Abstract
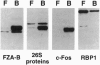
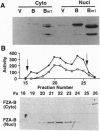
In a search for regulatory proteins that interact with the leucine zipper motif of c-Fos in the yeast two-hybrid screen, we have identified a protein (FZA-B) that has extensive sequence similarity to SUG1 of Saccharomyces cerevisiae. Here we show that FZA-B can functionally substitute for SUG1 in yeast and that FZA-B interacts with Fos proteins in vitro through their leucine zippers. In rat liver and in HeLa cells, FZA-B is present in the 26S proteasome complex, as is c-Fos. Immobilized antibody raised against an FZA-B-specific peptide depleted peptidase activity, proteasomal proteins, FZA-B, and c-Fos from a 26S proteasome preparation. FZA-B is found predominantly in the nuclear fraction of COS cells expressing an FZA-B transgene and in the nuclear 26S proteasome of HeLa cells. We conclude that FZA-B is the mammalian homolog of SUG1 (mSug1) and that it is present in the nuclear 26S proteasome of cells. Our results suggest that mSug1 may be involved in the degradation of c-Fos and other transcription factors.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akiyama K., Yokota K., Kagawa S., Shimbara N., DeMartino G. N., Slaughter C. A., Noda C., Tanaka K. cDNA cloning of a new putative ATPase subunit p45 of the human 26S proteasome, a homolog of yeast transcriptional factor Sug1p. FEBS Lett. 1995 Apr 17;363(1-2):151–156. doi: 10.1016/0014-5793(95)00304-r. [DOI] [PubMed] [Google Scholar]
- Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. Multiple ubiquitin-conjugating enzymes participate in the in vivo degradation of the yeast MAT alpha 2 repressor. Cell. 1993 Jul 30;74(2):357–369. doi: 10.1016/0092-8674(93)90426-q. [DOI] [PubMed] [Google Scholar]
- Chevray P. M., Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu-Ping M., Vu J. H., Proske R. J., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20 S proteasome. J Biol Chem. 1994 Feb 4;269(5):3539–3547. [PubMed] [Google Scholar]
- Curran T., Peters G., Van Beveren C., Teich N. M., Verma I. M. FBJ murine osteosarcoma virus: identification and molecular cloning of biologically active proviral DNA. J Virol. 1982 Nov;44(2):674–682. doi: 10.1128/jvi.44.2.674-682.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curran T., Teich N. M. Candidate product of the FBJ murine osteosarcoma virus oncogene: characterization of a 55,000-dalton phosphoprotein. J Virol. 1982 Apr;42(1):114–122. doi: 10.1128/jvi.42.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMartino G. N., Moomaw C. R., Zagnitko O. P., Proske R. J., Chu-Ping M., Afendis S. J., Swaffield J. C., Slaughter C. A. PA700, an ATP-dependent activator of the 20 S proteasome, is an ATPase containing multiple members of a nucleotide-binding protein family. J Biol Chem. 1994 Aug 19;269(33):20878–20884. [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Pratt G., Rechsteiner M. Subunit 4 of the 26 S protease is a member of a novel eukaryotic ATPase family. J Biol Chem. 1992 Nov 15;267(32):22699–22702. [PubMed] [Google Scholar]
- Dubiel W., Ferrell K., Rechsteiner M. Peptide sequencing identifies MSS1, a modulator of HIV Tat-mediated transactivation, as subunit 7 of the 26 S protease. FEBS Lett. 1993 Jun 1;323(3):276–278. doi: 10.1016/0014-5793(93)81356-5. [DOI] [PubMed] [Google Scholar]
- Eytan E., Ganoth D., Armon T., Hershko A. ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc Natl Acad Sci U S A. 1989 Oct;86(20):7751–7755. doi: 10.1073/pnas.86.20.7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Ghislain M., Udvardy A., Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993 Nov 25;366(6453):358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- Gordon C., McGurk G., Dillon P., Rosen C., Hastie N. D. Defective mitosis due to a mutation in the gene for a fission yeast 26S protease subunit. Nature. 1993 Nov 25;366(6453):355–357. doi: 10.1038/366355a0. [DOI] [PubMed] [Google Scholar]
- Goyer C., Lee H. S., Malo D., Sonenberg N. Isolation of a yeast gene encoding a protein homologous to the human Tat-binding protein TBP-1. DNA Cell Biol. 1992 Oct;11(8):579–585. doi: 10.1089/dna.1992.11.579. [DOI] [PubMed] [Google Scholar]
- Kadonaga J. T., Carner K. R., Masiarz F. R., Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987 Dec 24;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994 May 20;77(4):599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Lee J. W., Ryan F., Swaffield J. C., Johnston S. A., Moore D. D. Interaction of thyroid-hormone receptor with a conserved transcriptional mediator. Nature. 1995 Mar 2;374(6517):91–94. doi: 10.1038/374091a0. [DOI] [PubMed] [Google Scholar]
