Abstract
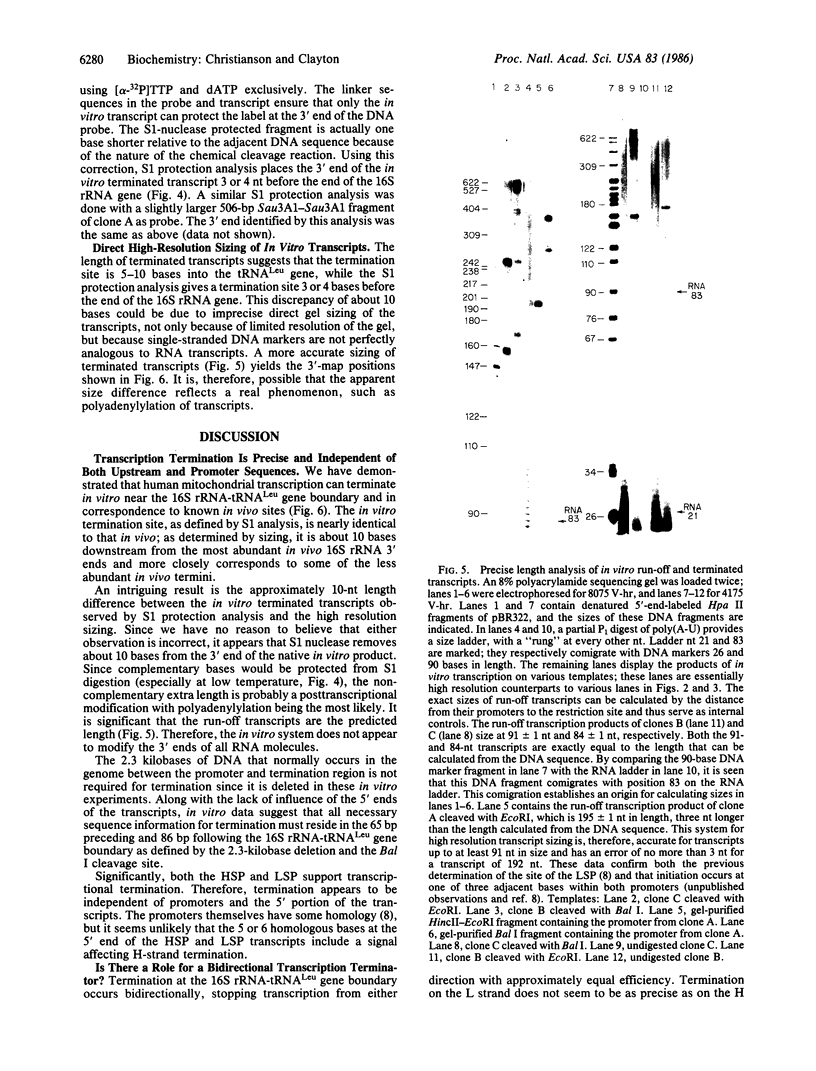
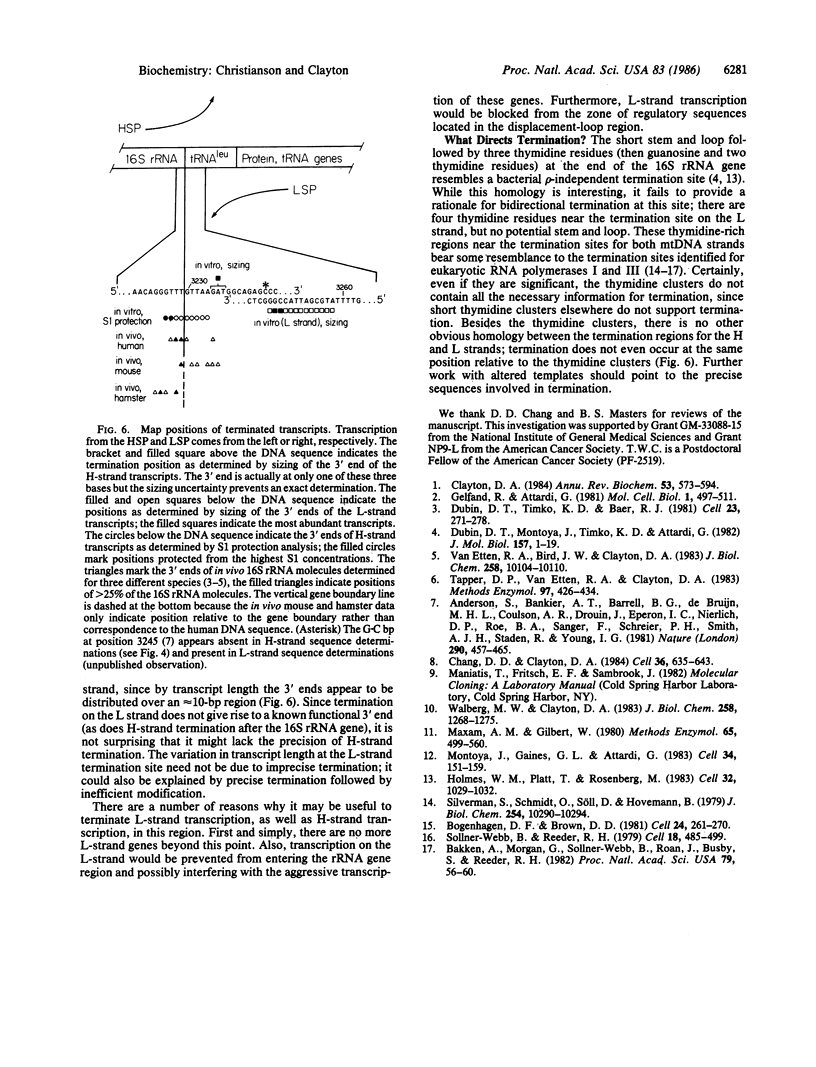
Mammalian mitochondrial genomes have a presumptive transcription termination site at the 16S rRNA-tRNALeu gene boundary. We have developed an in vitro system from human KB cells that terminates transcription at this gene boundary. By S1 nuclease protection, the 3' ends of terminated transcripts were mapped 3 and 4 base pairs upstream of the 16S rRNA-tRNALeu gene boundary, in agreement with in vivo data. By high-resolution sizing of transcripts, the 3' end was mapped 7 +/- 1 base pairs downstream from the gene boundary. Termination occurs with equal efficacy from transcriptional initiation at the heavy- or light-strand promoter. All template nucleotide sequence information necessary for termination appears to be located near the termination site itself. An unexpected observation is that the termination region functions bidirectionally.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bakken A., Morgan G., Sollner-Webb B., Roan J., Busby S., Reeder R. H. Mapping of transcription initiation and termination signals on Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1982 Jan;79(1):56–60. doi: 10.1073/pnas.79.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Brown D. D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981 Apr;24(1):261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- Chang D. D., Clayton D. A. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984 Mar;36(3):635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Clayton D. A. Transcription of the mammalian mitochondrial genome. Annu Rev Biochem. 1984;53:573–594. doi: 10.1146/annurev.bi.53.070184.003041. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Montoya J., Timko K. D., Attardi G. Sequence analysis and precise mapping of the 3' ends of HeLa cell mitochondrial ribosomal RNAs. J Mol Biol. 1982 May 5;157(1):1–19. doi: 10.1016/0022-2836(82)90510-1. [DOI] [PubMed] [Google Scholar]
