Abstract
Spindle orientation defines the plane of cell division and, thereby, the spatial position of all daughter cells. Here, we develop a live cell microscopy-based methodology to extract spindle movements in human epithelial cell lines and study how spindles are brought to a pre-defined orientation. We show that spindles undergo two distinct regimes of movements. Spindles are first actively rotated toward the cells’ long-axis and then maintained along this pre-defined axis. By quantifying spindle movements in cells depleted of LGN, we show that the first regime of rotational movements requires LGN that recruits cortical dynein. In contrast, the second regime of movements that maintains spindle orientation does not require LGN, but is sensitive to 2ME2 that suppresses microtubule dynamics. Our study sheds first insight into spatially defined spindle movement regimes in human cells, and supports the presence of LGN and dynein independent cortical anchors for astral microtubules.
Keywords: mitosis, spindle orientation, microtubule
Introduction
At the cortex of polarized cells, the PAR3 module (PAR3-PAR6-aPKC) localizes asymmetrically. This polarized module recruits macromolecular machines (Gαi-LGN-NuMA-dynein/dynactin) that pull at astral microtubules and orient the mitotic spindle (reviewed in refs. 1 and 2). In non-polarized mouse NRK cells3 and amphibian and fish embryos,4 cell shape cues can orient the spindle along the long-axis of the cell in a dynein-dependent manner. However, the temporal evolution of spindle movements in cultured human cells is not fully understood, partly due to the apparent cell-to-cell variability in the nature of spindle movements.
The importance of interphase cell shape-dependent changes in spindle orientation is best observed in precancerous lesions of intestines5 and the epidermis of a developing embryo,6 highlighting the importance of cell shape-associated spindle orientation in tissue organization.
Cell shape is intertwined with cell–cell and cell–substrate adhesion. In cells that lack neighbor cell contact, the memory of interphase cell shape is carried through mitosis in the form of cell–substrate adhesion that determines spindle orientation.7,8 This form of “mitotic cell polarity” is dependent on subcortical actin.9,10 The molecular details of how cell–cell and cell–substrate adhesion may jointly influence the dynamic movements of the spindle are poorly understood.
In human cells, spindle positioning (parallel to the substratum) and orientation (along a predefined axis in XY plane) require a diverse and somewhat non-overlapping set of proteins. While spindle positioning requires the microtubule-associated proteins EB1, APC, MAP4, CHICA, and HMMR, the motors Dynein and Myosin-X, the kinases PAK2, PI(3)K, LIMK1, and Abl1, and intracellular signaling regulators β1-integrin and Cdc42 GTPase, spindle orientation along a predefined axis requires Dynein, LGN, the centrosomal proteins, CPAP and STIL, and CLASP1.3,11-20 To elucidate how spindle positioning and orientation mechanisms may communicate with each other, we require a framework to systematically extract spindle movements in cells that maintain neighbor cell interactions.
Here, we use monolayer cultures of human cell lines for developing a methodology to study interphase cell shape-associated spindle orientation in cells that retain neighbor cell interactions. We developed an automated spindle pole tracking software, Spindle3D, and using this software, we studied the temporal evolution of spindle pole movements in individual cells of a population and extracted statistically significant patterns of spindle movements with respect to interphase cell shape. We find that the spindle-axis is initially directed toward the cell’s long-axis and then maintained close to the long-axis, revealing 2 spatially distinct regimes of spindle movements in non-polarized cells. By combining RNAi-mediated protein depletion and live cell imaging, we show that spindle rotation toward the long-axis is an early mitotic event that is reliant on LGN. However, mechanisms that maintain spindle alignment close to the long-axis are independent of LGN, suggesting the presence of LGN-independent mechanisms for orientation maintenance. Orientation maintenance is selectively sensitive to suppression of microtubule dynamics, but not increase in microtubule length. We propose that mitotic spindle movements are subjected to spatially distinct molecular mechanisms that first mediate spindle rotation toward the long-axis and then facilitate orientation maintenance along the long-axis.
Results
HeLa and RPE1 cells orient their spindle along the interphase long-axis
To study interphase cell shape-associated spindle orientation in cells that retained cell–cell contact, we studied human epithelial cells grown on a glass surface. Using time-lapse microscopy, we followed chromosome movements in over a hundred HeLa and RPE1 cells expressing the chromosome markers His2B-GFP (HeLaHis2B-GFP) and His2B-RFP (RPE1His2B-RFP), respectively, by imaging once every 4 min for a period of 5 h. In this assay, a line orthogonal to the segregating chromatid sets was used to approximate the final spindle-axis (Fig. 1A). The long-axis of the cell was determined 12 min before mitosis onset (Nuclear Envelope BreakDown [NEBD]). The angle between the final spindle-axis at metaphase–anaphase transition and the interphase long-axis was used to register the final orientation of the spindle (Fig. 1A). We binned the final spindle orientation angles into three sets, with 0–30 degrees corresponding to the least deviation of spindle-axis from the cell’s long-axis. This population analysis revealed that around 90% of cells aligned within 30 degrees, demonstrating a clear bias in spindle orientation in HeLa and RPE1 cells (Fig. 1B and C). An unbiased random orientation mechanism is expected to yield not more than 33% of cells in each of the 30 degrees spindle-axis bins. We conclude that the final orientation of the mitotic spindle is strongly correlated with the interphase cell’s long-axis in both HeLa and RPE1 cells grown on glass substrates, as shown for NRK cells.3 Although this system does not disentangle the effect of cell shape and cell–cell or cell–substrate adhesion on spindle orientation, it allows us a means to predict spindle orientation in human epithelial cell cultures that retained neighbor cell interactions.
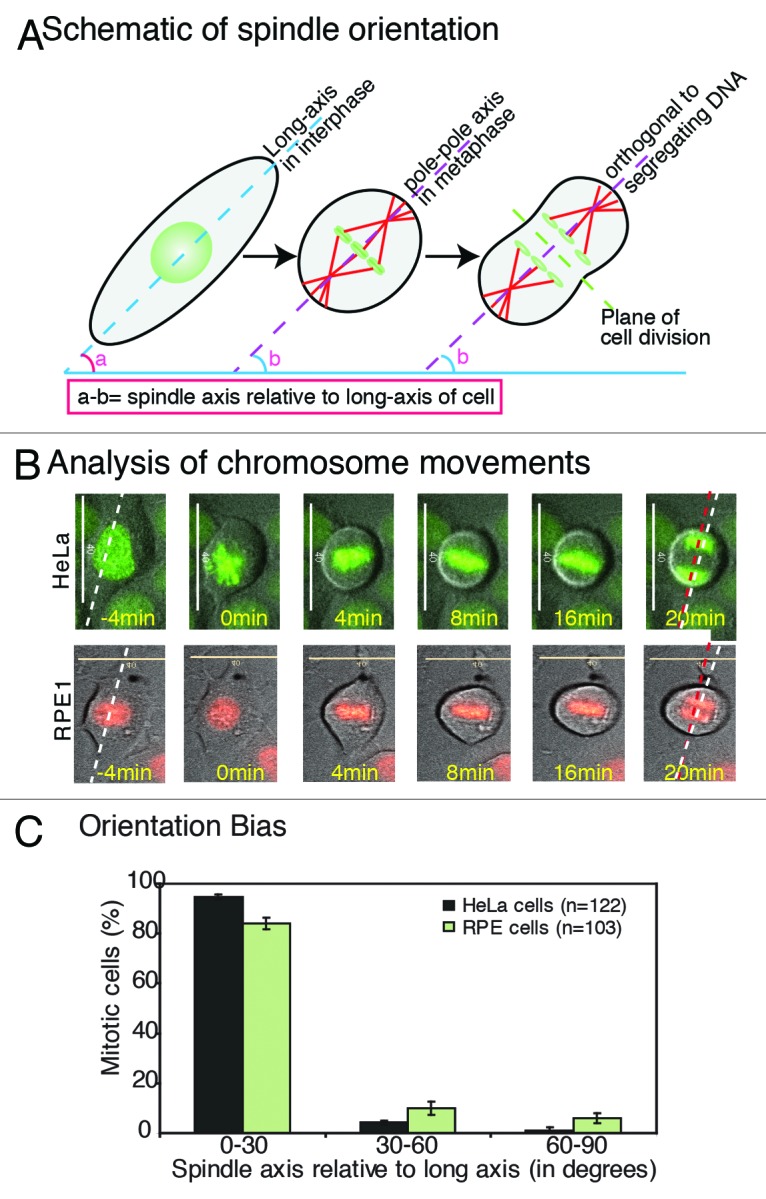
