Abstract
Most physiological processes in mammals display circadian rhythms that are driven by the endogenous circadian clock. This clock is comprised of a central component located in the hypothalamic suprachiasmatic nucleus and subordinate clocks in peripheral tissues. Circadian rhythms sustain 24-hour oscillations of a large number of master genes controlling the correct timing and synchronization of diverse physiological and metabolic processes within our bodies. This complex regulatory network provides an important communication link between our brain and several peripheral organs and tissues. At the molecular level, circadian oscillations of gene expression are regulated by a family of transcription factors called “clock genes”. Dysregulation of clock gene expression results in diverse human pathological conditions, including autoimmune diseases and cancer. There is increasing evidence that the circadian clock affects tooth development, salivary gland and oral epithelium homeostasis, and saliva production. This review summarizes current knowledge of the roles of clock genes in the formation and maintenance of oral tissues, and discusses potential links between “oral clocks” and diseases such as head and neck cancer and Sjögren’s syndrome.
Keywords: autoimmune diseases, salivary glands, clock genes, tooth, oral cancer, Sjögren’s syndrome
The Molecular Structure of the Circadian Clock
Biological rhythms can be generated within an organism by endogenous clocks, or driven by cyclic events in the environment such as the light/dark cycle. Circadian rhythms are controlled by the body’s biological clocks. The “master” or central clock is located in the hypothalamus and is called the suprachiasmatic nucleus (SCN) (Weaver, 1998), whereas clocks in other parts of the body are referred to as peripheral clocks. The central clock is light-responsive and can be entrained by light/dark cycles. The peripheral clocks cannot perceive light but can be entrained by the central clock or independently by other physiological stimuli, such as feeding. The mechanism of interactions through which the central clock entrains peripheral clocks is unclear (Dibner et al., 2010; Mohawk et al., 2012).
The molecular structure of the circadian clock has been extensively studied by systems biologists, who are applying experimental, theoretical, and computational techniques to the study of circadian rhythms at all physiological scales. The mechanisms of rhythm generation in many organisms require signaling pathways with interlocked transcription–translation feedback loops (TTFLs) (Novak and Tyson, 2008). In mammals, the core network generating circadian rhythms has 2 interlocked negative feedback loops (Fig. 1), which are part of the TTFL network. In the core negative feedback loop, PER and CRY proteins form a hetero-multimeric complex with another core circadian regulator, casein kinase I δ, translocate into the nucleus, and suppress Per, Cry, and Rev-Erbα transcription by directly inhibiting the activity of BMAL1/CLOCK heterodimers. The second negative feedback loop controls the expression of the activators BMAL1 and CLOCK, which inactivate their own transcription through the Rev-Erb genes (Preitner et al., 2002). This second negative feedback loop, controlled by Rev-Erbα, inhibits Bmal1 transcription through a retinoic-acid-related-orphan-receptor (ROR) response element in the Bmal1 promoter (Raspe et al., 2002).
Figure 1.
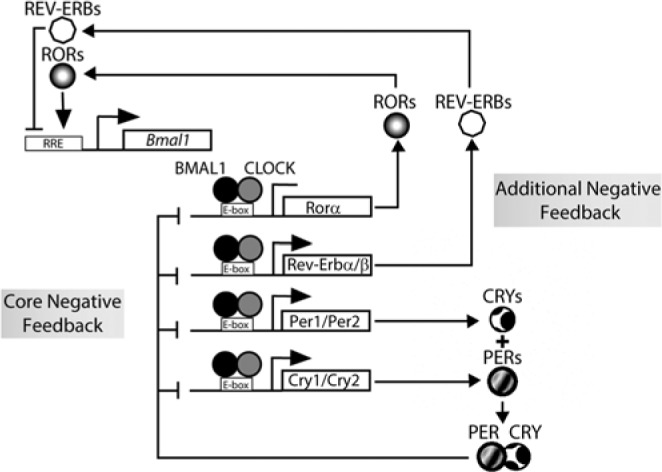
Schematic representation of the central mammalian circadian clock network. The oscillator consists of 2 interlocked negative feedback loops. In the core negative feedback loop, the repressors (PER and CRY proteins) inactivate the activators (BMA and CLOCK proteins). In the other negative feedback loop, the activators repress their own transcription expression through an inhibition mediated by REV-ERB and ROR proteins.
