Abstract
In the fruit fly Drosophila melanogaster, as in mammals, acute exposure to a high dose of ethanol leads to stereotypical behavioral changes beginning with increased activity, followed by incoordination, loss of postural control, and eventually, sedation. The mechanism(s) by which ethanol impacts the CNS leading to ethanol-induced sedation and the genes required for normal sedation sensitivity remain largely unknown. Here we identify the gene apontic (apt), an Myb/SANT-containing transcription factor that is required in the nervous system for normal sensitivity to ethanol sedation. Using genetic and behavioral analyses, we show that apt mediates sensitivity to ethanol sedation by acting in a small set of neurons that express Corazonin (Crz), a neuropeptide likely involved in the physiological response to stress. The activity of Crz neurons regulates the behavioral response to ethanol, as silencing and activating these neurons affects sedation sensitivity in opposite ways. Furthermore, this effect is mediated by Crz, as flies with reduced crz expression show reduced sensitivity to ethanol sedation. Finally, we find that both apt and crz are rapidly upregulated by acute ethanol exposure. Thus, we have identified two genes and a small set of peptidergic neurons that regulate sensitivity to ethanol-induced sedation. We propose that Apt regulates the activity of Crz neurons and/or release of the neuropeptide during ethanol exposure.
Introduction
Alcohol is a commonly abused drug, capable of altering the function of the CNS in humans and numerous animals. Studies indicate a strong genetic component to the risk for alcohol use disorders (AUDs), although few genes contributing to this risk have been identified (Goldman et al., 2005; Mayfield et al., 2008). Individuals with reduced sensitivity to the sedative effects of alcohol show increased risk for developing AUDs (Schuckit and Smith, 1996; Schuckit, 2000). Thus, identifying genes that influence alcohol sedation is critical for understanding the molecular basis for how alcohol impacts the CNS, as well as possibly mediating AUDs. In the fruit fly Drosophila melanogaster, as in mammals, acute exposure to high doses of alcohol initiate a series of behavioral changes beginning with increased locomotion, followed by incoordination and eventual sedation (Moore et al., 1998; Singh and Heberlein, 2000). Using genetic and neuroanatomical approaches, we identify two genes and a small set of peptidergic neurons that regulate ethanol-induced sedation: the transcriptional regulator apontic (apt), the neuropeptide corazonin (crz), and Crz-expressing neurosecretory cells.
Apt was first identified in Drosophila as a transcription factor involved in embryonic tissue morphogenesis (Eulenberg and Schuh, 1997; Gellon et al., 1997; Su et al., 1999). More recently, Apt was shown to promote synaptic function at the neuromuscular junction (Takasu-Ishikawa et al., 2001), and as a feedback inhibitor of the Jak/Stat pathway (Baeg et al., 2005; Starz-Gaiano et al., 2008). The human protein with the highest sequence homology to Apt is Fibrinogen silencer binding protein (FSBP), a negative regulator of transcription of the gamma chain of fibrinogen (Mizuguchi et al., 1995; Starz-Gaiano et al., 2008).
Crz is a neuropeptide that in insects and crustaceans is highly conserved with respect to structure and neuronal expression (Boerjan et al., 2010). However, the physiological functions of Crz vary considerably; for example, while cardiostimulatory effects have been observed in cockroaches (Veenstra, 1989), Crz induces cuticle pigmentation during the gregarious phase in locusts (Tawfik et al., 1999). In Drosophila, adult flies lacking Crz show altered trehalose homeostasis and triglyceride levels (Lee et al., 2008; Zhao et al., 2010), implicating Crz in fly metabolism and energy allocation. Crz function has also been linked to the fly's stress response, as crz transcription is inhibited by various stressors (Zhao et al., 2010). Additionally, Crz is thought to be the mammalian homolog of gonadotropin-releasing hormone (GnRH) (Cazzamali et al., 2002), which is directly regulated by stress hormones (Nikolarakis et al., 1986; Tellam et al., 1998; Li et al., 2004).
We identify Apt as a regulator of sensitivity to ethanol sedation. We show that Apt function in a small number of neurons, those expressing the neuropeptide Crz, is both necessary and sufficient for normal sensitivity to ethanol sedation. Ethanol sensitivity is also dependent on the activity of Crz neurons and the expression of the neuropeptide. We postulate that Apt and Crz, and/or the neurons in which they function, may be among the initial targets of ethanol that regulate sensitivity to its sedative effects.
Materials and Methods
Animals.
Flies were raised on standard cornmeal/molasses food at 25°C and 70% relative humidity and were grown in constant light, unless otherwise noted. Flies used for behavioral testing were backcrossed for five generations to the w1118 Berlin genetic background; exceptions are noted below. All behavioral experiments used 2- to 4-d-old males tested at ∼25°C; exceptions are noted below. For behavioral testing, each different genotype was tested over multiple days, from at least two independent sets of crosses.
Fly stocks.
apt13–66 flies were among the collection of ∼2000 mutant strains carrying P-element (PGawB; contains a GAL4 enhancer trap) insertions generated in the Heberlein lab. apt167/CyO, aptPΔ3/CyO, aptPΔ3/CyO, and UAS-apt (III) flies were obtained from Denise Montell (The Johns Hopkins University, Baltimore, MD; Eulenberg and Schuh, 1997; Starz-Gaiano et al., 2008). The apt167/CyO, aptPΔ3/CyO, and aptPΔ4/CyO flies were in the w1118 background on the X and III chromosomes. The aptGS11714 flies were obtained from the Drosophila Genetic Resource Center in Kyoto, Japan. elavC155-GAL4 (elav-GAL4), crz-GAL4, tub-GAL80ts, UAS-dcr2, UAS-Kir2.1, UAS-dTrpA1, UAS-aptRNAi2, and UAS-CrzRNAi2 flies were obtained from the Drosophila Stock Center in Bloomington, Illinois. UAS-aptRNAi (v4289) and UAS-crzRNAi (v106876) were obtained from the Vienna Drosophila RNAi Center (Dietzl et al., 2007). Both of the apt RNAi constructs (UAS-aptRNAi and UAS-aptRNAi2) have several predicted off-target effects. Ethanol sedation sensitivity was observed expressing UAS-aptRNAi with several pars intercerebralis (PI)-specific GAL4 drivers (12–10, 10–227, 10–260, and 7–56) of unknown cellular identities; these GAL4 drivers were generated in our laboratory. The PI is a region of the brain containing several peptidergic neurons (Siegmund and Korge, 2001).
Ethanol sedation assays.
Ethanol sedation assays were performed as previously described (Corl et al., 2009), with minor modifications. Briefly, 20 2- to 4-d-old males of each genotype raised at 25°C were allowed to equilibrate to room temperature (21−23°C) for 30 min, then introduced into “booze” tubes (long clear tubes) and placed into the booz-o-mat chamber. After 2–5 min exposure to humidified air to equilibrate the flies to the apparatus, a continuous stream of ethanol vapor (110 U ethanol/40 U air; 73% ethanol) was applied to the tubes for 30 min, unless noted otherwise. During the 30 min exposure period, flies were mechanically stimulated (by twirling the tubes) at 5 min intervals (seven time points). At each time point, the numbers of immobile flies were counted. The time to 50% sedation (ST50) was calculated as previously described (Rothenfluh et al., 2006).
All sedation assays were performed at ∼21−23°C; the only exceptions to this were experiments with TrpA1 activation and inactivation performed at 29 and 22°C, respectively. To avoid acute neuronal activation in the TrpA1 experiments, both the experimental and control flies were raised at 18°C and only removed from this temperature-controlled environment for ∼5–10 min before behavioral testing.
Real-time quantitative RT-PCR.
Quantitative PCR (qPCR) was performed as previously described (McClure et al., 2011). Primers used to amplify the five alternative apt transcripts, apt-RA, apt-RB, apt-RC, apt-RD, and apt-RE were as follows: apt-RA, 5′-TGCTTACACAACCAAGAAACCA-3′ and 5′-GGTGGCCGAATACTGTTTGT-3′; apt-RB, 5′-CAGTTCCTTTGGTTCTTGGAA-3′ and 5′-GGTGGCCGAATACTGTTTGT-3′; apt-RC, 5′-ATTCAAAAGGCTCGCGAATA-3′ and 5′-GCGGCAATCTTCATAAAAGC-3′; apt-RD, 5′-ACTTTTGCCGGAATTGTGTC-3′ and 5′-CCGAAAAGCGAGAGTTTTTG-3′; apt-RE, 5′-AGCTGCCAACCCCTTTTTAT-3′ and 5′-GGTGGCCGAATACTGTTTGT-3′. apt (common to all five transcripts), crz, and rp49 probe and primer sets (Dm01820107_g1, Dm02135830_g1, and Dm02151827_g1, respectively) were obtained from Applied Biosystems.
Molecular biology.
Genomic DNA flanking the apt13–66 and aptGS11714 insertions was isolated using inverse PCR. Comparison with the Drosophila genome sequence on Flybase (www.flybase.org) revealed that the insertions were located 295 and 180 base pairs upstream, respectively, of the apt-RA transcription start site. The P-element in apt13–66 flies was precisely excised using the Δ2–3Sb transposase (Robertson et al., 1988). Primers used to confirm the precise excision were as follows: 5′-AGCGAGAAAAGCAAAGACGA-3′ and 5′-TTCGCACTGCTTCACGTTTA-3′.