Figure 1. Epithelial cells cultured on glass orient their spindle along the interphase long-axis (A) Schematic describing cell shape-associated spindle orientation. The interphase long-axis (angle a; dashed blue line), metaphase spindle-axis (angle b; dashed purple line) and plane of cell division (dashed green line) orthogonal to spindle-axis are highlighted. (B and C) Spindle-axis at metaphase–anaphase transition shows biased alignment along the long-axis of the interphase cell. (B) Time-lapse analysis of chromosome movements to extract long-axis of the cell (dashed white line), spindle-axis (dashed red line) in HeLaHis2B-GFP and RPE1His2B-RFP cells. Representative frames of time-lapse movies are shown as a merge of DIC (cell shape) and either GFP/RFP (chromosome alignment) (C) Frequency distribution of final orientation angles (spindle-axis relative to long-axis) in HeLaHis2B-GFP and RPE1His2B-RFP. Error bars represent SEM from 3 independent experiments. Scale bar = 40 μm
Mitotic spindles align along the cell’s long-axis during prometaphase
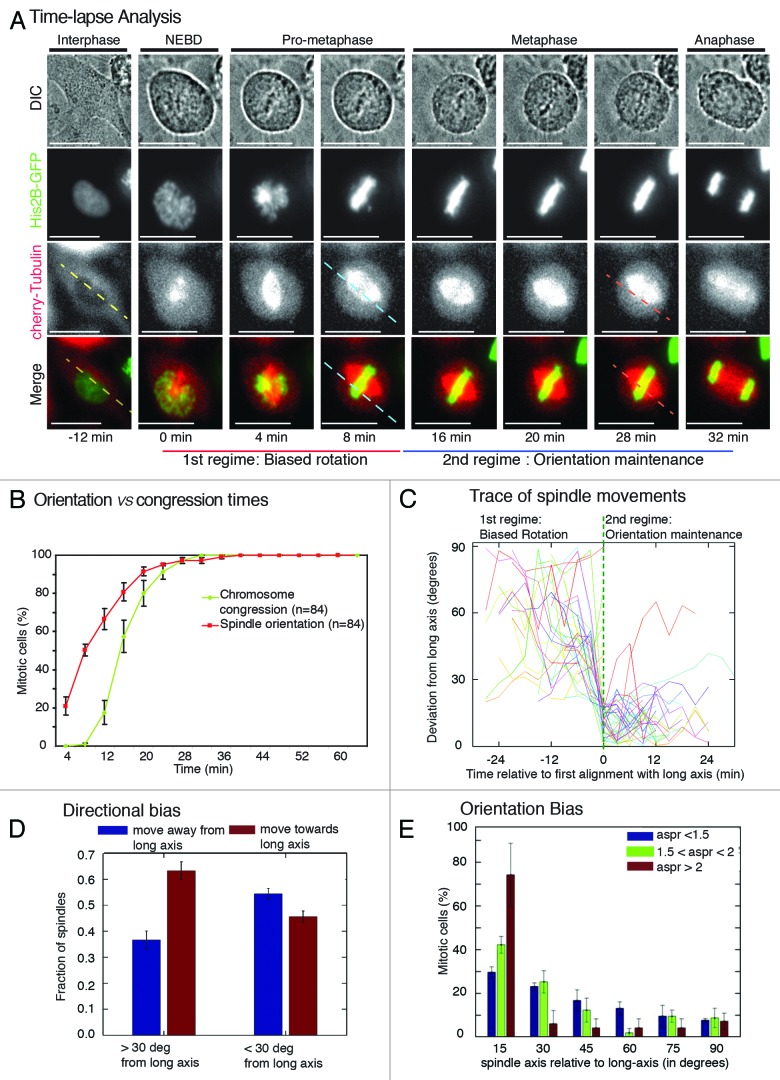
In frog and fish embryos,4 spindle orientation is achieved by pre-positioning of the centrosomes prior to mitosis, instead of active spindle rotation during mitosis.21,22 In non-polarized cells, centrosomes are known to separate both before and after nuclear envelope breakdown (reviewed in ref. 23); however, the extent to which prepositioning of centrosomes biases spindle orientation in HeLa cells is not known. To address this, we studied the kinetics of spindle orientation relative to chromosome congression. We generated a HeLa cell line expressing markers for both chromosomes (His2B-GFP) and microtubules (mCherry-Tub), HeLaHis2B-GFP;mCherry-Tub, to mark mitotic phases and define spindle-axis (pole to pole axis), respectively. We first confirmed that the introduction of mCherry-Tub did not perturb mitosis progression or spindle assembly, as no discernible difference in mitotic timing, bipolarity extent or segregation accuracy was observed between HeLaHis2B-GFP; mCherry-Tub and HeLaHis2B-GFP cell lines (Fig. S1). Time-lapse movies of HeLaHis2B-GFP;mCherry-Tub cells showed that in early prophase the spindles first formed at random orientations with respect to long-axis, and by metaphase, the spindles had completed orienting their pole-to-pole axis along the long-axis of the cell (Fig. 2A; Movies 1 and 2). We compared the time consumed to orient spindle along long-axis (spindle orientation time) and to congress chromosomes onto the metaphase plate (chromosome congression time). Four minutes after NEBD, more than 80% of cells had their spindle-axis oriented away from the long-axis, and nearly all cells displayed uncongressed chromosomes, confirming incorrect orientation and incomplete congression in early prophase (Fig. 2B). However, 12 min after NEBD, nearly 70% of cells had completed orienting their spindle-axis along the long-axis, but only 20% of cells had completed chromosome congression, indicating correct orientation and incomplete congression in prometaphase. The kinetics of orientation and congression showed two important findings: first, the completion of chromosome congression is not a prerequisite for spindle orientation along the long-axis. Second, final spindle orientation is not achieved by pre-positioning of centrosomes along the long-axis in the vast majority of cells, highlighting the usefulness of the system to study spindle movements that ensure the proper orientation of the spindle.
Figure 2. Two spatially distinct regimes of spindle movements facilitate the process of cell shape-dependent spindle orientation (A) Time-lapse images of a HeLaHis2B-GFP; mCherry-Tub cell showing the positions of spindle and chromosomes from nuclear envelope breakdown (NEBD) through anaphase. Dashed lines are drawn parallel to the interphase long-axis (yellow), first alignment along long-axis (blue), and final spindle orientation axis (pink). Scale bar = 15 μm. (B) Cumulative frequency distributions of the first time point when spindle-axis aligns with long-axis and the time point of completion of chromosome congression. Only cells that properly oriented along long-axis were considered for the analysis. (C) Trace of spindle-axis through time of a randomly chosen set of 20 cells. The vertical axis measures the angle between the spindle and the cell’s long-axis (0 corresponds to perfect alignment), each curve has been horizontally shifted, so that the time of first alignment (< 20 degrees) is at time 0. Dashed green vertical line indicates spindles’ first alignment with long-axis. (D) Bar graph showing direction bias for spindles rotating away from and toward the long-axis. Spindles were segregated into 2 sets according to their position (> or < 30 degrees from long-axis). (E) Frequency distribution of final orientation angles (spindle-axis relative to long-axis) in HeLaHis2B-GFP; mCherry-Tub cells that exhibited high (>2), medium (between 1.5 and 2) or low aspect ratio (<1.5) in interphase, 20 min prior to NEBD. Aspect ratio (aspr) is defined as major axis:minor axis of ellipse fitted to interphase cell shape, 20 min prior to NEBD. Error bars represent SEM from 3 independent experiments.
Spindle movements are directed toward the long-axis and are subsequently maintained proximal to the long-axis
Examining the dynamics of spindle movements has allowed the dissection of orientation mechanisms in model systems.4,24-26 To study the dynamics of spindle movements in cell cultures, we developed Spindle3D software (Fig. S2A), which automatically identifies spindle pole positions and quantifies the displacement of the spindle poles in time-lapse images. In this automated image analysis approach, the long-axis of the cell was determined by fitting an ellipsoid to the shape of the interphase cell 20 min prior to NEBD. We first confirmed that the final orientation angles were comparable in both Spindle3D automated analysis and manual analysis, in 2 different experiments (Fig. S2B). In both Spindle3D and manual analyses, final spindle orientation bias was slightly reduced in HeLaHis2B-GFP; mCherry-Tub cell line compared to HeLaHis2B-GFP cell line (Fig. S2B; Fig. 1C), presumably owing to increased precision in identifying spindle pole positions. Nevertheless, a prominent bias in orienting the spindle along long-axis was observed in HeLaHis2B-GFP; mCherry-Tub cell populations, highlighting the combined benefit of the spindle reporter cell line and automated Spindle3D analysis.
