Abstract
Gold nanorods absorb and scatter light strongly in the near-infrared portion of the electromagnetic spectrum, making them ideal tissue contrast agents for imaging techniques such as optical coherence tomography (OCT). Strong interactions occur at the nano-bio interface, such as proteins binding to gold nanorods forming a ‘corona.’ To fulfill the promise of nanorods for applications such as contrast agents, we must better understand the intrinsic interactions of these nanomaterials with biological systems at the molecular, cellular and tissue level. In this paper, we briefly review the nanorod-protein interface. We then present some new fast relaxation imaging (FReI) measurements of how the presence of strongly-absorbing gold nanorods affects protein binding and folding, taking into account inner filter effects and the strong quenching effect of nanorods on fluorescent-labeled proteins. Next we show that two-photon photoluminescence of the gold nanorods can be used to image the nanorods in tissue constructs, allowing us to independently study their tissue distribution so they can be used successfully as contrast agents in optical coherence microscopy.
Keywords: Fluorescence spectroscopy, nanoparticle, protein folding, tissue engineering, two photon microscopy, green fluorescent protein, optical coherence tomography, nanorod, temperature jump, fast relaxation imaging, kinetics
1. INTRODUCTION
The brilliant optical properties of metallic nanoparticles have been known for centuries [1]. However, only recently has interest in metallic nanotechnology at the molecular level gained momentum. The field has witnessed advances in synthetic routes to prepare different metallic nanoparticles with excellent control over size, shape, and surface chemistry [2–4]. Moreover, metallic nanoparticles have unique optical and electronic properties compared to their bulk material. The basis for the spectral properties of metallic nanoparticles, especially gold, is that they are smaller than the wavelength of light, allowing a coherent oscillation of electrons in the conduction band of the metal to occur in response to incoming electromagnetic radiation. This coherent oscillation gives rise to intense and tunable plasmon absorption and scattering through the visible into the near-infrared region of the electromagnetic spectrum, also known as localized surface plasmon resonance or LSPR (Fig. 1). LSPR depends on the shape, size, state of aggregation, and the local refractive index of the media surrounding the nanoparticles [3, 5]. With improved protocols for synthesis established, researchers are now exploring potential applications of metallic nanoparticles in biomedicine. This interest stems from the size of the nanoparticles (comparable to proteins and viruses) and from their biological applications in chemical sensing, drug delivery, biomedical imaging, and therapy [6, 7].
Fig. 1.
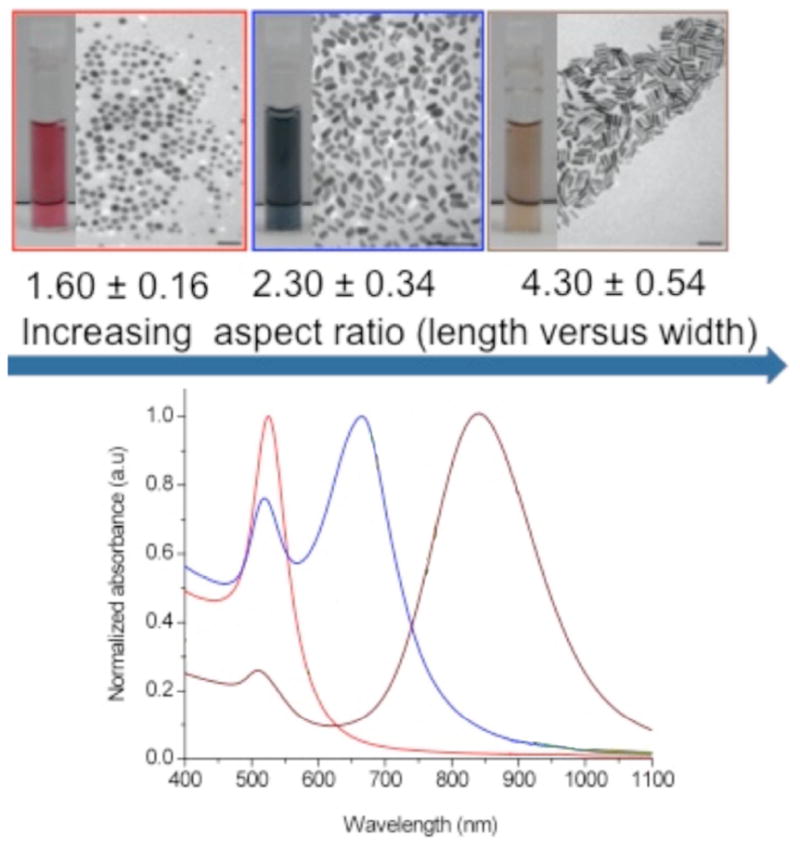
Transmission Electron Microscopy (TEM) of different aspect ratio gold nanorods, color-coded by shape, and their respective electronic absorbance spectra showing the red shift in the longitudinal plasmon band as the aspect ratio is increased.