Immunohistochemistry.
Whole-mount brains were fixed at room temperature in 4% paraformaldehyde in PBS. The neuropile-specific antibody NC82 was used at 1:50 (The Jackson Laboratory), mouse-anti-Fasciclin II was used at 1:200 (Drosophila Hybridoma Bank), rabbit-anti-GFP was used at 1:200 (Clontech), mouse-anti-GFP was used at 1:200 (Roche), rabbit-anti-Apontic was used at 1:100 (Eulenberg and Schuh, 1997), and rabbit-anti-Crz was used at 1:500 (Veenstra and Davis, 1993).
Ethanol concentration assays.
Internal ethanol concentrations were measured in homogenized adult extracts from wB and apt13–66 flies exposed to a sedating dose of ethanol (73% ethanol). Animals were exposed to vaporized ethanol for 0, 5, 10, 20, and 30 min then snap-frozen in liquid nitrogen and assayed for ethanol content using a colorimetric enzymatic kit (Diagnostic Chemicals) (Moore et al., 1998).
Statistics.
Statistics were performed using JMP 7.0.1 2007 (SAS Institute). Statistical significance was established using one-way ANOVA, followed by Tukey–Kramer post hoc tests for multiple comparisons. Error bars indicate SEM. Statistical significance (p < 0.05) is given when the experimental group differs both from the GAL4 and UAS transgene controls. In all graphs, ***p < 0.0001, **p < 0.001 and, *p < 0.01.
Results
13–66 flies show reduced sensitivity to ethanol sedation
We screened a collection of mutant strains, each carrying individual P-element insertions, for altered sensitivity to ethanol-induced sedation (data not shown). Flies exposed to a high concentration of ethanol (73% ethanol vapor) lose postural control and exhibit a loss of righting reflex (LORR) (Rothenfluh et al., 2006). To determine the fly's ethanol sedation sensitivity, we measured the time needed for 50% of the flies to lose their LORR (time to 50% sedation, ST50). One mutant fly strain, 13–66, displayed a dramatic reduction in ethanol sedation sensitivity, with a significantly increased ST50 compared with control flies (Fig. 1A,B; F(1,51) = 42.59, p < 0.0001). The reduced sensitivity to ethanol sedation displayed by 13–66 flies was not due to altered ethanol pharmacokinetics, as control and mutant flies contained similar internal ethanol concentration when measured after different times of ethanol exposure (data not shown).
Figure 1.
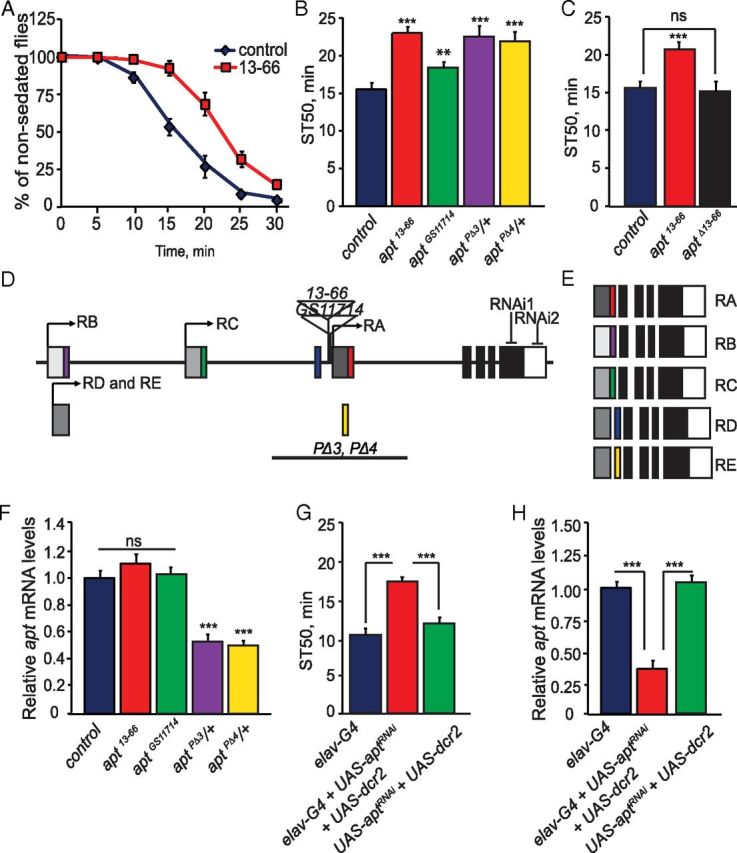
apt mutants display reduced ethanol sedation sensitivity. A, Control (wB) and apt13–66 flies were exposed to a sedating dose of ethanol (73% ethanol vapor) for 30 min. The number of sedated flies was measured over the course of the exposure. B, Quantification of ethanol sedation sensitivity in different apt mutant strains. The time at which 50% of the flies were sedated, the mean sedation time (ST50), was calculated by linear interpolation. ST50 values of apt mutant strains differed significantly from control (wB) flies (one-way ANOVA, Tukey–Kramer post hoc, n = 16). C, Precise excision of the P-element in apt13–66 flies (aptΔ13–66) completely reverted the ethanol sedation sensitivity phenotype. (Student's t test, n = 12) D, E, Schematic of the apt gene. The apt gene is predicted to encode for five alternative transcripts, producing five distinct protein isoforms. Transcription start sites for each of the transcripts are indicated by arrows; the same transcription start site is used for both the apt-RD and apt-RE transcripts. Translated regions are indicated in colors and black, while untranslated regions are in shades of gray. Locations of the P-element insertions in apt13–66 and aptGS11714 mutant flies are indicated; both insertions are upstream of the apt-RA transcript. Locations of the inverted-repeat sequences used to generate RNAi knockdown of apt and the location of sequences deleted in the aptPΔ3 and aptPΔ4 mutant flies are also shown. F, Expression of apt in control flies (wB) and apt13–66, aptGS11714, aptPΔ3/+, and aptPΔ4/+ mutant flies as quantified by qPCR. No difference in apt expression was observed between control (wB) and apt13–66 and aptGS11714 mutant flies, while decreased apt expression to that of 50 and 55% was observed in aptPΔ3/+ and aptPΔ4/+ mutant flies, respectively. (Student's t test, n = 3). G, RNAi knockdown of apt specifically in neurons reduces ethanol sedation sensitivity. ST50 values of flies coexpressing dcr2 and aptRNAi using pan-neuronal driver elavC155-GAL4 (elavC155-GAL4;UAS-dcr2/+;UAS-aptRNAi/+) differs significantly from the GAL4 (elavC155-GAL4) and UAS (UAS-dcr2/+;UAS-aptRNAi/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 16). H, apt expression upon RNAi knockdown. Pan-neuronal expression of aptRNAi (elavC155-GAL4;UAS-dcr2/+;UAS-aptRNAi/+) reduces apt expression by 65% compared with the GAL4 (elavC155-GAL4) and UAS (UAS-dcr2/+;UAS-aptRNAi/+) controls. One-way ANOVA, Tukey–Kramer post hoc, n = 3). For this figure and all other figures, error bars indicate SEM and p values are ***p < 0.0001, **p < 0.001, and *p < 0.01.
Molecular characterization of the apt locus and apt mutants
The P-element in 13–66 flies was mapped by inverse PCR and DNA sequence analysis to the gene apontic (apt) (Fig. 1D). The P-element insertion in 13–66 flies is responsible for the reduced sedation sensitivity, since precise excision of the transposon caused complete reversion to the wild-type phenotype (Fig. 1C). Several additional alleles of apt also displayed reduced ethanol sedation sensitivity, including another P-element insertion, aptGS11714 (F(1,42) = 13.05, p = 0.0008), and two alleles carrying genomic deletions in the gene, aptPΔ3/+ (F(1,43) = 54.52, p < 0.0001) and aptPΔ4/+ (F(1,42) = 35.90, p < 0.0001); the latter were tested as heterozygotes since homozygous mutant flies are not viable as adults (Fig. 1B) (Eulenberg and Schuh, 1997). To determine whether apt function is required in neurons for normal ethanol sedation sensitivity, we downregulated neuronal-specific expression of apt by RNA interference (RNAi). Pan-neuronal expression (using elav-GAL4) of a transgene (UAS-aptRNAi) that targets apt by RNAi, along with UAS-dicer2 (UAS-dcr2), a component of the RNAi machinery shown to increase RNAi efficacy (Dietzl et al., 2007), caused a significant reduction in ethanol sedation sensitivity relative to control flies (Fig. 1G; F(2,27) = 46.96, p < 0.0001). Importantly, apt transcript levels in flies with pan-neuronal expression of aptRNAi (elav-GAL4;UAS-dcr2/+;UAS-aptRNAi/+) were reduced by ∼65% compared with control flies expressing the individual transgenes (Fig. 1H; F(2,27) = 49.06, p < 0.0001). In addition, pan-neuronal expression of a second, nonoverlapping aptRNAi2 transgene without dcr2 also caused the same phenotype (data not shown; Fig. 1D). These data demonstrate that neuronal expression of apt is required for normal ethanol sedation sensitivity.