Because population averages might obscure important dynamic characteristics of spindle movements that are unsynchronized between cells, we included the analysis of spindle movements in individual cells. To our knowledge, human spindle movements have not been analyzed at this temporal and numerical resolution so far. Analyzing spindle movements in relation to long-axis revealed a biphasic trend in movement before and after the spindle’s first alignment with the long-axis (Fig. 2C). Prior to first alignment of spindle-axis with long-axis, the spindle-axis underwent directed movement toward the long-axis. After the first alignment, spindle-axis remained within 30 degrees of the long-axis, suggesting a mechanism that prevents the spindles from moving away from the long-axis. We conclude that two distinct regimes of spindle movements exist: (1) a directed movement that rotates the spindle-axis toward the long-axis and (2) a restrained movement that maintains the spindle position within 30 degrees of the long-axis.
We next studied dynamic switching in the direction of spindle movements during the period when spindle-axis was either within or outside of 30 degrees of long-axis. For this, we quantified the occurrence of 2 possible directions of spindle movement: spindles moving toward or away from the long-axis. When the angle between the spindle-axis and the long-axis was greater than 30 degrees, movement toward the long-axis was at least 1.5-fold more frequent than movement away from the long-axis. We refer to this 1.5-fold bias as “directional bias”. No such directional bias was observed in spindles that were aligned within 30 degrees of the long-axis (Fig. 2D). We conclude that the directional bias is specific to spindles oriented away from the long-axis.
The speed of spindle rotation was reduced one-fourth in the second regime compared with the first regime spindle rotation speed in degrees/frame: pre-align 13.1+/−0.7°; post-align 9.9+/−0.5° (n = 123 cells). Although speed values are susceptible to frame rates, this result, together with directional bias differences, show the existence of spatially distinguishable regimes of mitotic spindle movements.
Accuracy of spindle orientation is dependent on the aspect ratio of the cell
While the majority of HeLaHis2B-GFP;mCherry-Tub cells aligned the spindle-axis within 30 degrees of the long-axis (Fig. S2B), a minority of cells failed to align the spindles properly. We hypothesized that there could be a specific “shape threshold” or threshold aspect ratio that is required to specify a sufficiently prominent longest axis for successful detection by the spindle orientation machinery. To quantitatively test this hypothesis, we binned cells with a high (>2), medium (between 1.5 and 2) or low (<1.5) aspect ratio by fitting cells to nearest ellipsoids and measuring the ratios of major and minor axes lengths. To carefully analyze orientation bias, we reduced spindle-axis bin size to 15 degrees. An increased bias in orienting spindles along long-axis was observed in cells with a high aspect ratio compared with cells with a medium aspect ratio (Fig. 2E). In cells with low aspect ratio, the bias in orienting spindles was noticeably reduced. Previous studies in RPE1 cells showed that an aspect ratio of 1.2 can bias the mitotic spindle toward long-axis.10 Thus, we conclude that aspect ratio influences orientation success along long-axis. Because aspect ratio is closely linked to cell–cell and cell–substrate interaction, these findings support the previously demonstrated role of cell–substrate interaction in orientation8 and suggest a possible role for regulators of cell–cell interaction in spindle orientation and movements.
LGN is required for spindle rotation toward the interphase long-axis
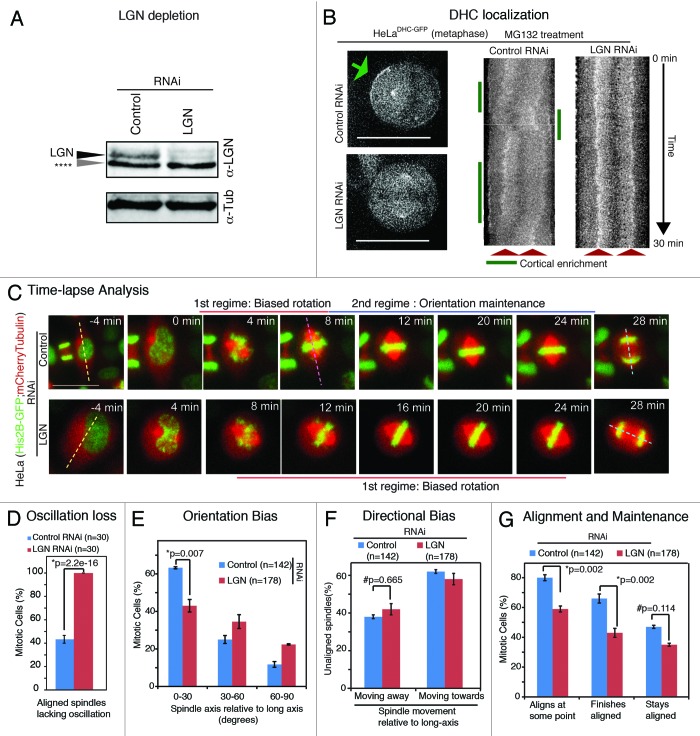
Cells grown on ECM substrate showed LGN’s role in orienting the spindle along a pre-defined axis; LGN is required for cortical localization of Dynein heavy chain (DHC) and spindle oscillations along pole-to-pole axis.19 We tested if LGN is required for spindle orientation in cells grown on glass substrates, and, if so, whether LGN is required for the 2 regimes of spindle movements. We used previously described LGN siRNA oligos19 and first confirmed siRNA-mediated depletion of LGN using immunoblotting (Fig. 3A). We next confirmed functional loss of LGN by studying spindle oscillations and the status of cortical DHC-GFP in HeLa cells expressing DHC-GFP (HeLaDHC-GFP). For this study, siRNA-treated and G2-M-synchronized cells were arrested in metaphase using MG132 treatment and then imaged once every minute to follow DHC-GFP signals at spindle poles and cell cortex. As reported,19 unlike control-depleted cells, LGN-depleted cells lacked cortical dynein, and spindles remained in the geometric center of the cell with no oscillation along the pole-to-pole axis (Fig. 3B), confirming the functional loss of LGN.
Figure 3. LGN controls spindle rotation speed but not directional bias in rotation (A) Immunoblot of cell lysates treated with LGN RNAi or control RNAi as indicated were probed with antibodies against LGN and tubulin. Grey arrow (****) refers to nonspecific band. (B) Single-plane live cell images (left) and their kymographs (right) of HeLaDHC-GFP cells treated with LGN or control siRNA as indicated. DHC-GFP at cell cortex marked with green arrow. Scale bar = 20 μm refers to indicated panel alone. Other images are cropped 40% to highlight the spindle. Kymographs showing DHC-GFP levels at spindle poles and cell cortex along pole-to-pole axis are generated from flattened time-lapse Z stacks, acquired once every minute, of cells treated with LGN or control siRNA. Spindle pole location (red arrows) and DHC-GFP enrichment at cell cortex (green lines) are marked. (C) Time-lapse images of control or LGN siRNA-transfected HeLaHis2B-GFP; mCherry-Tub cells undergoing cell division. Dashed lines indicate interphase long-axis (yellow), first alignment along long-axis (pink) and final spindle orientation axis (blue). Scale bar = 20 μm. (D) Graph of percentage of mitotic cells with no spindle oscillation in cells treated with control or LGN siRNA as assessed from time-lapse movies of HeLaHis2B-GFP; mCherry-Tub cells. (E) Frequency distribution of final orientation angles (spindle-axis relative to long-axis) in cells treated with control or LGN siRNA (n = 3 experiments). (F) Bar graph showing percentage of spindle movements toward or away from the long-axis, when spindle-axis was at least 30 degrees away from long-axis, in cells treated with siRNA as indicated. (G) Bar graph showing percentages of spindles that aligned within 30 degrees of long-axis at some point, at anaphase and maintained alignment after first alignment in cells treated with control or LGN siRNA. Error bars represent SEM from 3 independent experiments. P values are obtained using proportion test. Symbols # and * refer to insignificant and significant differences, respectively.
