Abstract
The measurement of lifespan pervades aging research. Because lifespan results from complex interactions between genetic, environmental and stochastic factors, it varies widely even among isogenic individuals. The action of molecular mechanisms on lifespan is therefore visible only through their statistical effects on populations. Survival assays in C. elegans provided critical insights into evolutionarily conserved determinants of aging. To enable the rapid acquisition of survival curves at arbitrary statistical resolution, we developed a scalable imaging and analysis platform to observe nematodes over multiple weeks across square meters of agar surface at 8 μm resolution. The method generates a permanent visual record of individual deaths from which survival curves are constructed and validated, producing data consistent with the manual method for several mutants in both standard and stressful environments. Our approach allows rapid, detailed reverse-genetic and chemical screens for effects on survival and enables quantitative investigations into the statistical structure of aging.
Introduction
Aging organisms exhibit functional declines at many levels of biological organization. These declines ultimately conspire to kill the organism and determine its lifespan. Because lifespans vary remarkably1 even within isogenic populations, useful information about aging mechanisms can be obtained by identifying their statistical signature in the response of lifespan distributions to genetic, chemical and physical interventions
C. elegans individuals live as self-fertile adults for a few weeks, producing large numbers of isogenic offspring. Early investigations revealed single point mutations in insulin/IGF-1 pathway components capable of doubling average lifespan2-5. These findings subsequently generalized to other organisms, including fruit flies and mice5, establishing C. elegans as a metazoan model for studying the genetics of aging. The acquisition of survival curves in C. elegans thus became an essential part of aging research. In routine practice, survival curves of animals cultured on solid agar in a Petri dish, fed by a bacterial lawn of E. coli, are acquired manually through daily observation with a low-power dissecting microscope6, 7. Death is recognized by the failure of an individual to react by motion to prodding with a wire, requiring labor-intensive, repetitive, and subjective observation. This protocol provides strong incentives to keep population sizes small, observations infrequent, and the number of replicates limited, thus curtailing the reproducibility and scope of data collected.
We present a scalable method for producing standardized survival curves at arbitrary statistical resolution, allowing “hands-free”, lifelong, and accurate observation of arbitrarily large nematode populations. Our method combines groups of flatbed scanners to function as a single spatially-extended microscope. Alone or in groups, these scanners produce time-lapse videos under highly controlled environmental conditions. We couple this imaging approach with an image-analysis pipeline that automatically identifies death times. By creating an auditable trail of images, the system allows rapid validation and refinement of processed data, and therefore supports a statistically rigorous analysis of aging and stress-resistance.
In several cases, notably in worms and flies, focused efforts have been made at acquiring high-resolution survival data8-15. Our method formalizes, standardizes, and routinizes such observations for nematodes, enabling the accumulation of a self-consistent body of quantitative demographic data tightly linking aging, genotype, and environment. No such comprehensive body of data exists for any metazoan to date, humans included. We refer to the integrated combination of scanner hardware and image-processing software as the “Lifespan Machine” (LM).
Results
A distributed microscope for large worm populations
Lifespan data are acquired over many scales, ranging from aging studies spanning multiple months to stress-resistance assays lasting less than a day16, involving targeted characterizations of only a few strains12 or screening projects of thousands of gene knock-downs2, 3, 5. Throughput is important for screening projects, while targeted studies benefit from large populations to provide statistical power. To suit all these purposes, our platform combines flatbed scanners in a scalable fashion to monitor arbitrarily large populations over a wide-range of timescales.
Our approach replaces human observation with an automated system that acquires and processes time-lapse images to identify the timing of death defined as the persistent cessation of spontaneous movement. Our current reference implementation can simultaneously monitor 30,000 animals across 800 plates, imaging 4.5 m2 of agar surface at a resolution of 8 μm (Supplementary Notes 1—4). This capacity is used to run many independent, single-scanner experiments in parallel with larger investigations involving ten scanners or more.
Young worms move quickly relative to the observation frequency attainable with scanners, precluding their tracking. However, as animals slow down with age, their motion can be quantified longitudinally, especially when it is limited to changes in posture at a fixed location.