The apt locus encompasses 35 kb and is predicted to encode five transcripts and five distinct protein isoforms (Fig. 1D,E). The five Apt proteins differ in their N termini, but share the same Myb/SANT motif, a DNA binding domain; previous work has shown that Apt binds DNA (Liu et al., 2003). Three of the apt transcripts use unique transcription start sites, while the apt-RD and apt-RE transcripts use a common start site (Fig. 1D). Locations of the P-element insertions in apt13–66 and aptGS11714 flies are 295 and 180 base pairs upstream, respectively, of the apt-RA transcript (Fig. 1D). The two apt genomic DNA deletion mutants, aptPΔ3/+ and aptPΔ4/+, remove an exon from the apt-RA, apt-RD, and apt-RE transcripts (Eulenberg and Schuh, 1997; Starz-Gaiano et al., 2008). To determine how these mutations affect apt expression, we measured apt transcript levels in all mutant and control strains by qPCR. Primer sets recognizing each of the five apt transcripts revealed that apt-RA is the sole apt transcript expressed in the adult fly head (data not shown). Using primers specific to the apt-RA transcript and a primer/probe set that amplifies all five apt transcripts, we found similar expression of apt in the adult heads of apt13–66 (t test, p = 0.1433), aptGS11714 (t test, p = 0.167), and wild-type control flies (Fig. 1F). However, the apt deletion mutants, aptPΔ3/+ and aptPΔ4/+, showed a near 50% reduction of apt expression compared with control flies (Fig. 1F; aptPΔ3/+, p = 0.00093 and aptPΔ4/+, p = 0.00089 by t test). The apparent lack of reduction of apt transcript levels in two of the apt mutant strains (apt13–66 and aptGS11714), both showing reduced ethanol sedation sensitivity phenotypes, may be explained by apt expression being affected in only a small subset of neurons in the brain, a difference that would be undetectable by qPCR (see below). Importantly, both apt13–66 and aptGS11714 fail to complement the embryonic lethality of a known null allele of apt, apt167 (Eulenberg and Schuh, 1997), indicating that apt13–66 and aptGS11714 indeed affect apt and are likely hypomorphic mutations.
In summary, we have shown that reduced apt expression correlates with decreased ethanol sedation sensitivity. To establish a causal relationship we performed phenotypic rescue experiments that are described below.
apt is required in neurons during both metamorphosis and adulthood for normal ethanol sedation sensitivity
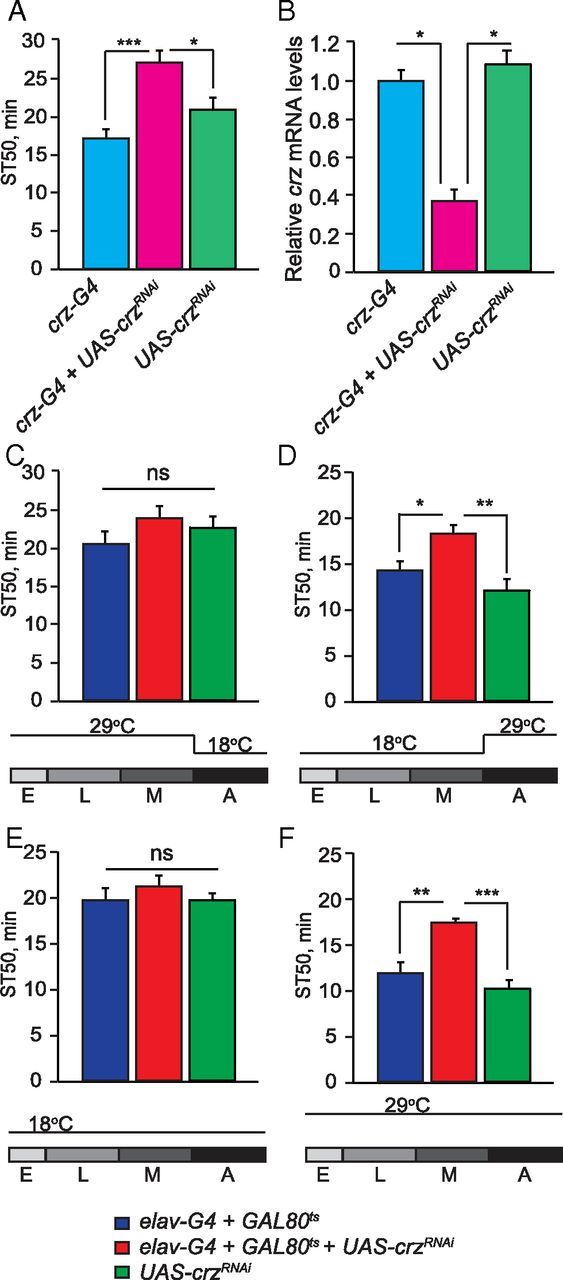
We have shown that neuronal expression of apt is required for normal ethanol sedation sensitivity (Fig. 1G). To determine when apt function is required in neurons to mediate this effect, we inhibited neuronal apt expression in a temporally specific manner using GAL80ts, which represses GAL4 function at 18°C but not at 29°C (McGuire et al., 2003). We used RNAi to inhibit apt expression specifically during development (by raising flies at 29°C and shifting them to 18°C after adult eclosion) or specifically during adulthood (by raising flies at 18°C and shifting them to 29°C after eclosion). Interestingly, both manipulations decreased sensitivity to ethanol sedation, suggesting that apt function during both development and adulthood is required for normal sedation sensitivity (Fig. 2A,B; development-specific inhibition of apt, F(2,28) = 14.08, p < 0.0001; adult-specific inhibition of apt, F(2,49) = 8.02, p = 0.001). Thus, apt may have a role in regulating the development of the adult nervous system as well as its function. To more precisely define the developmental period during which apt function is required, we inhibited apt expression during specific stages of development (Fig. 2C,D). Neuronal inhibition of apt expression solely during metamorphosis, a period of intense remodeling of the developing adult fly CNS (Truman, 1990), caused a striking decrease in ethanol sedation sensitivity (Fig. 2D; F(2,32) = 25.55, p < 0.0001). Remodeling of the adult fly CNS occurs mainly during metamorphosis; however, changes to the nervous system continue after adult eclosion (Truman, 1990). To determine whether apt function is specifically required during remodeling of the adult CNS (both during development and after eclosion) and/or in the mature adult CNS, we inhibited its expression during development and the initial 3 d after adult eclosion (by raising flies at 29°C and shifting them to 18°C 3 d after eclosion); we also inhibited apt expression during days 3–7 of adulthood (by raising flies at 18°C and shifting them to 29°C on the third day after eclosion). Both manipulations caused decreased sensitivity to ethanol sedation, suggesting that apt function in sedation sensitivity is required during remodeling of the adult CNS and in the mature adult CNS (Fig. 2E,F; inhibition of apt during CNS remodeling, F(2,24) = 6.08, p = 0.0034; inhibition of apt during adulthood, F(2,24) = 5.03, p = 0.01). Importantly, flies in which apt expression was presumably normal (maintained at 18°C during both development and adulthood) showed normal sedation sensitivity (Fig. 2G; F(2,25) = 1.58, p = 0.23), while flies with continuous neuronal inhibition of apt expression (maintained at 29°C throughout development and adulthood) had significantly reduced ethanol sedation sensitivity (Fig. 2H; F(2,28) = 22.95, p < 0.0001). From these results we conclude that apt function is required in neurons during two distinct life stages, metamorphosis and adulthood, to promote normal sensitivity to ethanol sedation.
Figure 2.