To study spindle rotation and orientation events following LGN depletion, control or LGN siRNA-treated and G2-M-synchronized HeLaHis2B-GFP;mCherry-Tub cells were imaged once every 4 min for 5 h (Fig. 3C). Analysis of time-lapse movies showed that in control-depleted cells, but not LGN-depleted cells, spindles that aligned along long-axis underwent oscillations along the pole-to-pole axis (Fig. 3D). Analysis of final spindle orientations using Spindle3D showed that the bias in orienting spindles along long-axis was significantly reduced in LGN siRNA-treated cells compared with control siRNA-treated cells (Fig. 3E), confirming LGN’s role in spindle orientation.
We tested if orientation failure in LGN siRNA-treated cells was due to a spindle rotation failure or orientation maintenance failure. Spindle rotation in both control- and LGN-depleted cells showed a comparable directional bias toward long-axis (Fig. 3F). However, the speed of spindle rotation prior to alignment along long-axis was reduced by half in LGN-depleted cells compared with control-depleted cells (spindle rotation speed in degrees/frame: control RNAi: pre-align 15.0 ± 0.8°, post-align 9.8 ± 0.4°, n = 142 cells; LGN RNAi: pre-align 8.7 ± 0.4°, post-align 7.7 ± 0.4°, n = 178 cells), revealing LGN’s role in spindle rotation. To test if the spindle orientation failure arose primarily from a spindle rotation defect, we analyzed the probability of alignment along long-axis based on the initial deviation of the spindle-axis from long-axis. In control cells, the probability of alignment was similar when spindle-axis was proximal (30–60 degrees) or distal (60–90 degrees) to the long-axis (Fig. S3). However, in LGN siRNA-treated cells, probability of alignment was significantly higher when spindle-axis was proximal (Fig. S3A), indicating a selective failure in orienting spindles that needed greater rotation to align along long-axis. Collectively, these observations together with the time-lapse sequences confirm that the spindle rotation defect leads to spindle orientation failure in LGN-depleted cells. Analysis of orientation maintenance showed that although a majority of the spindles failed to arrive at the long-axis, those spindles that were assembled close to the long-axis were successfully maintained along long-axis in LGN-depleted cells (Fig. 3G), indicating that LGN is dispensable for orientation maintenance. We conclude that LGN is specifically essential for normal spindle rotation, but not for the directional bias during rotation or for orientation maintenance along long-axis. This suggests the presence of LGN- and cortical dynein-independent cortical anchors for astral microtubules to tether and maintain spindle orientation.
2ME2 disrupts mechanisms that maintain orientation along the long-axis
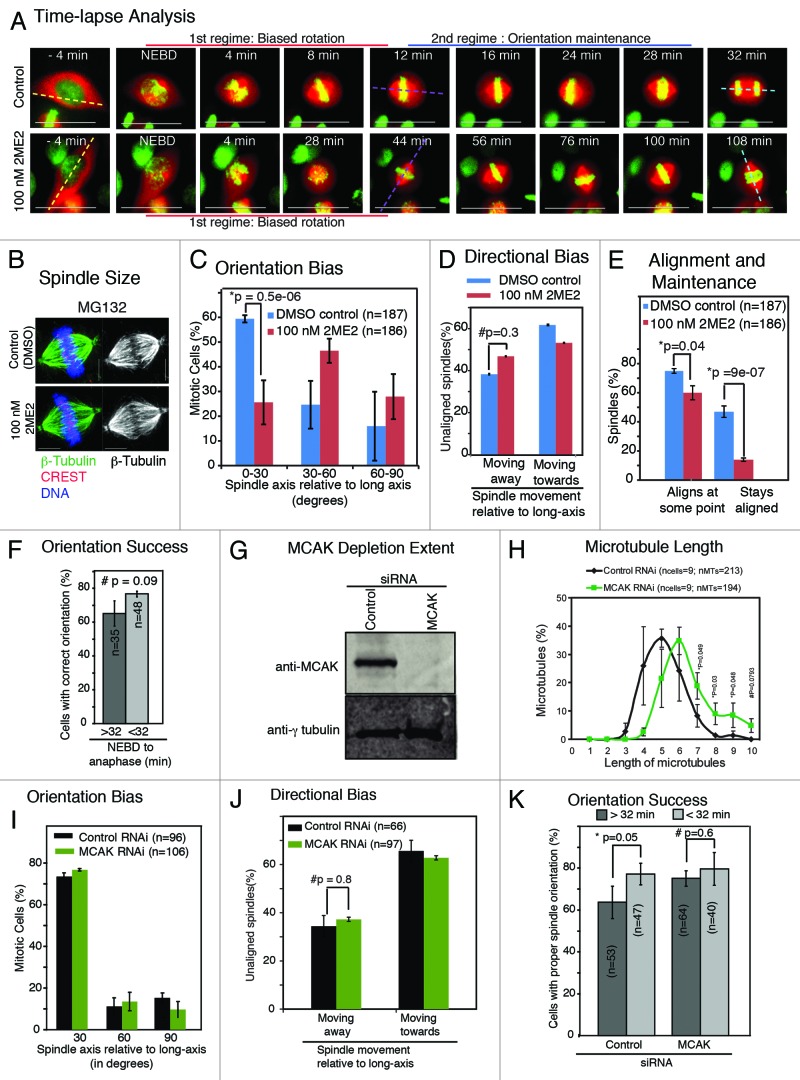
We investigated the extent to which subtle perturbation to microtubule dynamics influenced the 2 regimes of spindle movements. This is a technically challenging experiment, as perturbation of microtubules can disrupt chromosome-microtubule attachments and block anaphase onset. Because low concentration of Nocodazole that caused spindle misorientation was reported to cause spindle shortening,3 we used a subtle microtubule perturbing agent 2-Methoxy Estradiol (2ME2) that suppresses microtubule dynamics in interphase MCF7 cells without changing bulk levels of microtubules.27 We chose 100 nM concentration of 2ME2 for our study, as it was the highest concentration that allowed mitotic exit (data not shown). We first tested the extent to which 100 nM 2ME2 treatment altered mitotic microtubule dynamics in HeLa cells expressing EB1-YFP (HeLaEB1-YFP), a marker of growing microtubule ends, using time-lapse microscopy. EB1-YFP comets were not visually different between 100 nM 2ME2-treated and control cells (Fig. S4A). However, 2ME2 treatment induced a significant reduction in growth and shrinkage rates compared with controls (Table 1), as assessed using plusTipTracker,28 a software that infers growth and shrinkage rates of microtubules from the tracks of EB1-YFP-decorated microtubule end. Microtubule growth rates in control mitotic cells were comparable to previously reported values in LLCPK cell line.29 2ME2 treatment did not change bipolar spindle size in live or fixed cells (Fig. 4A and B) or mitotic microtubule length (Fig. S4B). These findings show that 100 nM 2ME2 reduces growth and shrinkage rate but does not reduce spindle size, indicating the drug’s usefulness for perturbing microtubule end behavior without altering spindle size.
Table 1. Effect of 2ME2 treatment on the dynamic instability of microtubules in mitotic cells.
| Mode* growth rate± SD(µm min−1) | Mode shrinkage rate ± SD (µm min−1) | Per cent time growing/shortening/paused | Pause frequency (s−1) | |
|---|---|---|---|---|
| Control (DMSO) |
14.25 ± 3.74 |
14.60 ± 9.01 |
37.4/37.4/25.1 |
0.118 |
| 2ME2 treatment | 4.860 ± 2.75 | 5.224 ± 2.25 | 38.8/36.3/24.9 | 0.130 |
* Mode values were chosen since the histograms of growth and shrinkage rates were not distributed normally. To define the mode in this table, we considered values within the peaks in the histogram and then calculated the mean and SD for the modes in individual cells. List of mode values of microtubule growth and shrinkage rates, percentage time microtubules spent growing or shrinking or pausing, and frequency of pause events (every s) observed in HeLaEB1-YFP cells that were treated with DMSO or 100 nM 2ME2. Eighty-four tracks of 5 control mitotic cells and 67 tracks of 5 2ME2-treated mitotic cells were used for analysis.