Depending on their chemical coating nanoparticles in general, and specifically gold nanorods, can exert unanticipated effects on proteins, cells or even tissue via a variety of physicochemical mechanisms. For example, the anisotropy of gold nanorods has been used as a probe of rotational diffusion in membranes [5]. Metallic nanoparticle optical properties can modulate the optical properties of biomolecules, for example through fluorescence quenching of biological chromophores such as tryptophan [6], or fluorescence enhancement by rod-assisted absorption or far field coupling of the emission [7]. Here we review the interaction of gold nanorods with biological systems, and illustrate with new spectroscopic, kinetic and imaging data how one can probe the interaction of gold nanorods with biological systems and their effects on biological events both in vitro and in vivo.
2. THE PROTEIN CORONA AND THE NANO-BIO INTERFACE
The presence of gold nanorods in physiological systems does not go unnoticed by cells, or even organisms. Nano-particle surfaces have a high surface to volume ratio, so they can interact strongly with proteins, lipids, and other biomolecules once introduced into biological media [8]. The interactions can range from a non-specifically adsorbed protein “corona” in rapid exchange with the medium, to very specific recognition by cell surface receptors.
Previous studies have found that as soon as spherical nanoparticles are placed in biological media they are immediately covered by proteins, and the same phenomenon is expected for other shapes [9, 10]. The composition of the adsorbed protein layer is dynamic and undergoes change during the nanoparticles transport inside the biological host system. This is the result of the large number of proteins (about 3700 different proteins in plasma) present, each having a specific affinity for the nanoparticle surface [8]. Thus, we now perceive the proteins associated with the nanoparticle as a corona instead of a fixed layer. The composition of the corona is determined by the concentrations of the different proteins present and the rate at which they adsorb/desorb from a specific nanoparticle at any point in time. This corona is what cells “see” as they interact with gold nanorods [11]. It tends to make nanorod charge and other properties more uniform than the naked nanorod [12]. The biological corona is to be distinguished from very specific in vitro preparation, where a protein can be more permanently linked to a nanorod (e.g. via biotin-streptavidin, or maleimide thiol reaction).
When nanomaterials interact with receptors present on cell surfaces, recognition can lead to membrane binding or even cellular uptake. For example, antibody-coated gold nanorods of aspect ratio 3.9 have been used to preferentially bind cancer cells for photothermal therapy [13]. The proteins or peptides exposed from the corona, along with their conformational distribution following adsorption, dictate their interaction with receptors at the cell surface and therefore the biological impact of the nanomaterials. Nanoparticles, such as PAA-GNP between 5–20 nm in hydrodynamic diameters, bound with proteins can result in specific physiological and pathological changes in a biological system including macrophage uptake, blood coagulation, protein aggregation, and complement activation [14].
The corona is only one part of a more complex nano-bio interface. The nano-bio interface has been described by Nel et al. in terms of three dynamic interfaces: the nanoparticle surface itself, a solid-liquid interface (where the nanoparticle merges into the corona), and a corona-media interface (the outer contact zone of nanorod-bound proteins with unbound biological components) [15].
The nanoparticle surface itself affects the properties of the nano-bio interface via surface functionality, shape (curvature, roughness), and surface charge. It has been found that a difference in spherical particle size as small as 10 nm can lead to a significant difference in the amount of protein adsorption [16]. Gold nanoparticles receptor-mediated endocytosis rates vary widely due to the size-dependence of nanopartice-receptor interactions at the surface of the cells [13, 17–19, 23]. Others have shown that the surface functionalization of spherical polystyrene nanoparticles strongly affects protein binding [17], and we expect surface chemistry of gold nanoparticles to play an important role in protein binding independently of its shape, for example via electrostatics.
The second important component of the nano-bio interface is the solid-liquid interface. As the nanoparticle enters the liquid media its surface becomes solvated and the large surface area tends to adsorb small molecules and biomolecules in order to reduce its surface energy [18]. It is susceptible to events such as surface hydration and dehydration, surface reconstruction, and buildup due to aggregation, all driven by a complex combination of the forces discussed above. The stability of the solid-liquid interface determines whether a nanoparticle will be stable in the biological environment.
The final component of the nano-bio interface, the corona-media boundary, is the region where the adsorbed protein layer communicates with cellular surfaces and intracellular regions within tissues and organs. It is influenced by membrane interactions, receptor-binding interactions, membrane wrapping, and biomolecule adsorption equilibria. Understanding the corona-media boundary will lead to better control of the cytotoxicity and uptake of nanoparticles. It will also enable the creation of more robust surface-specific receptors for biosensing and controlling the interaction of nanoparticles with cell membranes and lipid bilayers for photothermal therapy, imaging, and delivery applications [19]. Tailoring the corona media boundary will be the key for more widespread applications of nanoparticles in biomedicine.