Each scanner monitors 16 Petri dishes (plates) sealed face down to a glass sheet by a rubber mat, with each plate containing a population of about 35 animals (Online Methods). A fluorescent lamp inside the scanner lid sends light through plates, agar, bacteria, and worms, to be captured by a sensor chip moving underneath the glass surface in synchrony with the light source (Fig. 1a, Supplementary Notes 1 and 3). A scanner thus acts as 16 camera-equipped dissecting microscopes, while costing significantly less and in a form factor suited for dense shelving inside temperature-controlled incubators.
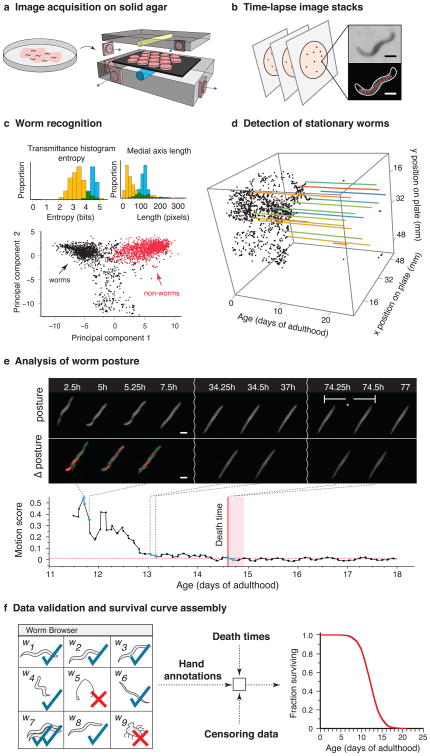
Figure 1.
Lifespan Machine workflow. (a) Agar plates with age-synchronized worms are placed face-down on the surface of flatbed scanners (Supplementary Note 1). (b) Each scanner captures a time series of images of each of its plates. Images are processed to identify foreground objects (Supplementary Note 2). Scale bar = 250 μm. (c) Various morphological features are quantified for each object. The population distributions of these features demonstrate that none permits by itself a discrimination of worms from non-worms. Based on many validated images of worm and non-worm objects, classifiers are constructed that are capable of categorizing objects based on their 65-dimensional feature vectors. The panel depicts a projection of the point cloud representing the validated set of feature vectors, indicating that differentiating worms (black) from non-worms (red) is possible (Supplementary Note 2). (d) Once worm objects have been identified, individual image stacks are analyzed to identify animals that have become stationary (Supplementary Note 5). (e) Stationary animals are further analyzed to detect posture changes, including head and tail movements. Death is identified by retrospective analysis as the final cessation of postural change (Supplementary Note 5). (f) The LM produces a time-lapse image record for each individual. These records can be used for visual validation or resolution of ambiguities employing a rapid data-inspection tool, the “Worm Browser” (Supplementary Video 6 and Note 4). Validated death times are combined with automatic censoring data (Supplementary Note 8) into a Kaplan-Meier survival curve (actual data shown) or into hazard rates using a statistics package.
Traditional protocols for monitoring large populations of nematodes over long periods of time move plates in and out of an incubator6, 7, 11, 12, exposing them to spatially and temporally heterogeneous conditions. Our apparatus maintains plates at a fixed position within a controlled environment for the duration of each experiment. This is important for accuracy, given the exquisite temperature sensitivity of C. elegans lifespan17 (Supplementary Note 1). We curbed temperature excursions by two systems of fans, one cooling each scanner, the other circulating air evenly throughout the enclosing incubator (Supplementary Note 3). Temperature fluctuations on a typical scanner surface were thus reduced from several degrees to less than 0.5 °C, while limiting differences between scanners to about 1 °C (Supplementary Note 1).
To improve image quality we repositioned a single fixed lens in the scanner's optical path, shifting the focal plane a few millimeters above the scanner glass, matching the agar surface inside plates. To ensure that the agar surface is consistent across experiments, we developed a robust method for the controlled drying of agar plates (Online Methods).
Automated identification of death times
Following an optimized scanning schedule (Online Methods), the LM captures 24 images of each plate per day. The image-processing pipeline identifies worm objects (Supplementary Note 2) and their position in every frame (Fig. 1b,c). Individual animals, once stationary (Fig. 1d), continue to change posture for a time, often moving their head or tail (Fig. 1e, Supplementary Videos 1-5 and Note 5), reminiscent of the “class C” animals described in studies of age-dependent locomotory decline18. Our software determines the final cessation of spontaneous postural movement through retrospective image analysis (Online Methods, Supplementary Note 5 and Video 6). Posture analysis of stationary animals is crucial to avoid underestimation of lifespan. We observed wild type animals spending on average 1.2 days of their life in this state (Fig. 1e), while certain mutants, for example age-1(hx546), persist in it more than twice as long (Supplementary Note 6), consistent with previous reports19.
