Scholar]
- Volbeda A., Charon M.‐H., Piras C., Hatchikian E.C., Frey M., Fontecilla‐Camps J.C. Crystal structure of the nickel‐iron hydrogenase from Desulfovibrio gigas. Nature. 1995;373:580–587. doi: 10.1038/373580a0. [DOI] [PubMed] [Google Scholar]
- Volbeda A., Garcin E., Piras C., De Lacey A.L., Fernandez V.M., Hatchikian E.C. Structure of the [NiFe] hydrogenase active site: evidence for biologically uncommon Fe ligands. J Am Chem Soc. 1996;118:12989–12996. et al. [Google Scholar]
- Watanabe S., Matsumi R., Arai T., Atomi H., Imanaka T., Miki K. Crystal structures of [NiFe] hydrogenase maturation proteins HypC, HypD, and HypE: insights into cyanation reaction by thiol redox signaling. Mol Cell. 2007;27:29–40. doi: 10.1016/j.molcel.2007.05.039. [DOI] [PubMed] [Google Scholar]
- Willenbrock H., Petersen A., Sekse C., Kiil K., Wasteson Y., Ussery D.W. Design of a seven‐genome Escherichia coli microarray for comparative genomic profiling. J Bacteriol. 2006;188:7713–7721. doi: 10.1128/JB.01043-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward J., Orr M., Cordray K., Greenbaum E. Enzymatic production of biohydrogen. Nature. 2000;405:1014–1015. doi: 10.1038/35016633. [DOI] [PubMed] [Google Scholar]
- Yang F., Hu W., Xu H., Li C., Xia B., Jin C. Solution structure and backbone dynamics of an endopeptidase HycI from Escherichia coli: implications for mechanism of the [NiFe] hydrogenase maturation. J Biol Chem. 2007;282:3856–3863. doi: 10.1074/jbc.M609263200. [DOI] [PubMed] [Google Scholar]
- Yi K.B., Harrison D.P. Low‐pressure sorption‐enhanced hydrogen production. Ind Eng Chem Res. 2005;44:1665–1669. [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. Enhanced hydrogen production from formic acid by formate hydrogen lyase‐overexpressing Escherichia coli strains. Appl Environ Microbiol. 2005;71:6762–6768. doi: 10.1128/AEM.71.11.6762-6768.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. Enhanced hydrogen production from glucose using ldh‐ and frd‐inactivated Escherichia coli strains. Appl Microbiol Biotechnol. 2006;73:67–72. doi: 10.1007/s00253-006-0456-9. [DOI] [PubMed] [Google Scholar]
- Yoshida A., Nishimura T., Kawaguchi H., Inui M., Yukawa H. Efficient induction of formate hydrogen lyase of aerobically grown Escherichia coli in a three‐step biohydrogen production process. Appl Microbiol Biotechnol. 2007;74:754–760. doi: 10.1007/s00253-006-0721-y. [DOI] [PubMed] [Google Scholar]
- Zhang J.W., Butland G., Greenblatt J.F., Emili A., Zamble D.B. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J Biol Chem. 2005;280:4360–4366. doi: 10.1074/jbc.M411799200. [DOI] [PubMed] [Google Scholar]
- Zilberman S., Stiefel E.I., Cohen M.H., Car R. Theoretical studies of [FeFe]‐hydrogenase: infrared fingerprints of the dithiol‐bridging ligand in the active site. Inorg Chem. 2007;46:1153–1161. doi: 10.1021/ic061432z. [DOI] [PubMed] [Google Scholar]
- Zirngibl C., Hedderich R., Thauer R.K. N5,N10‐Methylenetetrahydromethanopterin dehydrogenase from Methanobacterium thermoautotrophicum has hydrogenase activity. FEBS Lett. 1990;261:112–116. [Google Scholar]
- Zirngibl C., Van Dongen W., Schwörer B., Von Bünau R., Richter M., Klein A., Thauer R.K. H2‐forming methylenetetrahydromethanopterin dehydrogenase, a novel type of hydrogenase without iron‐sulfur clusters in methanogenic archaea. Eur J Biochem. 1992;208:511–520. doi: 10.1111/j.1432-1033.1992.tb17215.x. [DOI] [PubMed] [Google Scholar]