Many forces sculpt the nano-bio interface. Determining the energetic contributions of electrostatics (e.g. salt bridges), hydrogen-bonding, polarizability, hydrophobicity (driven by changes in solvent entropy) and steric factors (e.g. lone-pair electrons) to the adsorption process will give a more comprehensive understanding of the nano-bio interface [20]. Properly defining and distinguishing different energetic contributions to stability is already a challenge for bulk systems, and will be even more demanding for the nano-bio interface. Even finding the right analytical techniques to do it has been difficult, despite a large body of work by many researchers. For example, literature binding constants for gold nanoparticle (spheres and rods) with proteins range over 7 orders of magnitude from 104 to 1011 M−1 [6, 21, 22]. This difference might be caused by technical difference between analytical techniques being employed, by salt concentration and other solvent effects, or by the exact nature of the nanoparticle shape and coatings. Nevertheless, there are certain common observations between research groups. For example, it is now becoming accepted that smaller particles with higher surface of curvature lead to weaker protein adsorption [23, 24].
3. NEW NANOROD-PROTEIN SPECTROSCOPY AND IMAGING RESULTS
Here we discuss some recent developments from our laboratories based on two techniques that enable the sensing or imaging of gold nanorod interaction with proteins, cells, and tissues. Two-photon photoluminescence can provide high spatial resolution and low backgrounds. Förster resonance energy transfer (FRET) has been used at the single molecule level [25], and could potentially reach the ‘single rod level’ if signals from multiple adsorbed fluorophores is collected. There are many publications based on FRET involving the study of protein folding either on flat surfaces, or diffusing freely in solution [25, 26]. FRET has also been used to probe biomolecular folding and diffusion behavior in cells [27–29]. There are currently very few publications using FRET between nanoparticles and proteins to look at protein folding and function [30], and there are none that have studied FRET within a dual-labeled protein in response to gold nanorod binding. We probe such an interaction via fast relaxation imaging, which combines fluorescence microscopy and temperature jumps to look at the dynamics involved when a protein binds to gold nanorods and simultaneously folds/unfolds. Development of such new experimental tools will be required to map out energetics of biomolecules near gold nanoparticles surfaces. We end by discussing two-photon luminescence imaging of gold nanorods in tissues.
3A. Protein Binding and Stability in the Presence of Nanorods
Association of proteins with nanostructures is of great interest due to potential applications in drug delivery. Nanorods could even be used to control the function of adsorbed proteins. For example, many signaling proteins are partly unfolded and fold when they bind (so-called intrinsically disordered proteins or IDPs [31]); some enzymes undergo large conformational changes that can be used to control their activity [32]. Protein stability, conformation and folding may be different in the nanorod corona than in the bulk.
We studied binding and folding of the yeast enzyme phosphoglycerate kinase (PGK) to gold nanorods. PGK produces ATP in the glycolytic cycle. PGK was labeled at the termini with a (FRET) pair consisting of two fluorescent proteins, green fluorescent protein (AcGFP, green donor) and mCherry (red acceptor). PGK-FRET carries a net positive charge of +7 at neutral pH, the enzyme unfolds easily (~39 °C melting temperature of the PGK mutant), while the FRET probes remain folded up to 70 °C. When PGK is folded, the FRET labels are in close proximity and thus in a state of high FRET efficiency (more red emission). When PGK unfolds, its two termini fray freely in solution, increasing the end-to-end distance such that the AcGFP and mCherry labels are in a low FRET state (more green emission). Thus the two fluorescent protein labels serve as a local probe for PGK folding.
To monitor protein stability, we thermally denatured 40 nM PGK-FRET solutions in the presence of a varying concentration of nanorods (Fig. 2). The PGK-FRET construct was incubated with aspect ratio 4:1 gold nanorods coated with an anionic polymer (polyacrylic acid) to facilitate binding. 15 nanorod concentrations ranging from 0 to 1.5 nM were tested. We investigated nanorod concentrations where the number of proteins per nanorod was ≥ 30 because simple geometric considerations suggest that it would take roughly 30–50 protein molecules to form a monolayer on the surface of these particular nanorods. At each nanorod concentration, the temperature was titrated from 23 to 44 °C in 8 steps. Fluorescence spectra from 510 to 700 nm were recorded at each of the 120 temperature/nanorod concentration combinations. The collected FRET spectra were then analyzed at every temperature by using singular value decomposition (SVD) [33]. SVD decomposes the fluorescence data into three matrices: 1) linearly independent basis spectra as a function of wavelength (Fig. 2A,B); 2) trend vectors showing how each basis spectrum contributes as nanorod concentration is increased (Fig. 2C); 3) singular values that rank the contribution of each basis spectrum to the observed spectrum (w in Fig. 2B).
Fig. 2.