We found that most animals exhibit a stereotyped morphological change at or near the time of their final posture change (Supplementary Note 7). Individuals first shrink by at least 10% and expand by more than that amount shortly thereafter. We never observed a change in posture after this expansion. The correlation between the cessation of postural motion and this morphological change suggests a physiological transition, perhaps corresponding to the animal's death, corroborating our death criterion.
A crucial component of our platform is a software package that allows rapid validation of death times through visual inspection (Supplementary Note 4). This quality control step (Fig. 1f, Supplementary Video 6, Online Methods) allows a user to determine whether plates were compromised (e.g. desiccation or fungal contamination) and need exclusion. In particular, it permits identification of aggregates containing multiple worms (Supplementary Note 8) and the censoring of incorrectly classified objects (e.g. features of the agar lawn) and non-aging related deaths (e.g. body rupture), thus enabling a visual validation of machine operation.
The LM produces accurate and precise survival curves
To evaluate whether the LM produces results consistent with those of a human observer, we placed roughly 8,000 age-synchronous wild type animals into a single liquid suspension, and distributed aliquots of approximately 35 individuals across 170 agar plates seeded with E. coli OP50 (Online Methods). 160 of these plates were distributed across ten scanners situated in an incubator at 25 °C. The remaining 10 plates were placed in a separate incubator and monitored daily by hand using a dissecting microscope.
Because we drew all animals from the same pool, any significant difference between survival curves of individual plates must be attributed to either measurement error or environmental variation, such as differences in plate temperature. We compared death times grouped by plate on a single scanner with those acquired by the manual procedure (Fig. 2a). In both methods, plate means varied within essentially the same range, between 10.7 and 13.1 days. No significant effect was observed between plates collected by the manual method (Log-rank test for homogeneity P =0.059). A small, significant difference was observed between plates collected by the automated method (Log-rank P =0.002), reflecting either inaccuracies in the estimation of death times or a slightly increased environmental variability among plates. In two independent replicates, the survival curves determined by the LM differed in mean lifespan by 2.5 hours or 0.2% (Log-rank P =0.01, Fig. 2b) and 11.7 hours or 3.2% (Log-rank P <0.001) from those of cohorts assayed using the conventional method. These small differences could be explained by practical limitations in our ability to measure and maintain temperature constant in both methodologies.
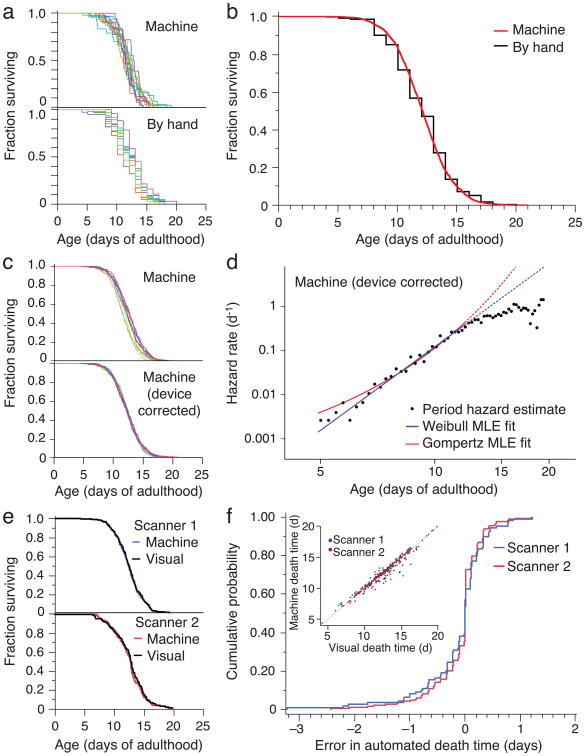
Figure 2.