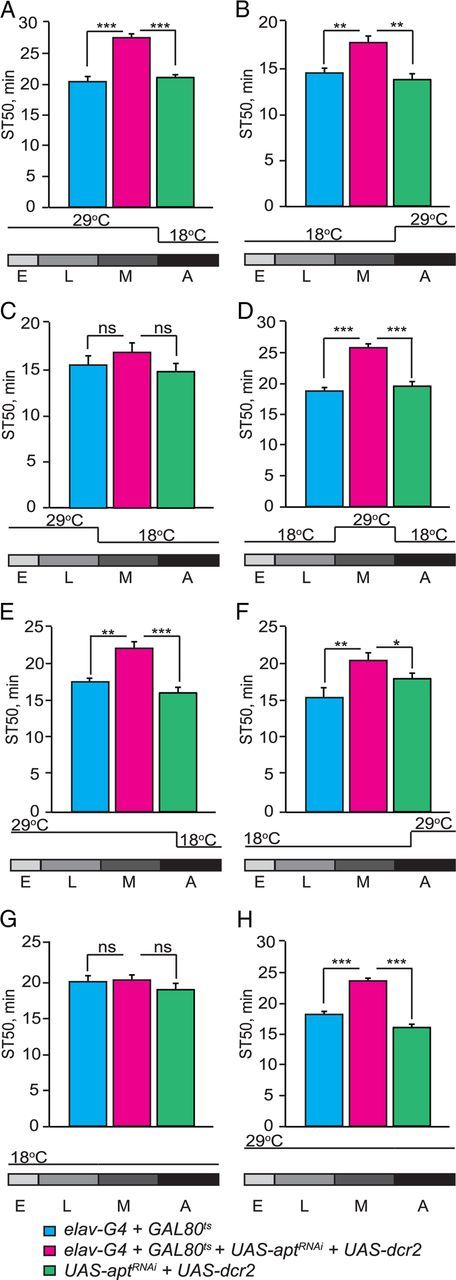
apt is required in neurons during both metamorphosis and adulthood for normal ethanol sedation sensitivity. A–D, Temporal knockdown of apt expression was achieved by coexpressing UAS-dcr2 and UAS-aptRNAi with elavc155-GAL4 and tub-GAL80ts. Flies were raised at 29 (GAL4 on, GAL80ts off) and 18°C (GAL4 off; GAL80ts on). ST50 values are shown for the experimental group with temporal inhibition of apt expression (elavc155-GAL4;UAS-dcr2/tub-GAL80ts;UAS-aptRNAi/+), the GAL4-GAL80ts (elavc155-GAL4; tub-GAL80ts /+), and UAS (UAS-dcr2/+;UAS-aptRNAi /+) controls. A, Neuronal inhibition of apt expression during development significantly reduces ethanol sedation sensitivity. Flies were raised at 29°C then shifted to 18°C upon adult eclosion until behavioral testing. B, Neuronal inhibition of apt expression during adulthood reduces ethanol sedation sensitivity. Flies were raised at 18°C then shifted to 29°C upon adult eclosion until behavioral testing. C–F, Temporal mapping of apt function during development. C, Neuronal inhibition of apt expression during embryogenesis and larval development does not alter ethanol sedation sensitivity (p > 0.05, n = 12). Flies were raised at 29°C then shifted to 18°C from the start of metamorphosis until behavioral testing. D, Neuronal inhibition of apt expression during metamorphosis reduces ethanol sedation sensitivity. Flies were raised at 18°C, then shifted to 29°C at the beginning of metamorphosis, and shifted back to 18°C upon adult eclosion until behavioral testing. E, Neuronal inhibition of apt expression during development and three initial days after eclosion reduces ethanol sedation sensitivity. Flies were raised at 29°C until the third day after eclosion then shifted to 18°C for 4 d. F, Neuronal inhibition of apt expression during adulthood, specifically days 3–7 of adulthood, decreases ethanol sedation sensitivity. Flies were raised at 18°C until the third day after eclosion, then shifted to 29°C for 4 d. G, H, Show temperature shift controls for mapping neuronal apt function. G, Raising flies at 18°C during both development and adulthood (GAL4 off) leads to normal ethanol sedation sensitivity (p > 0.05, n = 12). H, Raising flies at 29°C during both development and adulthood (GAL4 on) leads to decreased ethanol sedation sensitivity. (For all parts shown, one-way ANOVA, Tukey–Kramer post hoc, n = 16.)
apt13–66-GAL4 and Apt expression in the adult fly brain
To begin identifying the apt-expressing cells that regulate ethanol-induced sedation, we examined the expression of Apt in the adult brain. The transposon inserted into the apt locus in apt13–66 flies contains the transcriptional activator GAL4; thus, the cellular expression of GAL4 in apt13–66 flies likely reflects endogenous Apt expression. Flies carrying apt13–66-GAL4 as well as GFP reporters that allow localization of nuclei and neuronal projections [UAS-GFP (nls), UAS-mCD8GFP], showed GFP expression in the adult CNS that was limited to antennal lobe interneurons (AL-in), ventral neurons in the subesophageal ganglion (SOG), neurons in the PI, pars lateralis (PL), and some lateral neurons (Fig. 3A,B). The apt13–66-GAL4 expression pattern in the brain partially overlapped with Apt immunoreactivity in the AL-in, SOG, PI, PL, and lateral neurons (Fig. 3C–F). However, the Apt antibody detected additional cells in all of these brain regions, indicating that the apt13–66-GAL4 expression pattern, although more restricted, reproduces in part the endogenous Apt expression pattern.
Figure 3.
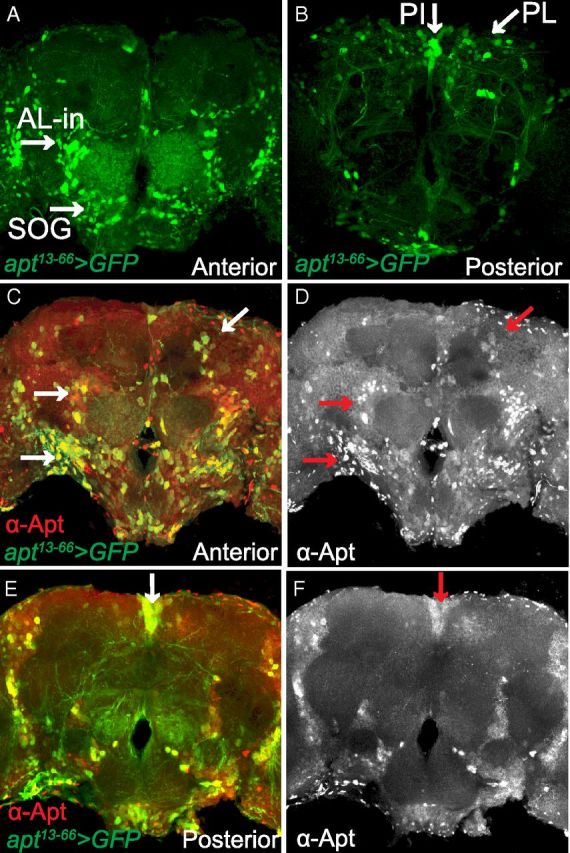
Apt expression in the adult CNS. A, B, Whole-mount brain from an adult male fly apt13–66-GAL4/UAS-GFP (nls);UAS-mCD8GFP/+ stained with anti-GFP. Confocal projections are from the anterior (A) and posterior (B) regions of the brain. A, In the anterior brain, apt13-66-GAL4 expression (green) is found in AL-in and neurons in the SOG. B, In the posterior brain, apt13–66-GAL4 expression is observed in neurons of the PL and PI. C–F, Whole-mount brain from an adult male fly apt13–66-GAL4/UAS-GFP (nls);UAS-mCD8GFP/+ stained with anti-GFP (green) and anti-Apt (red). Projections are from the anterior (C, D) and posterior (E, F) regions of the brain. C, E, There is colocalization of apt13–66-GAL4 (green) and Apt immunoreactivity (red) in the AL-in, SOG, PL, and PI (yellow; white arrows). D, F, Anterior and posterior projections showing the expression pattern of Apt in the Al-in, SOG, PL, and PI (red arrows).
To examine the brain anatomy of apt13–66 mutant flies we used the NC82 antibody, which labels synapses and thus neuropil (Laissue et al., 1999), and an antibody that recognizes Fasciclin II, a cell adhesion molecule that labels neurons of the ellipsoid body and mushroom body α/β and γ lobes (Cunningham et al., 1987; King et al., 2011). The gross morphology of the CNS in apt13–66 flies was normal compared with control flies (data not shown).
apt is required in corazonin-expressing neurons for normal ethanol sedation sensitivity
To define specific neurons in which apt function is required to regulate ethanol-induced sedation, we screened a collection of 22 GAL4 lines using UAS-aptRNAi. We selected GAL4 lines with expression limited to one or more of the following brain regions observed in the apt13–66-GAL4 and Apt expression patterns: AL-in; ventral neurons in the SOG; and neurons in the PI, PL, and lateral neurons. Since both the apt13–66-GAL4 and the Apt antibody reveal prominent expression in the PI and PL (Fig. 3), regions containing several peptidergic neurons (Siegmund and Korge, 2001), we screened additional GAL4 lines that drive expression in various classes of neuropeptidergic cells. A significant reduction in ethanol sedation sensitivity was observed when expressing aptRNAi using several PI- and PL-specific drivers (see Materials and Methods) of unknown cellular specificity. More intriguingly, however, ethanol sedation sensitivity was dramatically reduced when expressing UAS-aptRNAi with the neuropeptidergic driver corazonin-GAL4 (crz-GAL4) (Fig. 4A; F(2,24) = 22.87, p < 0.0001), indicating that Apt function is required in crz-expressing neurons.
Figure 4.
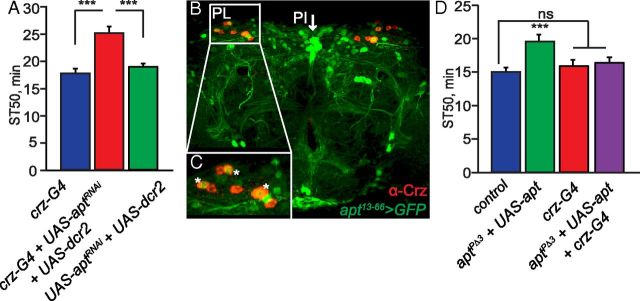
Requirement of apt function in crz-expressing neurons for normal ethanol sedation sensitivity. A, Inhibition of apt expression by RNAi in crz-expressing neurons reduces ethanol sedation sensitivity. ST50 values of flies with knockdown of apt expression in crz-expressing neurons (UAS-dcr2/+;crz-GAL4/UAS-aptRNAi) differ significantly from GAL4 (crz-GAL4/+) and UAS (UAS-dcr2/+;UAS-aptRNAi/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 16). B, C, Whole-mount brain from adult male fly apt13–66-GAL4/UAS-GFP (nls) stained with anti-GFP and anti-Crz antibodies. B, In the adult fly brain, Crz (red) is expressed in 6–8 neurons per lobe in the PL (Lee et al., 2008). In each lobe, there is colocalization of apt13–66-GAL4 (green) and Crz (red) in approximately three of the Crz-expressing neurons (yellow). C, Enlargement of region marked in B (white box) showing colocalization between apt13–66-GAL4 and cells labeled with the Crz antibody. D, Phenotypic rescue of the reduced ethanol sedation sensitivity in aptPΔ3/+ mutant flies by driving UAS-apt in crz-expressing neurons. ST50 values differ significantly between aptPΔ3/+ mutant flies with UAS-apt (aptPΔ3/+;UAS-apt/+) and aptPΔ3/+ mutant flies driving UAS-apt in crz-expressing neurons (aptPΔ3/+;UAS-apt/crz-GAL4) (one-way ANOVA, Tukey–Kramer post hoc, n = 12). ST50 values of aptPΔ3/+ mutant flies driving UAS-apt in crz-expressing neurons (aptPΔ3/+;UAS-apt/crz-GAL4) does not differ significantly from control (wB) or GAL4 (crz-GAL4/+) alone control flies (p > 0.05, n = 12).