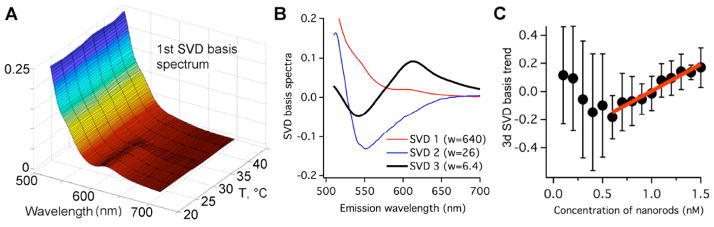
(A) 1st SVD basis spectrum as a function of temperature, showing that PGK unfolds (575–650 nm FRET peak disappears) as the temperature increases to 44 °C. (B) The 3 largest SVD basis spectra averaged over temperature and 3-point smoothed. #1 is the average FRET emission; 2 accounts for inner filter absorption as nanorod concentration is increased; 3 reveals FRET increase at higher nanorod concentration because acceptor fluorescence (575–650 nm) increases upon addition of nanorods, while donor fluorescence (<575 nm) decreases. (C) contribution of the 3d basis spectrum to the overall spectrum as a function of nanorod concentration; below 0.5 nanomolar, the spectral changes of the protein fluorescence cannot be determined reliably by SVD, hence the large uncertainty. Error bars at each nanorod concentration correspond to one standard deviation of the average of the 3d basis spectra at each temperature.
SVD was critical to deconvolute the large non-uniform absorption of gold nanorods from FRET, reducing the inner filter effect on the FRET signal from PGK. We found that the first SVD basis spectrum reflected the average FRET spectrum (Fig. 2A), the second basis function mainly reflected the decrease in AcGFP fluorescence due to the absorbance of green light by gold nanorods. The third SVD basis function represented an anti-correlated change of the AcGFP (<575 nm) vs. mCherry (575–650 nm) emission due to changing FRET. Increasing the nanorod concentration increased FRET (Fig. 2C). We attribute this to increased stability of PGK at the nanorod surface (binding and folding) and/or an increase of intermolecular FRET when PGK is confined to the surface of the nanorods (binding only). The third SVD eigenvalue w3 (Fig. 2B) had a similar magnitude relative to the first at all temperatures. This indicates that at least some of the FRET change is a result of confinement upon binding, otherwise w3/w1 would be largest near the denaturation midpoint at 38 °C, where binding should have the largest effect on folding.
3B. Protein Binding and Folding Kinetics in the Presence of Nanorods
Protein folding kinetics in the presence of nanorods in cells can be monitored via fluorescence based imaging techniques such as Fast Relaxation Imaging (FReI) [29]. In the FReI methodology, cells are transfected with a fluorescence labeled protein and then imaged via fluorescence microscopy. To probe the kinetics of binding and folding, which are temperature sensitive, a temperature jump is induced in the living cells by infrared laser heating. Fluorescence (e.g. FRET efficiency) is then imaged by a fast sensitive CCD camera, yielding a movie of folding inside the cell.
As a step towards live cell studies, we report kinetics of nanorods interacting with AcGFP-PGK. Unlike in section 3A, this version of PGK has only a single label. The green AcGFP fluorophore is quenched in the presence of nanorods, and thus sensitive to binding rather than folding. Even without a red acceptor label, folding still could have an indirect effect on the green signal if the binding affinity of folded and unfolded proteins to the gold nanorods differs. Although strong quenching by nanorods presents a challenge for measuring kinetics, our initial kinetic studies of protein-nanorod binding demonstrate the promise of the FReI system for studying protein folding in the presence of nanorods.
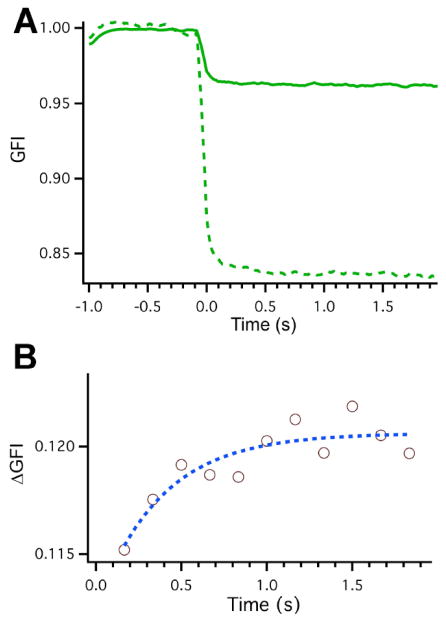
Fig. (3) shows temperature jump kinetics of the easily unfolded AcGFP-PGK mutant (39 °C melting point of PGK) in the presence of PAA-coated nanorods in aqueous solution. In these experiments, the 2000 nm heating laser was switched on at t=0 s, thus initiating a sudden temperature jump from 39 to 43 °C across the PGK unfolding transition. Although there was a loss of fluorescence intensity due to the uniform absorption of the gold nanorods, there is sufficient intensity to resolve protein-nanorod binding kinetics. Two kinetic phases are evident: one < 100 ms, and a slower ca. 300 ms phase.
Fig. 3.