Figure 4. Orientation maintenance is exquisitely sensitive to suppression of microtubule dynamics. (A) Time-lapse images showing spindle and chromosome movements in HeLaHis2B-GFP; mCherry-Tub cells treated with or without 100 nM 2ME2. Long-axis, first spindle alignment axis, and final spindle orientation axis are marked in yellow, purple, and blue dashed lines, respectively. Scale bar = 30 μm. (B) Immunofluorescence images showing spindle size and architecture in 100 nM 2ME2- or DMSO- (control-) treated HeLa cells arrested in metaphase using MG132 and immunostained with antibodies against β-tubulin and CREST (kinetochore marker) and stained with DAPI for DNA. Scale bar = 5 μm. (C) Frequency distribution of final orientation angles (spindle-axis relative to long-axis) in cells treated with 100 nM 2ME2. (D) Graph showing percentage of spindle movements either toward or away from the long-axis in unaligned spindles oriented at least 30 degrees away from long-axis, in cells treated with DMSO or 2ME2. (E) Graph showing percentage of mitotic cells that aligned spindles at some point and maintained alignment, within 30 degrees of the long-axis, in cells treated with DMSO or 2ME2. (F) Graph showing percentage of mitotic cells that oriented spindles within 30 degrees of the long-axis in unperturbed HeLaHis2B-GFP; mCherry-Tub cells with an NEBD to anaphase onset time greater or lesser than 32 min. (G) Immunoblots of cell lysates treated with siRNA oligos and immunostained with antibodies as indicated to assess the extent of MCAK depletion following MCAK RNAi. (H) Graph showing distribution of microtubule lengths (μm) in monopolar spindles of cells treated with siRNA as indicated and exposed to monastrol for 3 h prior to immunostaining with antibodies against β-tubulin. (I) Graph of distribution of spindle orientation angles in MCAK depleted HeLaHis2B-GFP; mCherry-Tub cells. (J) Graph showing percentage of spindle movements either toward or away from the long-axis in unaligned spindles oriented at least 30 degrees away from long-axis, in cells treated with control or MCAK siRNA. (K) Graph of percentage of mitotic cells that oriented spindles properly, within 30 degrees of the long-axis, in HeLaHis2B-GFP; mCherry-Tub cells treated with siRNA as indicated with an NEBD to anaphase onset time greater or less than 32 min. Error bars represent SEM from 3 independent experiments. n = number of cells. P values are obtained using proportion test, except for (C) where Mann–Whitney U test, was used. Symbols # and * refer to insignificant and significant differences, respectively.
HeLaHis2B-GFP;mCherry-Tub cells treated with 100 nM 2ME2, but not control cells (treated with DMSO alone), displayed congression defects and delayed anaphase onset (Fig. 4A; Fig. S4C). Despite the congression defect and delay in anaphase onset, 100 nM 2ME2-treated cells underwent anaphase and chromosome segregation (Fig. 4A; n = 43 cells). However, spindle orientation along long-axis was perturbed following 2ME2 treatment (Fig. 4A). Quantifying the final orientation bias using Spindle3D confirmed that only 25% of 2ME2-treated cells could orient spindles within 30 degrees of the long-axis (Fig. 4C). However, the directional bias in rotating spindles toward long-axis was not lost following either control or 2ME2 treatment (Fig. 4D), indicating the presence of 2ME2-resistant interphase cell shape memory that guides the first regime of spindle movements toward the pre-defined axis.
The speed of rotation toward long-axis was reduced by one-third following 2ME2 treatment (spindle rotation speed in degrees/frame: control: pre-align 13.1 ± 0.7°, post-align 9.9 ± 0.5°, n = 123 cells; 2ME2: pre-align 9.1 ± 0.3°, post-align 8.6 ± 0.3°, n = 100 cells). This speed reduction was not the cause of orientation failure, because time-lapse sequences showed that even spindles that aligned at long-axis failed to remain near the long-axis (Fig. 4A). In agreement, Spindle3D analysis showed that the percentage of cells that aligned along long-axis, at some point, was only slightly reduced in 2ME2-treated cells compared with control cells (Fig. 4E). However, the percentage of cells that maintained orientation within 30 degrees of long-axis was drastically reduced following 2ME2 treatment compared with controls (Fig. 4E). This shows that 2ME2 treatment induced orientation defect arises at least in part from a failure in the second regime of spindle movements required for orientation maintenance. We conclude that molecular mechanisms that maintain orientation are more sensitive to microtubule perturbations compared with mechanisms that govern rotation toward long-axis.
The orientation maintenance failure in 2ME2-treated cells could hypothetically be an indirect consequence of the anaphase delay caused by congression defects. We excluded this possibility, since control cells that by chance underwent delayed congression and an associated anaphase delay did not show a spindle orientation failure (Fig. 4F). We conclude that changes to microtubule dynamics can disrupt orientation maintenance mechanisms severely, without abrogating directional bias in spindle rotation. 2ME2-induced failure in orientation maintenance may explain the spindle orientation failure observed in mouse uterine tissues exposed to estradiol.30
MCAK is dispensable for cell shape-associated spindle orientation
Because suppression of microtubule dynamics using 2ME2 abrogated orientation maintenance, we tested the extent to which spindle orientation will be perturbed in response to increase in astral microtubule length. For addressing this, we depleted the evolutionarily conserved microtubule depolymerizing kinesin, MCAK/Kif2C that is required for microtubule shrinkage.31,32 To deplete MCAK in HeLaHis2B-GFP;mCherry-Tub cells (Fig. 4G), we used previously described siRNAs. Analysis of time-lapse movies showed a delay in chromosome congression and anaphase onset in MCAK-depleted cells compared with control-depleted cells (Fig. S4C), as reported,33 confirming a functional loss of MCAK. Mitotic spindle microtubule lengths, as assessed in monastrol-arrested prometaphase cells, were longer in MCAK-depleted cells compared with control-depleted cells (Fig. 4H), in agreement with previously reported long astral microtubules in MCAK-depleted cells.31
Analysis of orientation bias showed that the percentage of cells that aligned along long-axis was not significantly different between control and MCAK-depleted cells (Fig. 4I), indicating proper spindle orientation in MCAK-depleted cells. The speed of rotation toward long-axis was slightly reduced following MCAK depletion (spindle rotation speed in degrees/frame: control RNAi: pre-align 16.0+/−1.3°, post-align 11.2+/−1.2°, n = 123 cells; MCAK RNAi: pre-align 12.8+/−0.8°, post-align 8.9+/−0.7°, n = 100 cells; Table 2). However, directional bias in the rotation of unaligned spindles was comparable in control and MCAK-depleted cells (Fig. 4J). Collectively these findings show that increase in microtubule length does not perturb final orientation bias or directional bias in rotation. Because MCAK-depleted cells orient spindles properly, even in cells that showed a delayed anaphase time of more than 32 min (Fig. 4K), we conclude that the memory of interphase shape cues is retained even during a prolonged delay in anaphase onset. Thus, our findings demonstrate that although the increase in astral microtubule length reduces spindle rotation speed, it is well-tolerated by cell shape-associated orientation process.
Table 2. Spindle rotation rates before and after alignment along long-axis.
| Treatment | Post-align speed (deg/frame) | S.E.M. | Pre-align speed (deg/frame) | S.E.M. | ncells | Post-align frames | Pre-align frames | Pre-align speed (deg/min) | Post-align speed (deg/min) |
|---|---|---|---|---|---|---|---|---|---|
|
DMSO (Control) |
9.8986 |
0.5326 |
13.0507 |
0.7360 |
123 |
365 |
369 |
3.2626 |
2.4746 |
|
100nM 2ME2 |
8.5838 |
0.3196 |
9.1253 |
0.2953 |
100 |
827 |
1196 |
2.2813 |
2.1459 |
|
Cntrl RNAi (72 h; syn) |
9.8077 |
0.4118 |
15.0410 |
0.7714 |
142 |
531 |
430 |
3.7602 |
2.4519 |
|
LGN RNAi (72 h; syn) |
7.7368 |
0.38547 |
8.6709 |
0.4061 |
178 |
410 |
509 |
2.1677 |
1.9342 |
|
Cntrl RNAi (48 h; asy) |
11.1944 |
1.1866 |
16.0160 |
1.3021 |
66 |
150 |
183 |
4.0040 |
2.7986 |
| MCAK RNAi (48 h; asy) | 8.8873 | 0.6562 | 12.7987 | 0.7690 | 97 | 253 | 271 | 3.1996 | 2.2218 |
Speeds were calculated using rotational displacement of spindle pole positions between two consecutive frames (T1 and T2) as determined using Spindle3D. Spindle angle at T1 was used to distinguish pre-align (> 30 from long-axis) and post-align (< 30 degrees from long-axis) positions. Frame refers to four minutes intervals. Columns marked “Pre-align frames” and “Post-align frames” indicate the number of frames used to calculate speed. Speed in degrees/min was calculated by quartering degrees/frame values. SEM refers to Standard Error of Mean. ncells refers to total number of mitotic cells used for speed calculations. Cntrl RNAi, syn and asy refer to Control RNAi, synchronous cultures and asynchronous cultures, respectively. Hours (h) in column 1 refers to transfection period. List of spindle rotation speeds from time-lapse movies extracted using spindle pole positions identified by Spindle3D. Speeds were binned according to positions occupied by the spindle, either before spindle alignment along long-axis (< 30 degrees) or after alignment along long-axis (> 30 degrees).