To verify that Apt functions in crz-expressing cells, we first examined whether Apt was expressed in these neurons in the adult brain. In the Drosophila adult CNS, the neuropeptide Crz is expressed in 6–8 bilaterally symmetric neurons in the PL (Fig. 4B), and in a cluster of four abdominal ganglion neurons present only in male flies (Lee et al., 2008). Together, the PI and PL comprise the central neuroendocrine system in the Drosophila brain (de Velasco et al., 2007). We used the apt13–66-GAL4 driver to express a GFP reporter that drives GFP expression in nuclei [UAS-GFP (nls)] and immunostained the adult brains with antibodies recognizing both GFP and Crz. We observed three apt13–66-GAL4-positive neurons, in the PL, which also express the neuropeptide Crz (Fig. 4B,C).
We previously showed that apt function is necessary in crz-expressing neurons to mediate normal ethanol sedation (Fig. 4A). To determine whether apt function in these cells is sufficient to regulate this behavior, we examined whether expression of UAS-apt specifically in crz-expressing neurons would rescue the sedation resistance of aptPΔ3/+ mutant flies (aptPΔ3/+;crz-GAL4/UAS-apt) (Fig. 4D). Indeed, these flies showed normal ethanol sedation sensitivity (Fig. 4D; F(3,61) = 10.84, p = 0.839), showing that increasing expression of apt in Crz neurons is sufficient for normal ethanol sensitivity. When introducing the UAS-apt transgene into aptPΔ3/+ mutant flies (aptPΔ3/+;UAS-apt/+), which lacks expression of GAL4, no behavioral rescue was achieved (Fig. 4D; F(3,61) = 10.84, p < 0.0001). These results imply that reduced expression of apt specifically in crz-expressing neurons is responsible for the reduced ethanol sensitivity observed in apt mutant flies.
Activity of crz-expressing neurons mediates normal ethanol sedation sensitivity
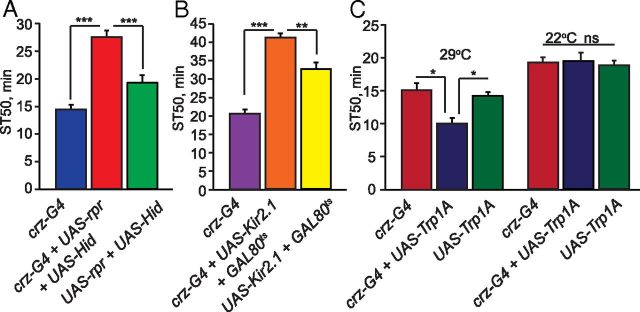
We next asked whether crz-expressing neurons are required for normal ethanol-induced sedation. First, we tested whether ablation of crz neurons, using transgenes encoding the pro-apoptotic genes reaper (rpr) and Head involution defective (Hid) (UAS-rpr and UAS-Hid), affected ethanol sedation. Flies in which these transgenes were driven by crz-GAL4 showed a complete loss of Crz immunolabeling in the adult brain (data not shown), and also displayed a significant reduction in sensitivity to ethanol sedation (Fig. 5A; F(2,28) = 75.45, p < 0.0001) indicating that crz-expressing neurons and possibly Crz signaling regulate sensitivity to ethanol sedation.
Figure 5.
Activity of crz-expressing neurons mediates normal ethanol sedation sensitivity. A, Ablation of crz-expressing neurons reduces ethanol sedation sensitivity. ST50 values of flies in which expression of the pro-apoptotic genes rpr and Hid is driven with crz-GAL4 (UAS-rpr;;UAS-Hid/crz-GAL4) differ significantly from GAL4 (crz-GAL4/+) and UAS (UAS-rpr;;UAS-Hid/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 12). B, Adult-specific silencing of crz-expressing neurons reduces ethanol sedation sensitivity. crz-expressing neurons were silenced specifically during adulthood by driving Kir2.1 with crz-GAL4;tub-GAL80ts. The experimental (tub-GAL80ts/+;UAS-Kir2.1/crz-GAL4), GAL80ts-UAS (tub-GAL80ts/+;UAS-Kir2.1/+), and GAL4 (crz-GAL4/+) alone control groups were raised at 18°C then shifted to 29°C at eclosion until behavioral testing. ST50 values of flies with adult-specific silencing of crz-expressing neurons using Kir2.1 and crz-GAL4;GAL80ts (tub-GAL80ts/+;UAS-Kir2.1/crz-GAL4) differ significantly from the GAL80ts-UAS (tub-GAL80ts/+;UAS-Kir2.1/+) and GAL4 (crz-GAL4/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 12). C, Adult-specific activation of crz-expressing neurons increases ethanol sedation sensitivity. The experimental flies (UAS-TrpA/Crz-GAL4) as well as the control GAL4 (Crz-GAL4/+) and UAS (UAS-TrpA) flies were raised at 18°C until behavioral testing, then shifted to 29°C, a temperature for which TrpA1 is activated, during the sedation assay. ST50 values of flies with adult-specific expression of TrpA1 in crz-expressing neurons (UAS-TrpA/Crz-GAL4), tested at 29°C, significantly increased ethanol sedation sensitivity compared with the GAL4 (crz-GAL4/+) and UAS (UAS-TrpA/+) alone control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 12). Temperature controls for TrpA activation in crz-expressing neurons (22°C) were used. The experimental and control flies were raised at 18°C until behavioral testing then shifted 22°C, a temperature for which TrpA is not activated, during the sedation assay. At this temperature the ST50 values of the experimental group (UAS-TrpA/Crz-GAL4) did not differ from the GAL4 (Crz-GAL4/+) and UAS (UAS-TrpA/+) alone control flies. (p > 0.05, n = 8).
To determine whether the activity of crz-expressing neurons regulates ethanol sedation sensitivity, we introduced transgenes driving the expression of either Kir2.1 or dTrpA1, which decrease or increase neuronal activity, respectively, to Crz neurons. Expression of UAS-Kir2.1, an inward rectifying K+-channel that causes neuronal hyperpolarization (Baines et al., 2001), driven by crz-GAL4, in adult flies (using GAL80ts) caused a marked reduction in ethanol sedation sensitivity (Fig. 5B; F(2,45) = 40.76, p < 0.0001). Conversely, acute activation of crz-expressing neurons using the temperature-gated cation channel dTrpA1 (Hamada et al., 2008) (crz-GAL4;UAS-dTrpA1) led to increased ethanol sedation sensitivity compared with controls; specifically, increased sensitivity was observed when TrpA1 was active at 29°C (F(2,39) = 6.69, p = 0.0033), but not when inactive at 22°C (F(2,24) = 0.52, p = 0.604) (Fig. 5C). Together, these experiments show that silencing and activating crz-expressing neurons causes opposite ethanol sedation sensitivity phenotypes, suggesting that the activity of crz-expressing neurons directly promotes ethanol sedation sensitivity.
The neuropeptide crz is required for normal ethanol sedation sensitivity
We next asked whether the neuropeptide Crz is involved in the regulation of ethanol sedation sensitivity. To do so, we inhibited the expression of crz by RNAi. We found that RNAi-mediated downregulation of crz in either crz-expressing neurons or pan-neuronally (using the crz-GAL4 and elav-GAL4 drivers, respectively), in the absence of dcr2, caused a significant reduction in ethanol sedation sensitivity (Fig. 6A; F(2,57) = 14.86, p < 0.0001; data not shown). Importantly, crz transcript levels in the heads of crz-GAL4/UAS-crzRNAi and elav-GAL4;UAS-crzRNAi/+ flies were reduced by ∼62 and 90% compared with control flies, respectively (Fig. 6B; F(2,12) = 10.97, p = 0.0039; data not shown). In addition, expression of a second, nonoverlapping CrzRNAi2 transgene in crz-expressing neurons caused the same decreased sedation sensitivity phenotype (data not shown; Fig. 6A). To determine the temporal requirements of crz function, we inhibited neuronal expression of crz by RNAi specifically during development or adulthood (using elav-GAL4 and GAL80ts) and found that only adult-specific inhibition of crz led to decreased sensitivity to ethanol sedation (Fig. 6C,D; F(2,43) = 9.91, p = 0.003), suggesting that Crz function during adulthood is required for normal sedation sensitivity. Flies with no manipulation of crz expression (maintained at 18°C during both development and adulthood) showed normal sedation sensitivity (Fig. 6E; F(2,30) = 1.54, p = 0.2313), while continuous neuronal inhibition of crz expression (flies maintained at 29°C throughout development and adulthood) caused significantly reduced ethanol sedation sensitivity (Fig. 6F; F(2,32) = 8.77, p = 0.005). These data suggest that Crz may play a role in regulating adult neuronal function during acute ethanol exposure. Moreover, ethanol-induced sedation is mediated in the adult by both the neuropeptide Crz and the activity of crz-expressing neurons.