- Dubin D. T., Timko K. D., Baer R. J. The 3' terminus of the large ribosomal subunit ("17S") RNA from hamster mitochondria is ragged and oligoadenylated. Cell. 1981 Jan;23(1):271–278. doi: 10.1016/0092-8674(81)90291-9. [DOI] [PubMed] [Google Scholar]
- Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981 Jun;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes W. M., Platt T., Rosenberg M. Termination of transcription in E. coli. Cell. 1983 Apr;32(4):1029–1032. doi: 10.1016/0092-8674(83)90287-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Montoya J., Gaines G. L., Attardi G. The pattern of transcription of the human mitochondrial rRNA genes reveals two overlapping transcription units. Cell. 1983 Aug;34(1):151–159. doi: 10.1016/0092-8674(83)90145-9. [DOI] [PubMed] [Google Scholar]
- Silverman S., Schmidt O., Söll D., Hovemann B. The nucleotide sequence of a cloned Drosophila arginine tRNA gene and its in vitro transcription in Xenopus germinal vesicle extracts. J Biol Chem. 1979 Oct 25;254(20):10290–10294. [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Tapper D. P., Van Etten R. A., Clayton D. A. Isolation of mammalian mitochondrial DNA and RNA and cloning of the mitochondrial genome. Methods Enzymol. 1983;97:426–434. doi: 10.1016/0076-6879(83)97153-7. [DOI] [PubMed] [Google Scholar]
- Van Etten R. A., Bird J. W., Clayton D. A. Identification of the 3'-ends of the two mouse mitochondrial ribosomal RNAs. The 3'-end of 16 S ribosomal RNA contains nucleotides encoded by the gene for transfer RNALeuUUR. J Biol Chem. 1983 Aug 25;258(16):10104–10110. [PubMed] [Google Scholar]
- Walberg M. W., Clayton D. A. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. J Biol Chem. 1983 Jan 25;258(2):1268–1275. [PubMed] [Google Scholar]