Discussion
Here we establish a methodology to study spindle movements that bring the spindles to a pre-defined axis. By extracting statistically relevant spindle movements in hundreds of cells displaying a continuum of shapes, we show the presence of 2 distinct regimes of spindle movements: first, spindles are actively directed toward the long-axis and then maintained along the long-axis. During the latter phase, spindles were found to oscillate along the pole-to-pole axis. We show that the first regime of spindle movement is dependent on LGN. The second regime of movements that maintain spindle orientation is independent of LGN, although spindle oscillations require LGN. We show that orientation maintenance mechanisms are sensitive to suppression of microtubule dynamics, but not increase in microtubule length (Fig. 5A). We propose that spindle orientation is controlled by spatially distinct factors that differently control spindle rotation and orientation maintenance movements.
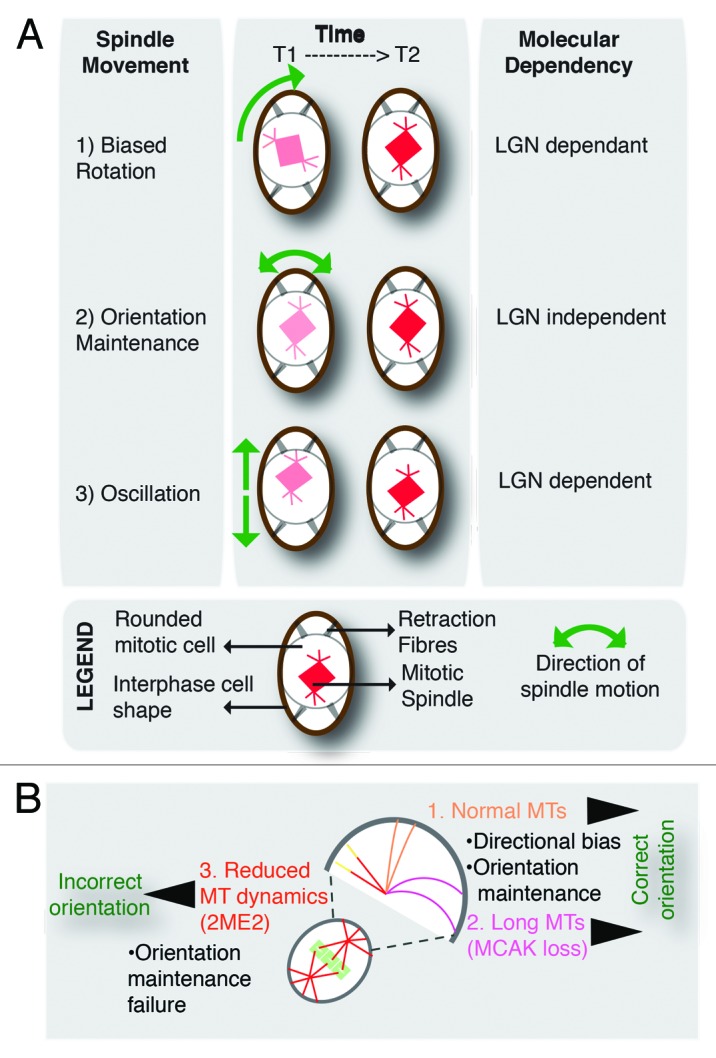
Figure 5. Model of two spatially regulated regimes of spindle movements that govern cell shape associated spindle orientation (A) Differential regulation of three distinct movements of the spindle. (1) Spindle rotation with a directional bias: Misoriented spindles are rotated toward the interphase cells' long-axis in an LGN-dependent manner. (2) Orientation maintenance: Spindle alignment maintained near the long-axis in an LGN-independent manner. (3) Spindle oscillation along pole-to-pole axis: During orientation maintenance phase, the spindle oscillates in an LGN-dependent manner. (B) Orientation maintenance is sensitive to suppression of microtubule dynamics but not increase in microtubule length. Illustration depicts spindle behavior in the two regimes. 2ME2 disrupts orientation by perturbing the maintenance of due to suppressed microtubule dynamics. However, an increase in astral microtubule (MT) length (MCAK-depleted cells) does not perturb directional bias or orientation maintenance.
Proper regulation of cortical dynein is important for the accurate positioning of spindles parallel to the substratum.12,13,34 Our study of LGN-depleted cells show that spindle rotation towards the final axis, alone, is reliant on cortical dynein. However, to maintain a stable orientation of the spindles, neither LGN nor cortical dynein is required. This shows that under certain conditions, loss of cortical dynein does not disrupt the stable positioning of the spindle parallel to the substratum (this study), consistent with previous findings of p150 dynactin depletions that did not perturb spindle positioning.34 Collectively, these findings imply that although cortical dynein is an important regulatory anchor for pulling at astral microtubules, LGN- and dynein-independent anchors are likely to exist for tethering astral microtubules to the cell cortex.
Microtubule interactions at the cell cortex that control spindle orientation are likely to be resilient to increase in microtubule length, since orientation is unperturbed following MCAK depletion, a condition that increases astral MT length (this work and ref. 31). This shows that increased contact between the microtubule wall and the cell cortex, which is likely to provide increased area for cortical dynein, is tolerated well by orientation mechanisms. In stark contrast, suppression of microtubule dynamics, as in 2ME2-treated cells, causes a severe spindle orientation defect; spindles are unable to maintain orientation. This finding shows the significance of microtubule dynamics-based mechanisms, rather than cortical dynein-based mechanism, for maintaining the orientation of spindles in a HeLa cell line that is non-polarized (Fig. 5B).
Whether orientation maintenance is achieved through spatially defined cues that: (1) actively retard spindle movements near the long-axis; or (2) establish mechanical equilibrium near the long-axis;7 or (3) both of the above mechanisms is beyond the scope of this work and are addressed through simulation studies (AC, VMD, and AD, unpublished work). The oscillation of the spindle during the orientation maintenance regime (this study) and enrichment of cortical dynein into crescents by metaphase35,36,37 are consistent with mechanisms that actively restrain spindle movements near the long-axis. Interestingly, biphasic movements of the spindle have been observed in chick embryos as well, wherein the spindle rapidly orients parallel to the apical surface (planar orientation) and is then maintained in this planar orientation.38
The quantitative framework we establish here to study spindle orientation in cells, maintaining cell–cell interaction, will allow future interrogation of the roles of junctional proteins, cell–cell adhesion, and microtubule networks39 in controlling spindle orientation. By defining parameters to distinguish the two regimes of spindle movements, this study sets up roads for measuring and modeling the complex movements of the mitotic spindle using a simple cell culture model system. Spindle orientation mechanisms we describe here may be relevant to cells that are normally polarized but have depolarized by virtue of epithelial–mesenchymal transformation.
Materials and Methods
Cell culture and G2-M synchronization
RPE-1 and HeLa cell cultures were grown on either glass coverslips or chambered glass coverslips (cover glass Lab-tek chambers, FISHER, http://www.fishersci.com/ecomm/servlet/fsproductdetail_10652_651183__-1_0) in DMEM supplemented with 10% FBS and antibiotics. For cell cycle synchronization, cells were treated for 24 h with Aphidicolin (1 μg/ml; Sigma, http://www.sigmaaldrich.com/catalog/product/fluka/89458), washed and then imaged 7 h later, when cells are in G2-M phase. Non-confluent monolayer cultures were chosen for imaging.