Figure 6.
The neuropeptide Crz mediates ethanol sedation sensitivity. A, RNAi knockdown of crz specifically in crz-expressing neurons reduces ethanol sedation sensitivity. ST50 values of flies in which UAS-crzRNAi is driven with crz-GAL4 (crz-GAL4/UAS-crzRNAi/+) differ significantly from the GAL4 (crz-GAL4/+) and UAS (UAS-crzRNAi/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 16). B, Expression of crz upon RNAi knockdown. Flies with neuronal knockdown of crz by RNAi, specifically in crz-expressing neurons (crz-GAL4/UAS-crzRNAi/+), showed a 62% reduction of crz transcript levels compared with the GAL4 (crz-GAL4/+) and UAS (UAS-crzRNAi/+) control flies (one-way ANOVA, Tukey–Kramer post hoc, n = 3). C, D, Temporal knockdown of crz expression was achieved by coexpressing UAS-crzRNAi with elavc155-GAL4;tub-GAL80ts. Flies were raised at 29 (GAL4 on, GAL80ts off) and 18°C (GAL4 off; GAL80ts on). ST50 values are shown for the experimental group with temporal inhibition of crz expression (elavc155-GAL4;tub-GAL80ts/UAS-crzRNAi), and the GAL4-GAL80ts (elavc155-GAL4;tub-GAL80ts /+), and UAS (UAS-crzRNAi /+) controls. C, Neuronal inhibition of crz expression during development does not alter ethanol sedation sensitivity. (p > 0.05, n = 12). Flies were raised at 29°C then shifted to 18°C at eclosion until behavioral testing. D, Neuronal inhibition of crz expression during adulthood reduces ethanol sedation sensitivity (one-way ANOVA, Tukey–Kramer post hoc, n = 12). Flies were raised at 18°C then shifted to 29°C from the time of adult eclosion until behavioral testing. E, F, Temperature shift controls. E, Raising flies at 18°C during both development and adulthood (GAL4 off) leads to normal ethanol sedation sensitivity. (p > 0.05, n = 12). F, Raising flies at 29°C during both development and adulthood (GAL4 on) leads to decreased ethanol sedation sensitivity (one-way ANOVA, Tukey–Kramer post hoc, n = 16).
Acute ethanol exposure increases transcription of both the apt and crz genes
To determine whether apt and/or crz expression is acutely regulated by ethanol, we examined apt and crz transcript levels in the heads of wild-type flies exposed to ethanol as in our sedation assay. We observed an 85% increase in apt transcript levels after 15 min of ethanol exposure (t test, p = 0.0028), and modestly elevated transcript levels by the end of the exposure period (Fig. 7A; t test, p = 0.0343). A similar pattern was observed with crz transcript levels: after 15 min of ethanol exposure crz expression increased by 25% (t test, p = 0.0432) and was slightly more elevated by the end of the 30 min exposure (t test, p = 0.0378) (Fig. 7B). Interestingly, both crz and apt transcript levels were still elevated after 1 h of recovery from the ethanol exposure (Fig. 7A,B; apt t test, p = 0.0279 and crz t test, p = 0.0321). In summary, acute ethanol exposure rapidly upregulates the transcription of both apt and crz, and these results are consistent with our findings that both genes function to promote ethanol sedation sensitivity.
Figure 7.
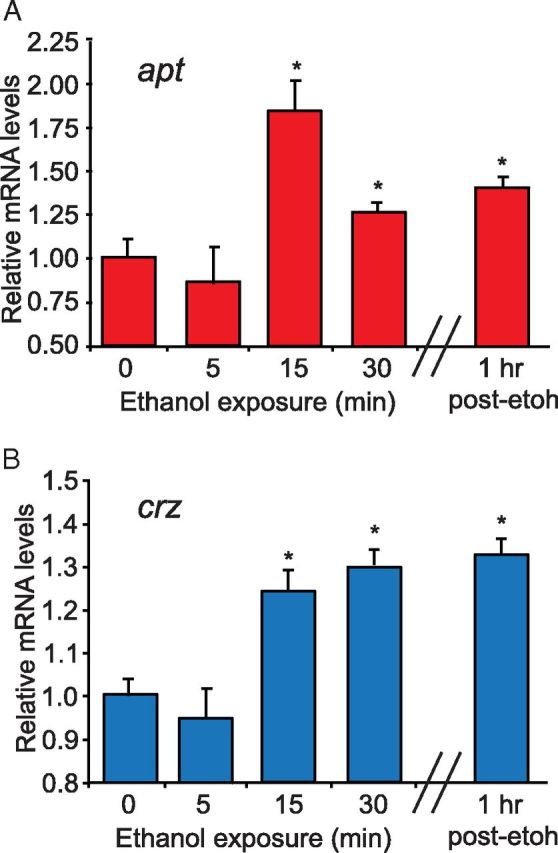
Acute ethanol exposure increases both apt and crz transcript levels. A, B, apt and crz transcript levels measured by qPCR in the heads of wild-type (wB) flies exposed to ethanol (73% ethanol vapor) for the times indicated. qPCR revealed that both apt and crz transcript levels increased after 15 min of ethanol exposure and remained elevated at the end of the exposure period as well as 1 h later. (Student's t test, n = 3).
Discussion
The work presented here identifies two genes, apt and crz, which mediate the fly's sensitivity to ethanol-induced sedation. Flies with reduced expression of either gene display dramatically decreased ethanol sedation sensitivity; thus, both apt and crz normally promote ethanol sedation. We show that normal sensitivity to ethanol sedation requires apt expression in neurons during two distinct life stages, metamorphosis and adulthood. Apt function in a subset of crz-expressing neurons (approximately 6 of the 12–16 crz-expressing cells) is necessary and sufficient for normal sensitivity to ethanol sedation. Acute manipulations of the activity of crz neurons led to altered ethanol sedation sensitivity, demonstrating that these neurons play an active role in regulating the behavioral response to ethanol-induced sedation. The neuropeptide Crz is also involved in ethanol sedation, as flies with reduced crz expression, specifically during adulthood, show dramatically decreased ethanol sedation sensitivity. Finally, in response to acute ethanol exposure the expression of both apt and crz are rapidly upregulated during ethanol exposure. We hypothesize that the Apt-Crz system, functioning in a very small group of neurosecretory cells, may be an early target of ethanol in the fly brain whose function is crucial for normal sensitivity to ethanol.
Mechanisms of Apt function
How does Apt regulate ethanol-induced sedation? Although Apt's role could be to regulate the expression of crz, we were unable to observe such a function. We thus postulate that Apt functions to regulate the activity of crz neurons and/or neuropeptide release. For example, Apt, acting as a transcription factor, could regulate the transcription of proteins required for synthesis, packaging, and/or release of Crz and possibly other neuropeptides. Alternatively, Apt could regulate the expression of proteins required for synapse formation. In support of these two possibilities, apt mutant embryos were observed to have defective synaptic transmission at the neuromuscular junction, as well as fewer numbers of active zones within motoneurons, indicating a presynaptic defect (Takasu-Ishikawa et al., 2001).
Another possible function for Apt in regulating ethanol sedation behavior may be found in its neuronal requirement during metamorphosis, a time of intense remodeling to construct the adult CNS (Truman, 1990). During embryogenesis, Apt functions in multiple morphogenetic processes, including tracheal, head, CNS, and heart morphogenesis, as well as border cell migration (Eulenberg and Schuh, 1997; Gellon et al., 1997; Su et al., 1999; Starz-Gaiano et al., 2008). It is therefore possible that during metamorphosis Apt establishes proper development and neuronal connectivity of the adult CNS, and in particular the Crz neurons. However, this possibility seems somewhat unlikely given that the adult CNS in apt13–66 flies appeared normal, as was the number and morphology of Crz-expressing neurons. Additionally, we found that adult-specific expression of apt in neurons was necessary for normal sedation sensitivity. However, there may be subtle defects in the adult CNS of apt mutant flies, which we were unable to detect, that could contribute to their altered sedation sensitivity.
Apt shows highest sequence conservation with the human FSBP, a negative regulator of transcription of the gamma chain of fibrinogen (Mizuguchi et al., 1995; Starz-Gaiano et al., 2008). Sequence conservation between apt and FSBP is observed within the DNA-binding domain (Starz-Gaiano et al., 2008). Interestingly, moderate alcohol consumption in humans has been known to exert a cardioprotective effect, in part by lowering levels of circulating Fibrinogen (Wang et al., 1999). The mechanism for how alcohol consumption regulates Fibrinogen is currently unknown, but in light of our findings we speculate that it may occur at the level of transcription. We found that ethanol exposure in flies acutely upregulates apt expression (Fig. 7A). We hypothesize a similar situation may occur in humans, whereby alcohol consumption could upregulate the transcription of FSBP, ultimately leading to negative regulation of the gamma chain of fibrinogen and lowered levels of circulating Fibrinogen, which in turn would provide cardioprotection.