- Liu F., Green M. R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994 Apr 7;368(6471):520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- Ma C. P., Slaughter C. A., DeMartino G. N. Identification, purification, and characterization of a protein activator (PA28) of the 20 S proteasome (macropain). J Biol Chem. 1992 May 25;267(15):10515–10523. [PubMed] [Google Scholar]
- McGuire M. J., Reckelhoff J. F., Croall D. E., DeMartino G. N. An enzyme related to the high molecular weight multicatalytic proteinase, macropain, participates in a ubiquitin-mediated, ATP-stimulated proteolytic pathway in soluble extracts of BHK 21/C13 fibroblasts. Biochim Biophys Acta. 1988 Nov 17;967(2):195–203. doi: 10.1016/0304-4165(88)90009-8. [DOI] [PubMed] [Google Scholar]
- Nelbock P., Dillon P. J., Perkins A., Rosen C. A. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990 Jun 29;248(4963):1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- Ohana B., Moore P. A., Ruben S. M., Southgate C. D., Green M. R., Rosen C. A. The type 1 human immunodeficiency virus Tat binding protein is a transcriptional activator belonging to an additional family of evolutionarily conserved genes. Proc Natl Acad Sci U S A. 1993 Jan 1;90(1):138–142. doi: 10.1073/pnas.90.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombella V. J., Rando O. J., Goldberg A. L., Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994 Sep 9;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Papavassiliou A. G., Treier M., Chavrier C., Bohmann D. Targeted degradation of c-Fos, but not v-Fos, by a phosphorylation-dependent signal on c-Jun. Science. 1992 Dec 18;258(5090):1941–1944. doi: 10.1126/science.1470918. [DOI] [PubMed] [Google Scholar]
- Peters J. M., Franke W. W., Kleinschmidt J. A. Distinct 19 S and 20 S subcomplexes of the 26 S proteasome and their distribution in the nucleus and the cytoplasm. J Biol Chem. 1994 Mar 11;269(10):7709–7718. [PubMed] [Google Scholar]
- Rechsteiner M., Hoffman L., Dubiel W. The multicatalytic and 26 S proteases. J Biol Chem. 1993 Mar 25;268(9):6065–6068. [PubMed] [Google Scholar]
- Rubin D. M., Coux O., Wefes I., Hengartner C., Young R. A., Goldberg A. L., Finley D. Identification of the gal4 suppressor Sug1 as a subunit of the yeast 26S proteasome. Nature. 1996 Feb 15;379(6566):655–657. doi: 10.1038/379655a0. [DOI] [PubMed] [Google Scholar]
- Schaefer T. S., Sanders L. K., Nathans D. Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc Natl Acad Sci U S A. 1995 Sep 26;92(20):9097–9101. doi: 10.1073/pnas.92.20.9097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw D. R., Ennis H. L. Molecular cloning and developmental regulation of Dictyostelium discoideum homologues of the human and yeast HIV1 Tat-binding protein. Biochem Biophys Res Commun. 1993 Jun 30;193(3):1291–1296. doi: 10.1006/bbrc.1993.1765. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Irie K., Ninomiya-Tsuji J., Goebl M., Taniguchi T., Matsumoto K. New human gene encoding a positive modulator of HIV Tat-mediated transactivation. Nature. 1992 Jun 25;357(6380):700–702. doi: 10.1038/357700a0. [DOI] [PubMed] [Google Scholar]
- Stancovski I., Gonen H., Orian A., Schwartz A. L., Ciechanover A. Degradation of the proto-oncogene product c-Fos by the ubiquitin proteolytic system in vivo and in vitro: identification and characterization of the conjugating enzymes. Mol Cell Biol. 1995 Dec;15(12):7106–7116. doi: 10.1128/mcb.15.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffield J. C., Bromberg J. F., Johnston S. A. Alterations in a yeast protein resembling HIV Tat-binding protein relieve requirement for an acidic activation domain in GAL4. Nature. 1992 Jun 25;357(6380):698–700. doi: 10.1038/357698a0. [DOI] [PubMed] [Google Scholar]
- Treier M., Staszewski L. M., Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994 Sep 9;78(5):787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Udvardy A. Purification and characterization of a multiprotein component of the Drosophila 26 S (1500 kDa) proteolytic complex. J Biol Chem. 1993 Apr 25;268(12):9055–9062. [PubMed] [Google Scholar]
- vom Baur E., Zechel C., Heery D., Heine M. J., Garnier J. M., Vivat V., Le Douarin B., Gronemeyer H., Chambon P., Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and the putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996 Jan 2;15(1):110–124. [PMC free article] [PubMed] [Google Scholar]