In vitro temperature jumps on the FReI apparatus of 2.4 μM PGK-GFP and 2.4 μM PGK-GFP plus 10 nM PAA coated gold nanorods in solution. The initial temperature for the experiments was 39 °C. The temperature jump initiated by application of the IR heating laser at t=0 s with duration of 2 s. (A) Normalized, smoothed, and with bleaching baseline-subtracted green fluorescence intensity (GFI) of PGK-GFP control (dashed line) and PGK-GFP plus nanorods (solid line). (B) Normalized and smoothed fluorescence intensity difference (ΔGFI) between PGK-GFP plus nanorods and PGK-GFP control for points after the jump initiation. Intensity increase during the jump was fitted to y = Aexp(−t/τ) (dotted blue line on plot) with τ−=0.37±.21 s.
The fast phase is partly due to the lower quantum yield of the AcGFP probe at higher temperature, which has nothing to do with binding or folding. The protein-only control shows a large rapid decrease of fluorescence intensity right after the jump (dotted curve in Fig. 3A). Of note is the much smaller loss of AcGFP-PGK fluorescence intensity in the presence of nanorods (Fig. 3A). This result suggests that PGK-GFP dissociates rapidly from the nanorods after the temperature jump, reducing the nanorod quenching of the AcGFP fluorophore and thus offsetting the lower quantum yield at higher temperature.
Difference traces between AcGFP-PGK + nanorod and AcGFP-PGK only samples revealed an additional slower phase (Fig. 3B). In the presence of nanorods, AcGFP fluorescence increases in ~370 ms. This behavior could be due to proteins that dissociate more slowly from the nanorods and see a reduction in quenching effects as they unbind. For example, the following hypothesis is consistent with the data: In the bulk, FRET-PGK is known to unfold in ~1500 ms [28]. If greater laser heating near the gold nanorod surface produces a larger local temperature jump, protein in the corona would unfold more rapidly, and then dissociate from the nanorod leading to an increase in fluorescence. In that case, rather than measure fast dissociation from nanorods, the 370 ms time scale would indirectly measure protein unfolding by tracking the dissociation of proteins from nanorods as the proteins unfold. Further in vitro and in vivo experiments are needed to explore these trends, but these initial results suggest that protein interactions with nanorods and their folding in the presence of nanorods can be probed very effectively by FReI.
3C. Imaging of the Biochemical Environment of Nanorods in Tissue Constructs
Nonlinear interferometric vibrational imaging (NIVI) provides high-speed acquisition of Raman spectra, as compared to confocal Raman microscopy [34]. Therefore, NIVI is suitable for label-free molecular imaging of dynamic processes in biological environments, such as the interaction of gold nanoparticles and biomolecules in cells. NIVI utilizes coherent anti-Stokes Raman scattering (CARS) with spectral interferometry detection [35]. The experimental setup and the CARS energy diagram are shown in Fig. (4). In a CARS process, molecular vibrations are coherently generated by pump and Stokes fields and the anti-Stokes signal is created by a probe field. The coherence enhancement of CARS makes it appealing for vibrational imaging. However, the four-wave-mixing non-resonant background can stymie the retrieval of Raman spectra. By using spectral interferometry, both amplitude and phase of the anti-Stokes field are measured for retrieving background-free Raman spectra.
Fig. 4.
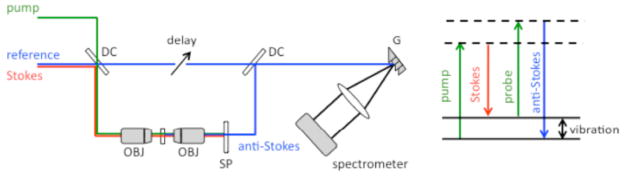
Schematic of NIVI setup (left) and CARS diagram (right). DC: dichroic mirror; OBJ: objective; SP: short-pass filter G: grating. Pump and Stokes fields generate the coherent vibration in sample molecules. Probe field scatters off the molecular vibration to create anti-Stokes field, which is then interfered with the reference beam for spectral interferometry detection.
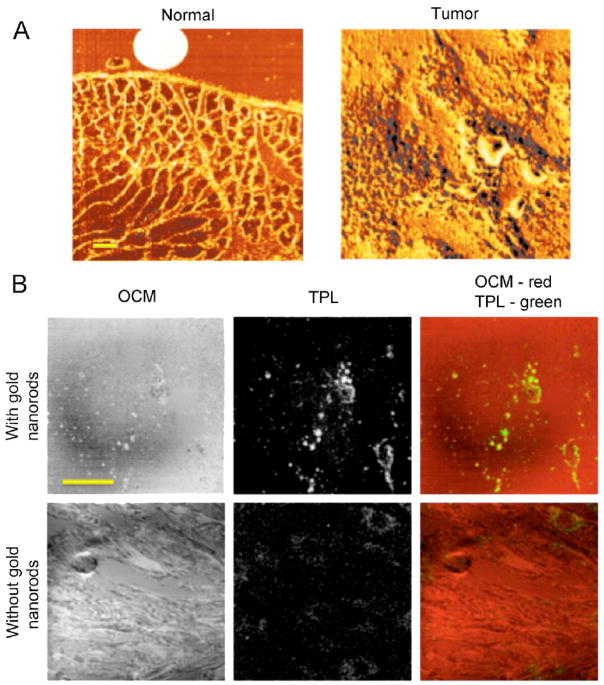
NIVI has been demonstrated to characterize chemical and biological samples and to identify molecular species within them [36–39]. NIVI images of a normal and a tumor mammary tissue sections are shown in Fig. (5A). The images can reflect the morphological changes while the spectral responses can reflect molecular composition. In a previous study, NIVI was used to differentiate normal and cancer tissues and to determine the tumor margin with greater than 99% confidence interval [36]. These studies demonstrate the potential of NIVI for real-time molecular imaging and clinical diagnosis.