Automated wild type survival data. (a) A survival curve was generated from 484 death times of wild type animals co-located on a single scanner (top) and 513 wild type animals observed using the manual method (bottom). (b) The entire population of 3,578 wild type animals over ten scanners was aggregated into a single curve, nearly indistinguishable from the manually-scored curve. (c) Scanner-specific microenvironments, mainly differences in temperature, affect lifespan (top). The effect was estimated using a categorical Accelerated Failure Time model, to obtain device-corrected lifespans registered to the grand mean (bottom), as detailed in Supplementary Note 10. (d) Hazard rates were estimated from lifespan data (Supplementary Note 11). The blue (Weibull) and red (Gompertz) lines represents a maximum likelihood fit of the parametric model using data up to median survival. A smaller automated assay performed at 20 °C on HT115 bacteria was consistent with the mortality kinetics at 25 °C on OP50 (Supplementary Note 11). (e) Image records of populations on two scanners were selected for validation using the Worm Browser to determine death times by eye (Supplementary Video 6 and Note 4). The resulting curves compare with those produced by the automated method (Log-rank P > 0.5 in both cases). (f) The cumulative LM error (i.e. the death time differences between automation and visual inspection) is shown for both scanners. Visual and automatic lifespans are highly correlated (R2 = 0.96), as shown in the inset, with the LM underestimating lifespan by an average of two hours.
Where experiments require comparison between populations much larger than 300 individuals, animals must be distributed across multiple scanners. With the statistical power (Supplementary Note 9) afforded by each scanner, even slight environmental differences between scanners translate into statistically significant differences between survival curves (Fig. 2c top). Indeed, scanner surface temperature correlated well with mean lifespan (R2 = 0.75; P=0.023; Supplementary Note 1).
Because devices produced a small effect on lifespan, we applied an accelerated failure time regression model with scanner identity as a categorical covariate to generate “device-corrected” lifespans (Fig. 2c, Supplementary Note 10). This procedure is useful for assessing whether data pooling is justified and for standardizing data against a desired baseline curve, for example, aligning large experiments to a single scanner.
The LM provides data in quantity and quality appropriate for estimating time-dependent hazard (mortality) rates, Supplementary Note 11. We observed a rapid increase and subsequent deceleration in mortality with time, as noted previously10-12,14. For the first 50% of deaths, mortality appears better fit by a power of time t (Weibull hazard, α/β (t/β)(α−1)) than an exponential in time (Gompertz hazard, a et/b), Fig. 2d and Supplementary Note 11, though for values of the Gompertz parameter a much smaller than 1, Gompertz and Weibull distributions behave very similarly for all but the lowest quantiles of survival, making disambiguation difficult. An analysis of the full hazard data, accounting for the deceleration phase, may require frailty models to incorporate phenotypic diversity potentially present even in isogenic populations20, 21. However, the observed hazard deceleration is not the result of environmental heterogeneity in our apparatus, as it persists after device-correction (data not shown).
The close agreement between automated and manual survival curves was surprising, as scanners expose animals to conditions that differ from the manual method, such as oscillating lights22 and temperature oscillations (Supplementary Note 1). The difference in lifespan between animals exposed and not exposed to the scanner environment, but scored by hand, was so small as to be explained by limitations in our ability to maintain the same temperature in both environments (Supplementary Note 12). Importantly, the LM and the manual procedure adopt different operational definitions of death—cessation of spontaneous vs. stimulated movement, respectively. We therefore sought a validation process other than comparison to the manual method by developing software to assist in the visual inspection of the image record generated by the LM for each experiment (Supplementary Note 4 and Video 6). Two survival curves constructed from 294 user-annotated death times based on images from two scanners were each statistically indistinguishable from curves produced by automated annotation (Log-rank P > 0.5), Fig. 2e-f.
The LM reproduces known lifespans of genetic perturbations
To test whether the LM could be used to characterize the effects of mutations and RNAi on lifespan, we divided wild type animals and daf-16(mu86) mutants3 between plates seeded with either an E. coli HT115(DE3) strain containing an RNAi construct targeting the insulin/IGF receptor daf-23, 22 or an empty vector. As expected, daf-16 mutants live shorter than wild type animals23, and knockdown of daf-2 by RNAi extended the lifespan of wild type animals but not daf-16 mutants (Fig. 3a)24. We found no significant difference between manual and automated survival curves for all conditions (Log-rank P > 0.3) except for those of wild type animals feeding on control bacteria (Log-rank P < 0.001), which appeared to live 21 hours longer in the manual assay. A subsequent replicate showed no significant difference (data not shown). These experiments were performed at 25 °C; we observed corresponding effects at 20 °C (Supplementary Note 13).