A systematic mathematical and computational analysis of biological rhythms has revealed that negative feedback signals create the oscillations, while positive feedback signals adjust the oscillations’ frequencies without changing the amplitude (Novak and Tyson, 2008). Kim and Forger (2012) developed a detailed mathematical model to investigate the molecular mechanisms of the circadian clock. They used their model to study the interplay between the positive and negative feedback loops to maintain robust oscillations. They showed that proper stoichiometric balance between activators (BMAL1/CLOCK and NPAS2) and repressors (PER and CRY proteins) is key to sustained circadian oscillations. They also discovered that an additional slow negative feedback loop, in which activators indirectly inactivate themselves, creates more robust oscillations with nearly a constant period over a large variation of gene expression levels. This additional feedback loop also improves the regulation of the stoichiometric balance. In addition, they demonstrated that the circadian rhythm oscillations are lost in the absence of tight binding between activators and repressors. An interesting finding made by Kim and Forger is that the interlocked negative feedback loop structure is suitable for biological clocks in which the maintenance of a fixed period is crucial, while a positive-negative feedback loop structure is suitable for oscillators that need to tune their period in biological systems (Kim and Forger, 2012). Therefore, defects in the maintenance of circadian rhythms leading to disease may result from changes in gene dosage or failure to restore gene dosage in the interlocked negative feedback loops (Lee et al., 2011).
The core molecular clock within cells keeps time, but it also responds to cues from the SCN through neural, hormonal, and activity-driven pathways, and signals from the local environment (Dibner et al., 2010). There is reciprocal control of the circadian clock by metabolic signaling. The core clock synchronizes through direct and indirect outputs with gluconeogenesis and oxidative metabolism. For example, PER interaction with REV-ERBa modulates transcription of gluconeogenic factor G6pase (Schmutz et al., 2010). Recent evidence (Bass and Takahashi, 2010) suggests that the core clock receives reciprocal input from nutrient signaling pathways, such as SIRT1 and AMPK, to couple circadian cycles to metabolic flux in peripheral tissues. The current view is that the circadian system in mammals is composed of a hierarchy of oscillators at the cellular, tissue, and physiological systems levels. Although there is a common molecular clock mechanism, it remains unclear how central and peripheral circadian clocks are coupled and synchronized to orchestrate normal physiological function in mammals. There are still major gaps in how the central clock in the SCN is connected to peripheral tissue clocks, the relationship between metabolism and circadian rhythms, and the general impact of circadian clocks in health and disease. Some of these gaps have been reviewed (Bass and Takahashi, 2010; Dibner et al., 2010; Mohawk et al., 2012).
In this review, we overview the key features of craniofacial tissue circadian clocks. We also summarize the current knowledge on the potential links between and among altered clock gene expression, autoimmune diseases, and cancer, focusing on Sjögren’s syndrome (SS) and oral cancer (OC).
Expression and Potential Roles of Circadian Clock Genes in Craniofacial Tissues
The initial discovery of clock genes and their expression in craniofacial tissues (Simmer et al., 2010; Zheng et al., 2011) (Fig. 2) opened a new field of research in the molecular clock era of mineralized tissue formation.
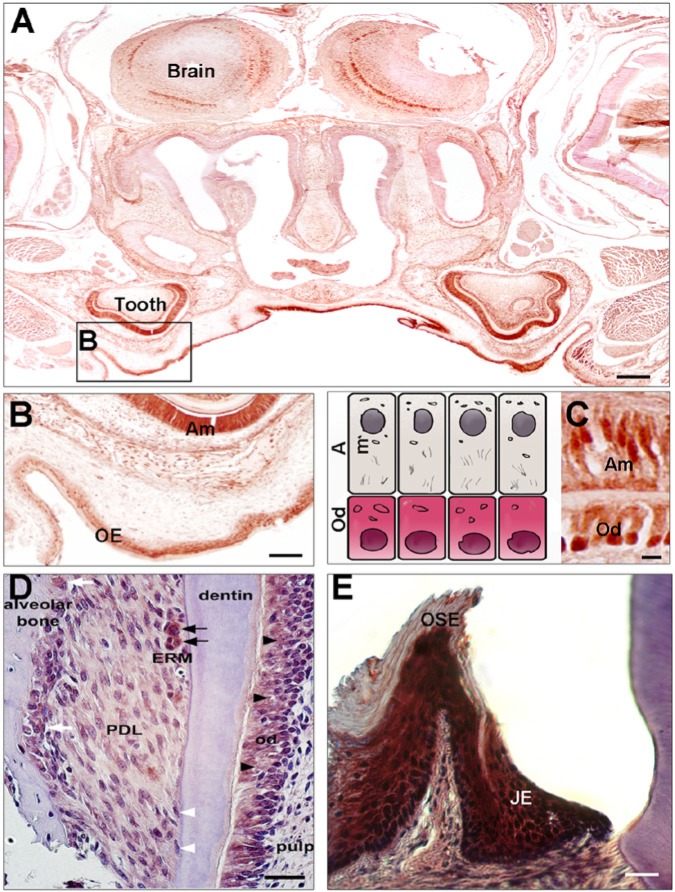
Figure 2.