Fig. 5.
(A) NIVI images of a normal and a tumor mammary tissue section (yellow scale bar: 100 μm). The morphological differences can be clearly distinguished, and the signal intensity is indicative of the lipid-protein molecular composition. (B) Optical coherence microscopy (OCM) and two-photon luminescence (TPL) images of HT-29 cells treated with and without gold nanorods (yellow scale bar: 50 μm). Both OCM and TPL can be used to visualize gold nanoparticles.
This high-speed microspectroscopic technique is ideal for investigating the dynamic processes of gold nanoparticles and the biomolecules in cells. The biomolecules adsorbed to gold nanoparticles can be revealed by studying the NIVI spectra, with the CARS signal further enhanced by the surface-enhanced CARS effect [40]. Along with other optical imaging techniques, like optical coherence microscopy (OCM) and two-photon luminescence (TPL) shown in Fig. (5B), one can visualize gold nanoparticles to monitor their interaction with biomolecules in real time [41, 42]. Understanding how gold nanoparticles interact with biomolecules and how cellular processes are altered are important to bring gold nanoparticles into biomedical applications.
4. CONCLUSIONS
In this paper we reviewed the nano-bio interface, and highlighted in particular new investigations of gold nanorods in biophysics and cell biology. The nano-bio interface is dynamic and it undergoes changes with time as it travels within a biological system and the environment evolves. Understanding the nature of the adsorbed layer of proteins at the surface of gold nanoparticles will help scientists predict the fate and biological effects of such particles in vivo.
However, this field needs new accurate techniques to probe the thermodynamics, kinetics, and structural changes occurring at the nano-bio interface. The different techniques being employed so far to probe the binding constant between nanoparticles and proteins have yielded a wide range of results, which might be inconsistent with each other. We have briefly described our attempts at obtaining such information using new spectroscopic techniques that allow more sensitive measurements both in vitro and in vivo. The greatest advantage of gold nanorods is also their greatest challenge: intense light absorption by gold nanoparticles makes possible local heating for therapeutic use, and enables sensitive detection, but it also complicates optical experiments. Using linear decomposition techniques such as SVD, represent a promising way to distinguish binding and folding effects from the gold nanorod optical absorption. We hope to expand these methods to in vivo measurements, and to use a global analysis of temperature and concentration trends to distinguish binding and folding. (Here, SVD analyzed the concentration trend separately at each temperature.)
We also highlighted the use of high-speed microspectroscopic techniques to look at the cell and tissue distribution of gold nanorods and the biomolecules they associate with, by combining the two-photon luminescence of gold nanorods with the Raman signal of these biomolecules. By using broadband laser sources and pulse shaping techniques [38], the NIVI spectra can be expanded to cover the molecular fingerprint region, and the Raman spectra retrieval can benefit from arbitrary pulse shapes. Spectra in fingerprint region are informative for identifying the biomolecules associated with gold nanoparticles. Also, the sensitivity can be improved with a cooled CCD camera, which can facilitate capturing signal from cellular samples. Therefore, this paper shows that further improvements in instrumentation will be critical to look at interaction of nanomaterials with biological media. More sensitive techniques specific to single protein-nanoparticle complex instead of the whole bulk will lead to more accurate data on the protein-nanoparticle interaction. The use of spectroscopic techniques such as those described in this paper can definitely provide an efficient tool to do so.
Acknowledgments
We dedicate this paper to Prof. Henrik Bohr, a pioneer in nano and quantum biology. This work was supported by funding from the Beckman Institute for Advanced Science and Technology at the University of Illinois. M.B.P. was a J. C. Bailar Fellow while part of this work was carried out. The authors would also like to acknowledge the contributions of Matthew Shulmerich and Prof. Rohit Bhargava for their help with the Raman spectroscopy imaging project. Additional support was provided by NIH R01 GM 093318 (M.G., M.B.P. and A.J.W.), NIH RC1 CA147096 (S.A.B.), and NSF 1011980 (C.J.M. and S.P.B.).
Footnotes
CONFLICT OF INTEREST
Declared none.