Live cell time-lapse imaging and analysis
Cells were imaged 48 h or 72 h after seeding in chambered glass coverslips (Cover glass Lab-tek Chambers, FISHER) in CO2-independent L15 medium (Invitrogen, http://products.invitrogen.com/ivgn/product/21083027?ICID=search-product) at 37 °C. For spindle pole-tracking movies, exposures of 0.1 s and at least 3 Z-planes, with 3 micron Z-steps, were acquired every 4 min for 4 h using a 40× NA 0.75 objective on an Applied Precision Deltavision Core microscope (http://www.api.com/deltavision.asp) equipped with a Xenon 100 W lamp, GFP-long pass filter and cherry-red filter (Chroma), phase-contrast filters, and Coolsnap HQ camera, and Cascade 2 EMCCD camera. The timing of anaphase onset was determined as previously described.11
siRNA transfection
LGN siRNA oligos (Dharmacon SMART POOL L-004092–00–005 GAACUAACAGCACGACUUA, CUUCAGGGAUGCAGUUAUA, ACAGUGAAAUUCUUGCUAA, UGAAGGGUUCUUUGACUUA) and MCAK siRNA oligos40 (GATCCAACGCAGTAATGGT) were used to transfect HeLa cells 48 h after seeding. Forty-eight hours after transfection, cells were imaged, and cell lysates were collected for testing the extent of protein depletion using immunoblotting.
Generation of the spindle-position reporter cell line, HeLaHis2B-GFP;mCherry-Tub and microtubule-end reporter cell line HeLaEB1-YFP
HeLaHis2B-GFP;mCherry-Tub cell line was generated by transfecting mCherry-Tubulin expressing eukaryotic plasmid vector into HeLaHis2B-GFP cells.11 HeLaEB1-YFP cell line was generated by transfecting EB1-YFP-expressing eukaryotic plasmid11 into HeLa cells. BAC transgenic HeLaDHC-GFP cell line expressing DHC-GFP from an endogenous promoter was obtained from Max Planck Institute.41 Fluorescent cells were enriched using FACSORT.
Image processing methods: Spindle 3D
Spindles are identified from the mCherry-Tub images by applying a difference of Gaussians (DoG) filter that selects for objects within a chosen size range. The filter size is tuned to detect each spindle pole as a bright spot. A spindle is identified as 2 such spots that are separated by less than a threshold distance. The noise (number of false positives) is reduced by simultaneously searching for rounded cells in the DIC channel and mitotic DNA in the His2B-GFP channel. Rounded cells are detected using a circular Hough transform on the gradient of the DIC channel. DNA acquires a more granular appearance and appears brighter due to condensation during mitosis, and this is detected again using a DoG filter. The filter response can be used to infer information about mitotic timings. From the combined data, a wide range of dynamic mitotic parameters can be automatically measured from live cell imaging experiments, including cell size, spindle orientation, spindle size, displacement of spindle from center of cell, and/or cortex, metaphase plate orientation, velocity of anaphase, speed of spindle movement/rotation, and the timing of spindle alignment relative to NEBD, establishment of metaphase plate and anaphase onset. The simplicity of the spindle identification is intuitive (i.e., the filtering parameters correspond directly to physical measurements of the spindle); however, to ensure that the dynamic properties measured are valid, a manual verification step is included to remove false positives and to ensure that spindle orientation and length measurements are accurate. Spindle3D work flowchart is presented in Figure S3. MATLAB code for Spindle3D can be downloaded from http://www.gen.cam.ac.uk/~vmd20/index.html
Shape detection
The cell boundary is detected by segmenting the stack of phase contrast images using the SePhace method (Ali et al, 2011) and a custom gradient watershed algorithm. Cell shape is measured 5 frames (where possible) prior to NEBD, and then automatically measured boundary is verified manually for each cell.
For measuring the long-axis and the shape aspect ratio, an ellipse-fitting method is employed. The eigenvectors and eigenvalues of the second moment tensor of the set of points on the perimeter of the cell are calculated. The principal eigenvector points along the long-axis, and the aspect ratio is given by the square root of the ratio of the eigenvalues. Conceptually, this corresponds to fitting an ellipse to the boundary of the cell; the long-axis of the ellipse lines up with the long-axis of the cell, and its elongation corresponds to the aspect ratio. The greater the aspect ratio, the more accurately the long-axis can be measured. In contrast, when the aspect ratio is close to unity the certainty with which the long-axis can be assigned is reduced. Therefore, we apply a minimum value threshold for the aspect ratio of 1.2 to include cells in our analysis.
Immunoflourescence and immunoblotting
For immunofluorescence studies, cells were fixed with methanol for 90 sec and then primary antibodies against tubulin (Abcam; http://www.abcam.com/tubulin-antibody-yl12-loading-control-ab6160.html), and secondary antibodies anti-rat Dylight -488 (Abcam; http://www.abcam.com/goat-polyclonal-secondary-antibody-to-rat-igg-h-l-dylight-488-pre-adsorbed-ab98420.html) and anti-rabbit Alexa Fluor 594 (Invitrogen; https://products.invitrogen.com/ivgn/product/A21207?ICID=search-a21207), anti-human Alexa Fluor 647 (https://products.invitrogen.com/ivgn/product/A21445?ICID=search-product) and CREST antisera (http://www.europa-bioproducts.com/Catalogue/product.aspx?product=ANA-Centromere%20CREST%20AutoAb%20Human&id=FZ90C-CS1058) were used. Images of immunostained cells were acquired using 100× NA 1.2 objective on a DeltaVision Core microscope equipped with CoolSnap HQ Camera (Photometrics). For immunoblotting, antibodies against MCAK42 and Tubulin (Sigma; http://www.sigmaaldrich.com/catalog/product/sigma/t4026?lang=en®ion=GB) were used. Immunoblots were developed using fluorescent secondary antibodies (LICOR) and analyzed using Odyssey (LI-COR).
EB1-YFP imaging and comet tracking
HeLaEB1-YFP cells in mitosis were imaged for 15 s, once every 300 ms. At each time point, we acquired 5 z-planes to facilitate deconvolution. A step size of 0.4 μm was optimized to ensure the counting of all comets in given region and to exclude the counting of the same comet in more than one Z step. Image planes were manually chosen to allow EB1 comet tracking of astral microtubule ends using the plusTipTracker software.28,43
Microtubule length measurements
Cells were treated with monastrol and then immunostained with antibody against α-tubulin, HEC1, and CREST antisera and stained with DAPI for DNA. Z stack images with 0.2 μm Z step were acquired. Length of microtubule from spindle pole to microtubule tip was measured using the SoftWorx “measure” tool. To exclude measurement artifacts, only those non-curled microtubules that were in the plane of imaging were considered for quantification. Microtubule lengths from 3 cells for each RNAi condition from 3 experimental repeats were combined to generate the length distribution graph.
Inhibitor treatment
One hundred mM 2ME2 (Sigma; http://www.sigmaaldrich.com/catalog/product/sigma/m6383) was prepared in DMSO and serially diluted in DMEM without FCS. During the filming process, 100 nM 2ME2 in DMEM was added to L15, a Co2- and serum-independent media. For fixed-cell studies, cells were treated with 2ME2 for an hour prior to fixation. Cells were treated with 10 μM Monastrol (Tocris; http://www.tocris.com/dispprod.php?ItemId=1911) for 3 h and 10 μM MG132 (Tocris; http://www.tocris.com/dispprod.php?ItemId=69832) for either 1 h (fixed-cell imaging) or up to 3 h (live cell imaging).
Statistical analysis
Error bars represent SEM values (standard error of mean). Values obtained across experiments, cells, or directional switches are as indicated in legend. This allowed us to compare differences in the spread of the means, in addition to differences in the mean values per se. P values representing significance were obtained using Mann–Whitney U test or proportion test (on percentage values). Symbols # and * refer to insignificant and significant differences, respectively as assessed using a 95% confidence interval.44 For growth and shrinkage rates, mode values were chosen, since histograms of growth and shrinkage rates did not distribute normally. Hence we avoided the use of mean values (average) to represent the value of the population Table 2.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by Cancer Research UK career development grant to VMD, a collaborative seed project grant awarded to AD (PI) and VMD (Co-PI), and a Royal Society joint project grant awarded to VMD (PI) and AF (Co-PI). We thank Draviam group members and Bernhard Strauss for comments on the manuscript, and Nigel Miller for FACSORT of HeLaHis2B-GFP;mCherry-Tub and HeLaEB1-YFP cells.