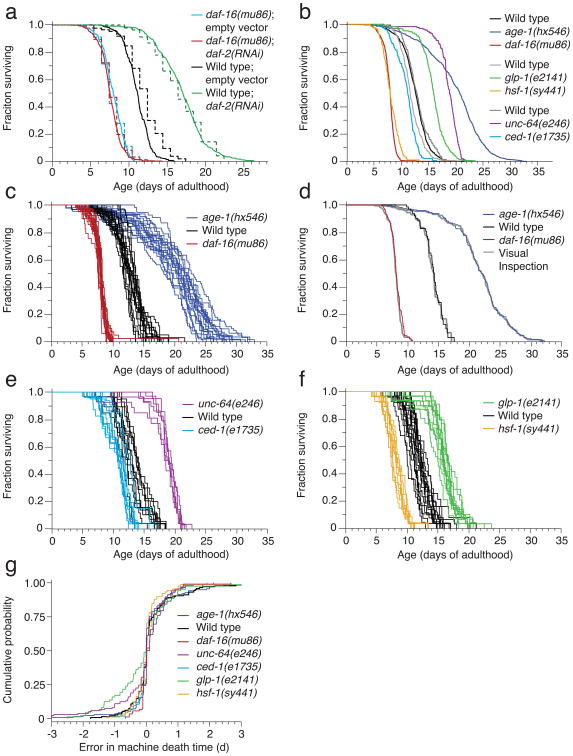
Figure 3.
Automated mutant survival data. The manual experiments required one hour per measurement once a day, whereas the automated method required no experimenter time to collect twenty-four daily measurements of significantly larger populations. (a) Wild type animals and daf-16(mu86) mutants were fed bacteria containing either an empty vector or a vector for daf-2(RNAi). These populations were observed either by the automated (solid, 2,015 animals) or manual (dashed, 541 animals) method. (b) In addition, six mutant populations were monitored by the LM. Their lifespan data are shown both aggregated and, in subsequent panels, grouped by plate. (c-h) Each panel pair refers to a separate scanner also running a wild type population as reference. The population sizes were (c,d) daf-16(mu86): 654, wild type: 594, age-1(hx546): 1109. (e,f) ced-1(e1735): 255, wild type: 314, unc-64(e246): 193. (g,h) hsf-1(sy441):234, wildype: 472, glp-1(e2141): 335 (d, f, h) Hazard rates were estimated as in Fig. 2d (Supplementary Note 11). (i) As in Fig. 2e, a comparison between survival curves determined from visually and automatically scored death times validates the LM operation. Comparisons for all other mutants are shown in Supplementary Note 13. (j) The cumulative LM error (see Fig. 2f) for each mutant population provides an alternative view of validation.
Consistent with previous reports2, 23, 25,5,26, mutations in the phosphoinositide 3-kinase age-1, the syntaxin homolog unc-64, and the Notch receptor glp-1 result in lifespan extension, whereas mutations in daf-16, the heat shock factor hsf-1, and the CD91 homolog ced-1 shorten lifespan. Our automated technique correctly identifies known genetic determinants of C. elegans lifespan (Fig. 3b—h). Although population sizes (Supplementary Table 1) were too small to compare parametric models, we used these data to estimate27 the hazard rates of each mutant (Fig. 3d,f,h, Online Methods).
Visual image inspection (Fig. 3i,j) validates the LM-acquired survival curves of mutants. We note that the machine performed well on the movement-defective mutants unc-64(e246), unc-4(e120), unc-50(e306), and unc-119(ed3) (Supplementary Note 13), even though the image analysis parameters were optimized for wild type movement. The analysis of glp-1(e2141) performed equally well as that of wild type, despite mutant animals being thinner from the absence of a germline.
The LM enables high-resolution stress resistance assays
Survival assays are widely used to evaluate the ability of individuals to withstand exogenous stresses. Exposure to high temperature (35 °C) or toxins dramatically shortens the lifespan of C. elegans. Mean survival ranges from several hours to a few days16, 28, making data collection at high frequency challenging for the manual approach. We evaluated the performance of the LM in two stress-resistance scenarios: exposure to high temperature16 and the oxidant tert-butyl hydroperoxide (t-BuOOH)28. The LM required no modifications beyond adjustments to a subset of parameters used by our image analysis software to quantify worm movement and identify worm death times (Online Methods).