Regulation of ethanol sedation by crz neurons
Our observations implicate crz-expressing neurons in the regulation of ethanol sedation behavior, a function not previously attributed to these neurons. We demonstrate that adult-specific silencing of crz neurons significantly reduced ethanol sedation sensitivity, while increasing their activity resulted in the opposite phenotype, an increase in ethanol sedation sensitivity (Fig. 5). Based on the observation that inhibiting crz expression also reduced ethanol sedation sensitivity (Fig. 6), we believe that the phenotypes associated with crz neuronal manipulations reflect changes in the release of the neuropeptide Crz and activation of its signaling pathway. However, a few Crz neurons also express the short Neuropeptide F (sNPF) (Nässel et al., 2008). sNPF is considered to be a multifunctional neuropeptide due to its broad expression in diverse neuronal types and we have not excluded its possible role in crz-expressing neurons and in ethanol sedation sensitivity. However, the observation that Apt function is required in a subset of crz neurons to promote ethanol sedation behavior, firmly establishes the importance of these neurons in mediating the behavioral response to ethanol. Our data also suggest that the function of both genes, crz and apt, overlaps in a small set of neurons likely located in the PL to mediate the behavioral response to ethanol-induced sedation.
Mechanisms of Crz function
It has recently been hypothesized that Crz is released in response to various types of stress in insects (Veenstra, 2009; Boerjan et al., 2010), and that this could explain its pleiotropic effects. This hypothesis was bolstered by a recent study showing that flies deficient in Crz are resistant to metabolic, osmotic, and oxidative stress, as measured by survival (Zhao et al., 2010). In addition, Crz plays a role in stress physiology through its association with well characterized stress hormones. For instance, crz-expressing neurons in the PL also express receptors for two diuretic hormones, DH44 and DH31 (Johnson et al., 2005). By virtue of receptor similarity, DH44 and DH31 are related to corticotrophin-releasing factor (CRF) and calcitonin-gene related peptide (CGRP), respectively, both of which mediate the mammalian physiological and behavioral responses to stress (Dunn and Berridge, 1990; Bale and Vale, 2004). Interestingly, both CRF and CGRP act to inhibit secretion of GnRH in the mammalian hypothalamus (Nikolarakis et al., 1986; Tellam et al., 1998; Li et al., 2004). This is significant because Crz is thought to be the homolog of mammalian GnRH, and suggests that analogous regulation occurs in Drosophila (Cazzamali et al., 2002). It is thus possible that in flies a stress signal or the animal's stress status may be relayed to Crz neurons and alters their function. Thus, based on its functional and molecular associations with stress physiology, it is tempting to speculate that the role of Crz signaling in ethanol sedation sensitivity is related to a stress response. A previous study has shown that stress, in the form of heat shock, induces tolerance to a subsequent ethanol exposure, and that ethanol tolerance relies on the gene hangover, a large nuclear zinc-finger protein, that mediates various other stress responses (Scholz et al., 2005). In addition, several genes related to stress responses have been shown to be upregulated by ethanol exposure in transcriptional profiling studies, including nearly half of all Drosophila heat shock protein genes, as well as genes involved in the regulation of oxidative stress and aging (Morozova et al., 2006; Kong et al., 2010; Awofala, 2011). Importantly, a maladaptive response to stress has been shown in humans to be a major and common element contributing to drug addiction (Nemeroff et al., 2005). Finally, an increase in ethanol self-administration has been observed in animal models with physical, social, and emotional stress (Higley et al., 1991; Mollenauer et al., 1993). In light of these findings, it will be interesting to further explore the role of Crz and its function in stress physiology and the regulation of ethanol-related behaviors.
Neuropeptides as regulators of ethanol-related behaviors
Neuropeptides are diverse signaling molecules that mediate a broad spectrum of physiological and behavioral processes (Altstein and Nässel, 2010). Several studies have linked neuropeptides to behavioral responses to ethanol. For instance, one of the first ethanol sensitivity mutants described in Drosophila, amnesiac, encodes a neuropeptide homologous to the vertebrate pituitary adenylate cyclase-activating peptide (Feany and Quinn, 1995; Moore et al., 1998). In addition, mice lacking either neuropeptide Y (NPY), a neuromodulator abundantly expressed in many regions of the CNS, or its Y1 receptor subtype, display increased ethanol consumption and resistance to ethanol sedation, whereas animals overexpressing NPY show the opposite behavioral phenotypes (Thiele et al., 1998, 2002; Thorsell et al., 2007). Neuropeptide F (NPF), the sole member of the NPY family in Drosophila, and its receptor NPFR1, has similarly been shown to mediate the fly's sensitivity to ethanol-induced sedation (Wen et al., 2005). Finally, flies with neuronal perturbations in the insulin signaling pathway displayed increased ethanol sedation sensitivity (Corl et al., 2005). These studies and our own, implicating the neuropeptide Crz in sensitivity to ethanol sedation, suggest that neuropeptides are important regulators of the behavioral response to ethanol, and it would therefore be interesting to survey all known Drosophila neuropeptides and their downstream signaling components for possible role(s) in ethanol-related behaviors.
Footnotes
This work was supported by grants from the National Institute on Alcohol Abuse and Alcoholism to K.D.M. and U.H. We thank members of the Heberlein lab for their critical comments on the research and this manuscript; the Bloomington Stock Center, Vienna Drosophila RNAi Center, and Drosophila Genetic Resource Center for fly stocks; the TRiP at Harvard Medical School (National Institutes of Health–National Institute of General Medical Sciences R01-GM084947) for providing transgenic RNAi fly stocks; J. Veenstra for antibodies against Corazonin; R. Schuh for antibodies against Apt; and D. Montell for fly stocks.
The authors declare no competing financial interests.
References
- Altstein M, Nässel DR. Neuropeptide signaling in insects. Adv Exp Med Biol. 2010;692:155–165. doi: 10.1007/978-1-4419-6902-6_8. [DOI] [PubMed] [Google Scholar]
- Awofala AA. Genetic approaches to alcohol addiction: gene expression studies and recent candidates from Drosophila. Invert Neurosci. 2011;11:1–7. doi: 10.1007/s10158-010-0113-y. [DOI] [PubMed] [Google Scholar]
- Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 2005;19:1861–1870. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines RA, Uhler JP, Thompson A, Sweeney ST, Bate M. Altered electrical properties in Drosophila neurons developing without synaptic transmission. J Neurosci. 2001;21:1523–1531. doi: 10.1523/JNEUROSCI.21-05-01523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL, Vale WW. CRF and CRF receptors: role in stress responsivity and other behaviors. Annu Rev Pharmacol Toxicol. 2004;44:525–557. doi: 10.1146/annurev.pharmtox.44.101802.121410. [DOI] [PubMed] [Google Scholar]
- Boerjan B, Verleyen P, Huybrechts J, Schoofs L, De Loof A. In search for a common denominator for the diverse functions of arthropod corazonin: a role in the physiology of stress? Gen Comp Endocrinol. 2010;166:222–233. doi: 10.1016/j.ygcen.2009.09.004. [DOI] [PubMed] [Google Scholar]
- Cazzamali G, Saxild N, Grimmelikhuijzen C. Molecular cloning and functional expression of a Drosophila corazonin receptor. Biochem Biophys Res Commun. 2002;298:31–36. doi: 10.1016/s0006-291x(02)02398-7. [DOI] [PubMed] [Google Scholar]
- Corl AB, Rodan AR, Heberlein U. Insulin signaling in the nervous system regulates ethanol intoxication in Drosophila melanogaster. Nat Neurosci. 2005;8:18–19. doi: 10.1038/nn1363. [DOI] [PubMed] [Google Scholar]
- Corl AB, Berger KH, Ophir-Shohat G, Gesch J, Simms JA, Bartlett SE, Heberlein U. Happyhour, a Ste20 family kinase, implicates EGFR signaling in ethanol-induced behaviors. Cell. 2009;137:949–960. doi: 10.1016/j.cell.2009.03.020. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- de Velasco B, Erclik T, Shy D, Sclafani J, Lipshitz H, McInnes R, Hartenstein V. Specification and development of the pars intercerebralis and pars lateralis, neuroendocrine command centers in the Drosophila brain. Dev Biol. 2007;302:309–323. doi: 10.1016/j.ydbio.2006.09.035. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, Couto A, Marra V, Keleman K, Dickson BJ. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- Dunn AJ, Berridge CW. Physiological and behavioral responses to corticotropin-releasing factor administration: is CRF a mediator of anxiety or stress responses? Brain Res Brain Res Rev. 1990;15:71–100. doi: 10.1016/0165-0173(90)90012-d. [DOI] [PubMed] [Google Scholar]
- Eulenberg KG, Schuh R. The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. EMBO J. 1997;16:7156–7165. doi: 10.1093/emboj/16.23.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany MB, Quinn WG. A neuropeptide gene defined by the Drosophila memory mutant amnesiac. Science. 1995;268:869–873. doi: 10.1126/science.7754370. [DOI] [PubMed] [Google Scholar]