Author Contribution
Manuscript was written by VMD and included editing comments from AD. Experiments were designed by VMD, performed and analyzed by RS, IZ, and AC. IZ generated HeLaHis2B-GFP;mCherry-Tub and NT generated HeLa EB1-YFP cell lines. Spindle3D software was conceived by AC, VMD and AD, developed primarily by AC, and adapted by BY and YL. Figure contributions are as follows: RS, Figures 1, 4, S2, and S4; IZ, Figures 3, 4, and S1; AC, Figures 2, S3 and Table 2; JP, Figures 3D and 4C, respectively; NH with AF, Table 1.
Supplemental Materials
Supplemental materials may be found here: http://www.landesbioscience.com/journals/cc/article/25671
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/25671
References
- 1.Mapelli M, Gonzalez C. On the inscrutable role of Inscuteable: structural basis and functional implications for the competitive binding of NuMA and Inscuteable to LGN. Open Biol. 2012;2:120102–5. doi: 10.1098/rsob.120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noatynska A, Gotta M, Meraldi P. Mitotic spindle (DIS)orientation and DISease: cause or consequence? J Cell Biol. 2012;199:1025–35. doi: 10.1083/jcb.201209015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–74. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wühr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. A model for cleavage plane determination in early amphibian and fish embryos. Curr Biol. 2010;20:2040–5. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quyn AJ, Appleton PL, Carey FA, Steele RJC, Barker N, Clevers H, et al. Spindle orientation bias in gut epithelial stem cell compartments is lost in precancerous tissue. Cell Stem Cell. 2010;6:175–81. doi: 10.1016/j.stem.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Luxenburg C, Pasolli HA, Williams SE, Fuchs E. Developmental roles for Srf, cortical cytoskeleton and cell shape in epidermal spindle orientation. Nat Cell Biol. 2011;13:203–14. doi: 10.1038/ncb2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Théry M, Jiménez-Dalmaroni A, Racine V, Bornens M, Jülicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–6. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 8.Théry M, Racine V, Pépin A, Piel M, Chen Y, Sibarita J-B, et al. The extracellular matrix guides the orientation of the cell division axis. Nat Cell Biol. 2005;7:947–53. doi: 10.1038/ncb1307. [DOI] [PubMed] [Google Scholar]
- 9.Fink J, Carpi N, Betz T, Bétard A, Chebah M, Azioune A, et al. External forces control mitotic spindle positioning. Nat Cell Biol. 2011;13:771–8. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 10.Fernandez P, Maier M, Lindauer M, Kuffer C, Storchova Z, Bausch AR. Mitotic spindle orients perpendicular to the forces imposed by dynamic shear. PLoS One. 2011;6:e28965. doi: 10.1371/journal.pone.0028965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Draviam VM, Shapiro I, Aldridge B, Sorger PK. Misorientation and reduced stretching of aligned sister kinetochores promote chromosome missegregation in EB1- or APC-depleted cells. EMBO J. 2006;25:2814–27. doi: 10.1038/sj.emboj.7601168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–50. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- 13.Dunsch AK, Hammond D, Lloyd J, Schermelleh L, Gruneberg U, Barr FA. Dynein light chain 1 and a spindle-associated adaptor promote dynein asymmetry and spindle orientation. J Cell Biol. 2012;198:1039–54. doi: 10.1083/jcb.201202112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaji N, Muramoto A, Mizuno K. LIM kinase-mediated cofilin phosphorylation during mitosis is required for precise spindle positioning. J Biol Chem. 2008;283:4983–92. doi: 10.1074/jbc.M708644200. [DOI] [PubMed] [Google Scholar]
- 15.Mitsushima M, Toyoshima F, Nishida E. Dual role of Cdc42 in spindle orientation control of adherent cells. Mol Cell Biol. 2009;29:2816–27. doi: 10.1128/MCB.01713-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumura S, Hamasaki M, Yamamoto T, Ebisuya M, Sato M, Nishida E, et al. ABL1 regulates spindle orientation in adherent cells and mammalian skin. Nat Commun. 2012;3:626. doi: 10.1038/ncomms1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toyoshima F, Matsumura S, Morimoto H, Mitsushima M, Nishida E. PtdIns(3,4,5)P3 regulates spindle orientation in adherent cells. Dev Cell. 2007;13:796–811. doi: 10.1016/j.devcel.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 18.Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–98. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;14:311–7. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitagawa D, Kohlmaier G, Keller D, Strnad P, Balestra FR, Flückiger I, et al. Spindle positioning in human cells relies on proper centriole formation and on the microcephaly proteins CPAP and STIL. J Cell Sci. 2011;124:3884–93. doi: 10.1242/jcs.089888. [DOI] [PubMed] [Google Scholar]
- 21.Rebollo E, Roldán M, Gonzalez C. Spindle alignment is achieved without rotation after the first cell cycle in Drosophila embryonic neuroblasts. Development. 2009;136:3393–7. doi: 10.1242/dev.041822. [DOI] [PubMed] [Google Scholar]
- 22.Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, González C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–74. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 23.Rosenblatt J. Spindle assembly: asters part their separate ways. Nat Cell Biol. 2005;7:219–22. doi: 10.1038/ncb0305-219. [DOI] [PubMed] [Google Scholar]
- 24.Gotta M, Ahringer J. Distinct roles for Galpha and Gbetagamma in regulating spindle position and orientation in Caenorhabditis elegans embryos. Nat Cell Biol. 2001;3:297–300. doi: 10.1038/35060092. [DOI] [PubMed] [Google Scholar]
- 25.Grill SW, Howard J, Schäffer E, Stelzer EHK, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–21. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 26.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr Biol. 2008;18:1986–92. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 27.Kamath K, Okouneva T, Larson G, Panda D, Wilson L, Jordan MA. 2-Methoxyestradiol suppresses microtubule dynamics and arrests mitosis without depolymerizing microtubules. Mol Cancer Ther. 2006;5:2225–33. doi: 10.1158/1535-7163.MCT-06-0113. [DOI] [PubMed] [Google Scholar]
- 28.Matov A, Applegate K, Kumar P, Thoma C, Krek W, Danuser G, et al. Analysis of microtubule dynamic instability using a plus-end growth marker. Nat Methods. 2010;7:761–8. doi: 10.1038/nmeth.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piehl M, Cassimeris L. Organization and dynamics of growing microtubule plus ends during early mitosis. Mol Biol Cell. 2003;14:916–25. doi: 10.1091/mbc.E02-09-0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunin AG, Mashin IN, Zakharov DA. Proliferation, mitosis orientation and morphogenetic changes in the uterus of mice following chronic treatment with both estrogen and glucocorticoid hormones. J Endocrinol. 2001;169:23–31. doi: 10.1677/joe.0.1690023. [DOI] [PubMed] [Google Scholar]
- 31.Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J Cell Biol. 2012;197:231–7. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–42. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kotak S, Busso C, Gönczy P. Cortical dynein is critical for proper spindle positioning in human cells. J Cell Biol. 2012;199:97–110. doi: 10.1083/jcb.201203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Busson S, Dujardin D, Moreau A, Dompierre J, De Mey JR. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr Biol. 1998;8:541–4. doi: 10.1016/S0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- 36.Collins ES, Balchand SK, Faraci JL, Wadsworth P, Lee WL. Cell cycle-regulated cortical dynein/dynactin promotes symmetric cell division by differential pole motion in anaphase. Mol Biol Cell. 2012;23:3380–90. doi: 10.1091/mbc.E12-02-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiyomitsu T, Cheeseman IM. Chromosome- and spindle pole- derived signals generate an intrinsic code for spindle position and orientation. Nat Cell Biol. 2012;12:311–7. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peyre E, Jaouen F, Saadaoui M, Haren L, Merdes A, Durbec P, et al. A lateral belt of cortical LGN and NuMA guides mitotic spindle movements and planar division in neuroepithelial cells. J Cell Biol. 2011;193:141–54. doi: 10.1083/jcb.201101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tamura N, Draviam VM. Microtubule plus-ends within a mitotic cell are ‘moving platforms’ with anchoring, signalling and force-coupling roles. Open Biol. 2012;2:120132. doi: 10.1098/rsob.120132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cassimeris L, Morabito J. TOGp, the human homolog of XMAP215/Dis1 is required for centrosome integrity, spindle pole organization, and bipolar spindle assemblys. Mol Biol Cell. 2012;15:1580–90. doi: 10.1091/mbc.E03-07-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poser I, Sarov M, Hutchins JRA, Hériché J-K, Toyoda Y, Pozniakovsky A, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–15. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, et al. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–7. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–84. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher RA. Statistical Methods and Scientific inference . Edinburgh : Oliver and Boyd ; 1956 . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.