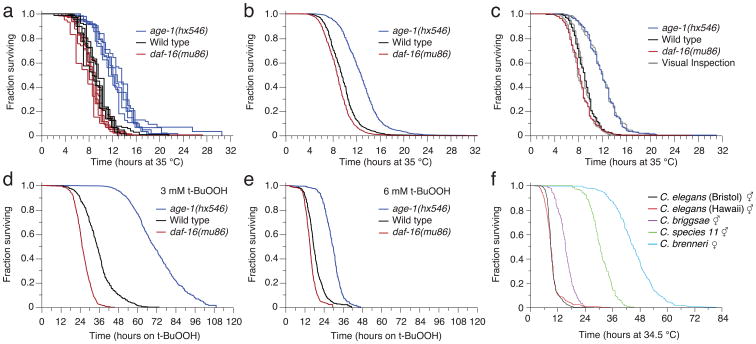
Age-synchronous wild type, age-1(hx546), and daf-16(mu86) mutants were grown under standard conditions at 25 °C and shifted on the second day of adulthood to one of four conditions: agar plates at 25 °C or 35 °C and agar plates with 3 mM or 6 mM t-BuOOH at 25 °C. While no wild type animals died during the first two days on control plates at 25 °C, their lifespan was shortened dramatically at 35 °C. In agreement with previous reports16, age-1(hx546) mutants lived longer than wild type and daf-16(mu86) mutants showed a small but statistically significant reduction in survival (Fig. 4a,b). The results obtained by visual and automated image analyses were highly correlated (Fig. 4c,d). We observed a similar, concentration-dependent effect of t-BuOOH on lifespan, consistent with previous studies28 (Fig. 4e,f). The LM is therefore well suited for assaying the effects of chemicals on survival in C. elegans.
Figure 4.
Automated stress-resistance assays. (a,b) Age-synchronous populations of 1,873 age-1(hx546) mutants, 2106 daf-16(mu86) mutants, and 1,726 wild type animals were transferred to 35 °C on their second day of adulthood and their death times were recorded by scanners operating at that temperature. Animals died over the course of thirty hours, as shown across the populations of individual plates (a), and in aggregate (b) for all animals of each genotype. (c,d) The LM operation is validated as in Figs. 2e,f and 3i,j (Log-rank P > 0.5 in all cases). (e,f) LM-acquired survival curves are shown for populations of age-1(hx546), wild type, and daf-16(mu86) animals that were transferred onto (e) 3 mM t-BuOOH (2,135 animals) and (f) 6 mM t-BuOOH (1,808 animals total) at 25 °C.
The hazard rate of animals under stress
We found the hazard rate function of wild type, age-1(hx546) and daf-16(mu86) at 35 °C to consist of an accelerating and decelerating phase (Fig. 5a), as at 25 °C, suggesting that the demographic characteristics of aging are preserved even under intense stress conditions. These features did not depend on the presence of live bacteria, as both were also observed in populations raised and assayed on UV-killed bacteria (Fig. 5b). To our knowledge, this is the first quantification of a hazard rate function for metazoans under extreme conditions.
The effect of thermal stress appears to consist primarily in rescaling time, while barely impacting the coefficient of variation (CV) of the lifespan distribution (Supplementary Table 1). Up to median lifespan and barring frailty effects, 1 hour for animals at 25 °C corresponds to 1.8 minutes for animals at 35 °C and the latter experience at every moment a 33-fold higher risk of death. Likewise, the effect of age-1(hx546) and daf-16(mu86) mutations relative to each other at 35 °C consists primarily in a change of time scale (Fig. 5a) with a marginal effect on the CV. This is a marked departure from the behavior at 25 °C (Fig. 3d) where DAF-16 increases and AGE-1 decreases lifespan variation disproportionately relative to the mean.
Previous studies in C. elegans have shown that a brief heat pulse induces a heterogeneous stress response, whose magnitude predicts survival upon a subsequent shift to high temperature16, 29. We evaluated the effect of pretreatment on thermotolerance and found a substantial stretching of the time scale (Fig. 5b).