- Gellon G, Harding KW, McGinnis N, Martin MM, McGinnis W. A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development. 1997;124:3321–3331. doi: 10.1242/dev.124.17.3321. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, Garrity PA. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454:217–220. doi: 10.1038/nature07001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higley JD, Hasert MF, Suomi SJ, Linnoila M. Nonhuman primate model of alcohol abuse: effect of early experience, personality, and stress on alcohol consumption. Proc Natl Acad Sci U S A. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Shafer OT, Trigg JS, Park J, Schooley DA, Dow JA, Taghert PH. A novel diuretic hormone receptor in Drosophila: evidence for conservation of CGRP signaling. J Exp Biol. 2005;208:1239–1246. doi: 10.1242/jeb.01529. [DOI] [PubMed] [Google Scholar]
- King I, Tsai LT, Pflanz R, Voigt A, Lee S, Jäckle H, Lu B, Heberlein U. Drosophila tao controls mushroom body development and ethanol-stimulated behavior through par-1. J Neurosci. 2011;31:1139–1148. doi: 10.1523/JNEUROSCI.4416-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong EC, Allouche L, Chapot PA, Vranizan K, Moore MS, Heberlein U, Wolf FW. Ethanol-regulated genes that contribute to ethanol sensitivity and rapid tolerance in Drosophila. Alcohol Clin Exp Res. 2010;34:302–316. doi: 10.1111/j.1530-0277.2009.01093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laissue PP, Reiter C, Hiesinger PR, Halter S, Fischbach KF, Stocker RF. Three-dimensional reconstruction of the antennal lobe in Drosophila melanogaster. J Comp Neurol. 1999;405:543–552. [PubMed] [Google Scholar]
- Lee G, Kim KM, Kikuno K, Wang Z, Choi YJ, Park JH. Developmental regulation and functions of the expression of the neuropeptide corazonin in Drosophila melanogaster. Cell Tissue Res. 2008;331:659–673. doi: 10.1007/s00441-007-0549-5. [DOI] [PubMed] [Google Scholar]
- Li XF, Bowe JE, Mitchell JC, Brain SD, Lightman SL, O'Byrne KT. Stress-induced suppression of the gonadotropin-releasing hormone pulse generator in the female rat: a novel neural action for calcitonin gene-related peptide. Endocrinology. 2004;145:1556–1563. doi: 10.1210/en.2003-1609. [DOI] [PubMed] [Google Scholar]
- Liu QX, Jindra M, Ueda H, Hiromi Y, Hirose S. Drosophila MBF1 is a co-activator for Tracheae Defective and contributes to the formation of tracheal and nervous systems. Development. 2003;130:719–728. doi: 10.1242/dev.00297. [DOI] [PubMed] [Google Scholar]
- Mayfield RD, Harris RA, Schuckit MA. Genetic factors influencing alcohol dependence. Br J Pharmacol. 2008;154:275–287. doi: 10.1038/bjp.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure KD, French RL, Heberlein U. A Drosophila model for fetal alcohol syndrome disorders: role for the insulin pathway. Dis Model Mech. 2011;4:335–346. doi: 10.1242/dmm.006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- Mizuguchi J, Hu CH, Cao Z, Loeb KR, Chung DW, Davie EW. Characterization of the 5′-flanking region of the gene for the gamma chain of human fibrinogen. J Biol Chem. 1995;270:28350–28356. doi: 10.1074/jbc.270.47.28350. [DOI] [PubMed] [Google Scholar]
- Mollenauer S, Bryson R, Robison M, Sardo J, Coleman C. EtOH self-administration in anticipation of noise stress in C57BL/6J mice. Pharmacol Biochem Behav. 1993;46:35–38. doi: 10.1016/0091-3057(93)90313-i. [DOI] [PubMed] [Google Scholar]
- Moore MS, DeZazzo J, Luk AY, Tully T, Singh CM, Heberlein U. Ethanol intoxication in Drosophila: genetic and pharmacological evidence for regulation by the cAMP signaling pathway. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- Morozova TV, Anholt RR, Mackay TF. Transcriptional response to alcohol exposure in Drosophila melanogaster. Genome Biol. 2006;7:R95. doi: 10.1186/gb-2006-7-10-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nässel DR, Enell LE, Santos JG, Wegener C, Johard HA. A large population of diverse neurons in the Drosophila central nervous system expresses short neuropeptide F, suggesting multiple distributed peptide functions. BMC Neurosci. 2008;9:90. doi: 10.1186/1471-2202-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff CB, Vale WW. The neurobiology of depression: inroads to treatment and new drug discovery. J Clin Psychiatry. 2005;66(Suppl 7):5–13. [PubMed] [Google Scholar]
- Nikolarakis KE, Almeida OF, Herz A. Corticotropin-releasing factor (CRF) inhibits gonadotropin-releasing hormone (GnRH) release from superfused rat hypothalami in vitro. Brain Res. 1986;377:388–390. doi: 10.1016/0006-8993(86)90887-5. [DOI] [PubMed] [Google Scholar]
- Robertson HM, Preston CR, Phillis RW, Johnson-Schlitz DM, Benz WK, Engels WR. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics. 1988;118:461–470. doi: 10.1093/genetics/118.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenfluh A, Threlkeld RJ, Bainton RJ, Tsai LT, Lasek AW, Heberlein U. Distinct behavioral responses to ethanol are regulated by alternate RhoGAP18B isoforms. Cell. 2006;127:199–211. doi: 10.1016/j.cell.2006.09.010. [DOI] [PubMed] [Google Scholar]
- Scholz H, Franz M, Heberlein U. The hangover gene defines a stress pathway required for ethanol tolerance development. Nature. 2005;436:845–847. doi: 10.1038/nature03864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuckit MA. Genetics of the risk for alcoholism. Am J Addict. 2000;9:103–112. doi: 10.1080/10550490050173172. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Smith TL. An 8-year follow-up of 450 sons of alcoholic and control subjects. Arch Gen Psychiatry. 1996;53:202–210. doi: 10.1001/archpsyc.1996.01830030020005. [DOI] [PubMed] [Google Scholar]
- Siegmund T, Korge G. Innervation of the ring gland of Drosophila melanogaster. J Comp Neurol. 2001;431:481–491. doi: 10.1002/1096-9861(20010319)431:4<481::aid-cne1084>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Singh CM, Heberlein U. Genetic control of acute ethanol-induced behaviors in Drosophila. Alcohol Clin Exp Res. 2000;24:1127–1136. [PubMed] [Google Scholar]
- Starz-Gaiano M, Melani M, Wang X, Meinhardt H, Montell DJ. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev Cell. 2008;14:726–738. doi: 10.1016/j.devcel.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Su MT, Venkatesh TV, Wu X, Golden K, Bodmer R. The pioneer gene, apontic, is required for morphogenesis and function of the Drosophila heart. Mech Dev. 1999;80:125–132. doi: 10.1016/s0925-4773(98)00197-x. [DOI] [PubMed] [Google Scholar]
- Takasu-Ishikawa E, Yoshihara M, Ueda A, Rheuben MB, Hotta Y, Kidokoro Y. Screening for synaptic defects revealed a locus involved in presynaptic and postsynaptic functions in Drosophila embryos. J Neurobiol. 2001;48:101–119. [PubMed] [Google Scholar]
- Tawfik AI, Tanaka S, De Loof A, Schoofs L, Baggerman G, Waelkens E, Derua R, Milner Y, Yerushalmi Y, Pener MP. Identification of the gregarization-associated dark-pigmentotropin in locusts through an albino mutant. Proc Natl Acad Sci U S A. 1999;96:7083–7087. doi: 10.1073/pnas.96.12.7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam DJ, Perone MJ, Dunn IC, Radovick S, Brennand J, Rivier JE, Castro MG, Lovejoy DA. Direct regulation of GnRH transcription by CRF-like peptides in an immortalized neuronal cell line. Neuroreport. 1998;9:3135–3140. doi: 10.1097/00001756-199810050-00003. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–369. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- Thiele TE, Koh MT, Pedrazzini T. Voluntary alcohol consumption is controlled via the neuropeptide Y Y1 receptor. J Neurosci. 2002;22:RC208. doi: 10.1523/JNEUROSCI.22-03-j0006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorsell A, Repunte-Canonigo V, O'Dell LE, Chen SA, King AR, Lekic D, Koob GF, Sanna PP. Viral vector-induced amygdala NPY overexpression reverses increased alcohol intake caused by repeated deprivations in Wistar rats. Brain. 2007;130:1330–1337. doi: 10.1093/brain/awm033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman JW. Metamorphosis of the central nervous system of Drosophila. J Neurobiol. 1990;21:1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Isolation and structure of corazonin, a cardioactive peptide from the American cockroach. FEBS Lett. 1989;250:231–234. doi: 10.1016/0014-5793(89)80727-6. [DOI] [PubMed] [Google Scholar]
- Veenstra JA. Does corazonin signal nutritional stress in insects? Insect Biochem Mol Biol. 2009;39:755–762. doi: 10.1016/j.ibmb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Veenstra JA, Davis NT. Localization of corazonin in the nervous system of the cockroach Periplaneta americana. Cell Tissue Res. 1993;274:57–64. doi: 10.1007/BF00327985. [DOI] [PubMed] [Google Scholar]
- Wang A, Barker TH, Fuller GM. Alcohol at moderate levels decreases fibrinogen expression in vivo and in vitro. Alcohol Clin Exp Res. 1999;23:1927–1932. [PubMed] [Google Scholar]
- Wen T, Parrish CA, Xu D, Wu Q, Shen P. Drosophila neuropeptide F and its receptor, NPFR1, define a signaling pathway that acutely modulates alcohol sensitivity. Proc Natl Acad Sci U S A. 2005;102:2141–2146. doi: 10.1073/pnas.0406814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Bretz CA, Hawksworth SA, Hirsh J, Johnson EC. Corazonin neurons function in sexually dimorphic circuitry that shape behavioral responses to stress in Drosophila. PloS One. 2010;5:e9141. doi: 10.1371/journal.pone.0009141. [DOI] [PMC free article] [PubMed] [Google Scholar]