References
- 1.Edwards PP, Thomas JM. Gold in a Metallic divided state—from Faraday to present-day nanoscience. Angew Chem Int Ed. 2007;46:5480–5486. doi: 10.1002/anie.200700428. [DOI] [PubMed] [Google Scholar]
- 2.Perez-Juste J, Pastoriza-Santos I, Liz-Marzan L, Mulvaney P. Gold nanorods: synthesis, characterization and applications. Coordin Chem Rev. 2005;249:1870–1901. [Google Scholar]
- 3.Murphy CJ, Sau TK, Gole AM, Orendorff CJ, Gao J, Gou L, Hunyadi SE, Li T. Anisotropic metal nanoparticles: synthesis, assembly, and optical applications. J Phys Chem B. 2005;109:13857–13870. doi: 10.1021/jp0516846. [DOI] [PubMed] [Google Scholar]
- 4.Jana N, Gearheart L, Murphy C. Seed-mediated growth approach for shape-controlled synthesis of spheroidal and rod-like gold nanoparticles using a surfactant template. Adv Mater. 2001;13:1389–1393. [Google Scholar]
- 5.Gu Y, Sun W, Wang G, Fang N. Single particle orientation and rotation tracking discloses distinctive rotational dynamics of drug delivery vectors on live cell membranes. J Am Chem Soc. 2011;133:5720–5723. doi: 10.1021/ja200603x. [DOI] [PubMed] [Google Scholar]
- 6.Iosin M, Toderas F, Baldeck PL, Astilean S. Study of protein-gold nanoparticle conjugates by fluorescence and surface-enhanced Raman scattering. J Mol Struct. 2009:924–26. 196–200. [Google Scholar]
- 7.Bardhan R, Grady NK, Cole JR, Joshi A, Halas NJ. Fluorescence Enhancement by Au nanostructures: nanoshells and nanorods. ACS Nano. 2009;3:744–752. doi: 10.1021/nn900001q. [DOI] [PubMed] [Google Scholar]
- 8.Lynch I, Cedervall T, Lundqvist M, Cabaleiro-Lago C, Linse S, Dawson KA. The nanoparticle - protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv Colloid Interfac. 2007;134–35:167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 9.Lundqvist M, Stigler J, Cedervall T, Berggard T, Flanagan MB, Lynch I, Elia G, Dawson K. The evolution of the protein corona around nanoparticles: a test study. ACS Nano. 2011;5:7503–7509. doi: 10.1021/nn202458g. [DOI] [PubMed] [Google Scholar]
- 10.Lynch I, Dawson KA. Protein-nanoparticle interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- 11.Lynch I, Salvati A, Dawson KA. Protein-nanoparticle interactions: What does the cell see? Nat Nanotechnol. 2009;4:546–547. doi: 10.1038/nnano.2009.248. [DOI] [PubMed] [Google Scholar]
- 12.Alkilany A, Nagaria P, Hexel C, Shaw T, Murphy C, Wyatt M. Cellular uptake and cytotoxicity of gold nanorods: molecular origin of cytotoxicity and surface effects. Small. 2009;5:701–708. doi: 10.1002/smll.200801546. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, El-Sayed IH, Qian W, El-Sayed MA. Cancer cell imaging and photothermal therapy in the near-infrared region by using gold nanorods. J Am Chem Soc. 2006;128:2115–2120. doi: 10.1021/ja057254a. [DOI] [PubMed] [Google Scholar]
- 14.Deng ZJ, Liang M, Monteiro M, Toth I, Minchin RF. Nanoparticle-induced unfolding of fibrinogen promotes Mac-1 receptor activation and inflammation. Nat Nanotechnol. 2010;6:39–44. doi: 10.1038/nnano.2010.250. [DOI] [PubMed] [Google Scholar]
- 15.Nel AE, Maedler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 16.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev J, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C, Sirirattanapan J, Mann W, Treuel L, Zellner R, Maskos M, Schild H, Stauber RH. Nanoparticle Size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS Nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 17.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. P Natl A Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang K, Xing B. Adsorption of organic compounds by carbon nanomaterials in aqueous phase: Polanyi theory and its application. Chem Rev. 2010;110:5989–6008. doi: 10.1021/cr100059s. [DOI] [PubMed] [Google Scholar]
- 19.Saha K, Bajaj A, Duncan B, Rotello VM. Beauty is skin deep: a surface monolayer perspective on nanoparticle interactions with cells and bio-macromolecules. Small. 2011;7:1903–1918. doi: 10.1002/smll.201100478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia XR, Monteiro-Riviere NA, Riviere JE. An index for characterization of nanomaterials in biological systems. Nature Nanotechnol. 2010;5:671–675. doi: 10.1038/nnano.2010.164. [DOI] [PubMed] [Google Scholar]
- 21.Wangoo N, Suri CR, Shekhawat G. Interaction of gold nanoparticles with protein: A spectroscopic study to monitor protein conformational changes. Appl Phys Lett. 2008;92:133104. [Google Scholar]