Despite a separation of 100 million years or more30, the Caenorhabditis species elegans, briggsae, brenneri, and species 11 are sufficiently similar in appearance for the LM to function correctly with no additional calibration at 35 °C. We confirmed previous reports31 that C. briggsae, and C. brenneri are more thermotolerant than C. elegans, and found this to be the case also for C. species 11. The thermotolerance profiles of these species correlate with the temperatures of their natural habitats30, with tropical species being more thermotolerant than temperate ones. As we observed for C. elegans mutants, the genetic background of Caenorhabditis species, too, causes predominantly a rescaling of time, while having a much smaller effect on the coefficient of variation of the lifespan distribution (Supplementary Table 1).
Discussion
By using standard nematode culture conditions, our method meaningfully extends, within limits (Supplementary Note 16), the existing experimental literature and can be checked against it. The modular design of the LM gives researchers the flexibility to expand an installation by simply adding scanners (Supplementary Notes 14 and 15). A single scanner is sufficient to compare the lifespan of a mutant population against a wild type control, detecting 10% differences in survival with 99% confidence and 99% power. Facilities of ten or more scanners provide sufficient throughput to systematically characterize the effects of many genes and environmental conditions on survival. At even larger scale, scope can be paired with depth by evaluating samples that are large enough to resolve and quantitatively compare hazard rate features, thus linking interventions at the molecular level with their statistical footprint at the organismic level.
Supplementary Material
Acknowledgments
We would like to thank Joy Alcedo (Wayne State University) for providing the hsf-1 and glp-1 mutant strains, Xavier Manière (Université Paris Descartes) for providing the NEC937 strain, Becky Ward and Debora Marks for critical reading of our manuscript and Catalina Romero, Debora Marks and members of the Fontana lab for helpful discussions and encouragement throughout this project. We would like to thank Tom Kolokotrones, Eric Smith, and Lee Jen Wei for discussions and statistical advice, and Mason Miranda, our departmental IT specialist, for patiently meeting our needs for data storage. This work was funded by the National Institute of Health through grants R03 AG032481, R03 AG032481-S1, and R01 AG034994.
Footnotes
Author Contributions: N.S. designed and implemented hardware and software. N.S. and B.U. constructed and calibrated equipment. N.S. and J.A. conceived and designed experiments. N.S., B.U., J.A., Z.N. and I.F.L.M. performed experiments. N.S. designed analytic tools. N.S., J.A. and W.F. provided guidance, analyzed data, interpreted results, and wrote the manuscript. J.A and W.F. are co-last authors.
Figure 5 Hazard of animals under thermal stress. (a) Hazard rates (dots) were estimated as in Fig. 2d for populations of 1,873 age-1(hx546), 2,106 daf-16(mu86) and 1,726 wild type animals. (b) An acquired thermotolerance experiment is shown from a hazard perspective. Roughly 4,000 C. elegans were grown on UV-killed bacteria at 20 °C. On the first day of adulthood, half of the animals were transferred to 33 °C for one hour and allowed to recover for twelve hours at 20 °C. Treated animals were then transferred a second time to 33 °C and their survival determined with the LM. The hazard rate curve of 2,022 pre-treated animals was compared to that of 2,459 untreated animals. (c) The panel shows LM-acquired survival curves obtained from populations of 1,306 C. elegans (N2 Bristol isolate), 255 C. elegans (Hawaiian isolate), 285 C. briggsae, 224 C. species 11, and 295 C. brenneri nematodes raised on live OP50 at 20 °C and transferred to 34.5 °C on the second day of adulthood. (d) Hazard rate plots for the Caenorhabditis species of panel (c).