- 22.Gomes I, Santos NC, Oliveira LMA, Quintas A, Eaton P, Pereira E, Franco R. Probing surface properties of cytochrome c at au bionanoconjugates. J Phys Chem C. 2008;112:16340–16347. [Google Scholar]
- 23.Li N, Zeng S, He L, Zhong W. Probing nanoparticle-protein interaction by capillary electrophoresis. Anal Chem. 2010;82:7460–7466. doi: 10.1021/ac101627p. [DOI] [PubMed] [Google Scholar]
- 24.Lacerda SHDP, Park JJ, Meuse C, Pristinski D, Becker ML, Karim A, Douglas JF. Interaction of gold nanoparticles with common human blood proteins. ACS Nano. 2010;4:365–379. doi: 10.1021/nn9011187. [DOI] [PubMed] [Google Scholar]
- 25.Deniz AA. Single-molecule protein folding: Diffusion fluorescence resonance energy transfer studies of the denaturation of chymotrypsin inhibitor 2. P Natl Acad Sci US A. 2000;97:5179–5184. doi: 10.1073/pnas.090104997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lipman EA. Single-molecule measurement of protein folding kinetics. Science. 2003;301:1233–1235. doi: 10.1126/science.1085399. [DOI] [PubMed] [Google Scholar]
- 27.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in fret-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen AW, Daugherty PS. Evolutionary optimization of fluorescent proteins for intracellular FRET. Nat Biotechnol. 2005;23:355–360. doi: 10.1038/nbt1066. [DOI] [PubMed] [Google Scholar]
- 29.Ebbinghaus S, Dhar A, McDonald J, Gruebele M. Protein folding stability and dynamics imaged in a living cell. Nat Methods. 2010;7:319–323. doi: 10.1038/nmeth.1435. [DOI] [PubMed] [Google Scholar]
- 30.Sarkar R, Narayanan SS, Pålsson LO, Dias F, Monkman A, Pal SK. Direct conjugation of semiconductor nanocrystals to a globular protein to study protein-folding intermediates. J Phys Chem B. 2007;111:12294–12298. doi: 10.1021/jp075239h. [DOI] [PubMed] [Google Scholar]
- 31.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 32.Dhar A, Samiotakis A, Ebbinghaus S, Nienhaus L, Homouz D, Gruebele M, Cheung MS. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. P Natl A Sci USA. 2010;107:17586–17591. doi: 10.1073/pnas.1006760107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henry E, Hofrichter J. Singular value decomposition: application to analysis of experimental data. Method Enzymol. 1992;210:129–192. [Google Scholar]
- 34.Marks DL, Boppart SA. Nonlinear interferometric vibrational imaging. Phys Rev Lett. 2004;92:123905. doi: 10.1103/PhysRevLett.92.123905. [DOI] [PubMed] [Google Scholar]
- 35.Lepetit L, Chériaux G, Joffre M. Linear techniques of phase measurement by femtosecond spectral interferometry for applications in spectroscopy. J Opt Soc Am B. 1995;12:2467. [Google Scholar]
- 36.Chowdary PD, Jiang Z, Chaney EJ, Benalcazar WA, Marks DL, Gruebele M, Boppart SA. Molecular histopathology by spectrally reconstructed nonlinear interferometric vibrational imaging. Cancer Res. 2010;70:9562–9569. doi: 10.1158/0008-5472.CAN-10-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benalcazar WA, Chowdary PD, Jiang Z, Marks DL, Chaney EJ, Gruebele M, Boppart SA. High-speed nonlinear interferometric vibrational imaging of biological tissue with comparison to Raman microscopy. IEEE J Select Topics Quantum Electron. 2010;16:824–832. doi: 10.1109/JSTQE.2009.2035537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benalcazar WA, Boppart SA. Nonlinear interferometric vibrational imaging for fast label-free visualization of molecular domains in skin. Anal Bioanal Chem. 2011;400:2817–2825. doi: 10.1007/s00216-011-4953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chowdary PD, Benalcazar WA, Jiang Z, Marks DM, Boppart SA, Gruebele M. High speed nonlinear interferometric vibrational analysis of lipids by spectral decomposition. Anal Chem. 2010;82:3812–3818. doi: 10.1021/ac100222c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ichimura T, Hayazawa N, Hashimoto M, Inouye Y, Kawata S. Local enhancement of coherent anti-Stokes Raman scattering by isolated gold nanoparticles. J Raman Spectrosc. 2003;34:651–654. [Google Scholar]
- 41.Graf BW, Jiang Z, Tu H, Boppart SA. Dual-spectrum laser source based on fiber continuum generation for integrated optical coherence and multiphoton microscopy. J Biomed Opt. 2009;14:034019. doi: 10.1117/1.3147422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H, Huff TB, Zweifel DA, He W, Low PS, wei A, Cheng JX. In vitro and in vivo two-photon luminescence imaging of single gold nanorods. P Natl Acad Sci US A. 2005;102:15752–15756. doi: 10.1073/pnas.0504892102. [DOI] [PMC free article] [PubMed] [Google Scholar]