References
- 1.Kirkwood TB, et al. What accounts for the wide variation in life span of genetically identical organisms reared in a constant environment? Mech Ageing Dev. 2005;126:439–443. doi: 10.1016/j.mad.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 4.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 5.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 6.Sutphin GL, Kaeberlein M. Measuring Caenorhabditis elegans life span on solid media. J Vis Exp. 2009 doi: 10.3791/1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson DS, Taylor RC, Dillin A. Analysis of aging in Caenorhabditis elegans. Methods Cell Biol. 2012;107:353–381. doi: 10.1016/B978-0-12-394620-1.00012-6. [DOI] [PubMed] [Google Scholar]
- 8.Curtsinger JW, Fukui HH, Townsend DR, Vaupel JW. Demography of genotypes: failure of the limited life-span paradigm in Drosophila melanogaster. Science. 1992;258:461–463. doi: 10.1126/science.1411541. [DOI] [PubMed] [Google Scholar]
- 9.Carey JR, Liedo P, Vaupel JW. Mortality dynamics of density in the Mediterranean fruit fly. Exp Gerontol. 1995;30:605–629. doi: 10.1016/0531-5565(95)00013-5. [DOI] [PubMed] [Google Scholar]
- 10.Vaupel JW, et al. Biodemographic trajectories of longevity. Science. 1998;280:855–860. doi: 10.1126/science.280.5365.855. [DOI] [PubMed] [Google Scholar]
- 11.Vanfleteren JR, De Vreese A, Braeckman BP. Two-parameter logistic and Weibull equations provide better fits to survival data from isogenic populations of Caenorhabditis elegans in axenic culture than does the Gompertz model. J Gerontol A Biol Sci Med Sci. 1998;53:B393–403. doi: 10.1093/gerona/53a.6.b393. discussion B404-398. [DOI] [PubMed] [Google Scholar]
- 12.Johnson TE, Wu D, Tedesco P, Dames S, Vaupel JW. Age-specific demographic profiles of longevity mutants in Caenorhabditis elegans show segmental effects. J Gerontol A Biol Sci Med Sci. 2001;56:B331–339. doi: 10.1093/gerona/56.8.b331. [DOI] [PubMed] [Google Scholar]
- 13.Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- 14.Baeriswyl S, et al. Modulation of aging profiles in isogenic populations of Caenorhabditis elegans by bacteria causing different extrinsic mortality rates. Biogerontology. 2009;11:53–65. doi: 10.1007/s10522-009-9228-0. [DOI] [PubMed] [Google Scholar]
- 15.Wu D, Rea SL, Cypser JR, Johnson TE. Mortality shifts in Caenorhabditis elegans: remembrance of conditions past. Aging Cell. 2009;8:666–675. doi: 10.1111/j.1474-9726.2009.00523.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 18.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 19.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaupel JW, Manton KG, Stallard E. The impact of heterogeneity in individual frailty on the dynamics of mortality. Demography. 1979;16:439–454. [PubMed] [Google Scholar]
- 21.Weitz JS, Fraser HB. Explaining mortality rate plateaus. Proc Natl Acad Sci U S A. 2001;98:15383–15386. doi: 10.1073/pnas.261228098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathew MD, Mathew ND, Ebert PR. WormScan: a technique for high-throughput phenotypic analysis of Caenorhabditis elegans. PLoS One. 2012;7:e33483. doi: 10.1371/journal.pone.0033483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. 2002;298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 25.Ailion M, Inoue T, Weaver CI, Holdcraft RW, Thomas JH. Neurosecretory control of aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1999;96:7394–7397. doi: 10.1073/pnas.96.13.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A. Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell. 2008;15:87–97. doi: 10.1016/j.devcel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller HG, Wang JL. Hazard rate estimation under random censoring with varying kernels and bandwidths. Biometrics. 1994;50:61–76. [PubMed] [Google Scholar]
- 28.Tullet JM, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rea SL, Wu D, Cypser JR, Vaupel JW, Johnson TE. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Genet. 2005;37:894–898. doi: 10.1038/ng1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiontke KC, et al. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol Biol. 2011;11:339. doi: 10.1186/1471-2148-11-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amrit FR, Boehnisch CM, May RC. Phenotypic covariance of longevity, immunity and stress resistance in the caenorhabditis nematodes. PLoS One. 2010;5:e9978. doi: 10.1371/journal.pone.0009978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong JX, Krzyzak A, Suen CY. Fast SVM training algorithm with decomposition on very large data sets. IEEE Trans Pattern Anal Mach Intell. 2005;27:603–618. doi: 10.1109/TPAMI.2005.77. [DOI] [PubMed] [Google Scholar]
- 34.Sall J SAS Institute. JMP start statistics: a guide to statistics and data analysis using JMP. 5th. SAS Institute; Cary, NC: 2012. [Google Scholar]
- 35.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2006. [Google Scholar]
- 36.Buckley J, James I. Linear regression with censored data. Biometrika. 1979;66:429–436. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.