Immunohistochemistry of CLOCK and PER2 proteins in post-natal days 4 and 21 mouse heads. (A) CLOCK protein expression is detected in the developing first molars and oral epithelium (OE) at the palate. (B,C) Higher magnifications showing that the nuclei (arrows) of ameloblasts (Am) and odontoblasts (Od) show strong CLOCK expression relative to the dental pulp cells. PER2 shows relatively weak expression in periodontal dental ligament (PDL) cells when compared with odontoblast expression (D). Epithelial rests of Malassez (ERM) show strong expression of the PER2 protein within the PDL space (D, black arrows). PER2 proteins are also detected in the nuclei of osteoblasts and osteoclasts in the alveolar bone (D, white arrows). No PER2 protein expression is detected in cementoblasts (D, white arrowheads). PER2 protein is also strongly detected in junctional epithelium (JE) but not in oral sulcular epithelium (OSE) (E). Scale bars = 200 μm in A, 100 μm in B, 20 μm in C, D, and E.
Teeth
Teeth are composed of 3 mineralized tissues: enamel, dentin, and cementum. These mineralized tissues form in extracellular spaces by matrix-mediated biomineralization. Several observations strongly suggest that dental mineralized tissue formation is under the control of complex biological clocks (Simmer et al., 2010). Indeed, enamel, dentin, and cementum form by additive modes of growth that preserve within the hard tissues short- and long-period lines of incremental growth. The information content in these periodic lines is increasingly being leveraged in many fields, including evolutionary biology, anthropology, archaeology, forensics, and dental development. Appropriate and full utilization of the information recorded in dental mineralized tissues requires understanding the developmental and circadian controls that cause teeth to form as they do. Understanding the circadian controls that govern tooth shape permits identification and characterization of the biological regulatory pathways that dictate dental cell differentiation (Athanassiou-Papaefthymiou et al., 2011; Zheng et al., 2013).
Oral Epithelium
Clock gene expression is detected in several epithelial craniofacial tissues, mainly in basal cells of oral epithelium, including palatal and junctional epithelia, but also in the epithelial rests of Malassez surrounding dental roots (Zheng et al., 2011) (Fig. 2). The role(s) of clock gene expression in oral epithelia remains to be elucidated.
Effects of the Circadian Clock in Salivary Glands and Saliva Flow
Salivary flow rate is known to follow circadian rhythms (Dawes, 1972) (Fig. 3). The secretory levels of saliva substances follow circadian rhythms, which, evidence suggests, are important for effective nutrition and self-defense. Studies suggest that salivary glands (SGs) may contain a circadian clock to regulate the type, amount, and content of saliva. Indeed, the existence of such a clock mechanism has been demonstrated for other vital organs such as the kidney (Stow and Gumz, 2011). SGs and the kidney share several physiological mechanisms, including a multitude of ion and water channel activities. Several genes that regulate fluid movement are highly expressed in both the kidney and SGs (Maeda et al., 2008). We have recently shown that clock genes are expressed in SG (Fig. 3), similar to the kidney (Zheng et al., 2012), supporting the idea of a peripheral clock in SGs. We also showed that clock genes regulate the water channel gene aquaporin-5 (Aqp5) (Zheng et al., 2012) (Fig. 3). Aqp5 plays an important role in salivary fluid secretion (Ishikawa and Ishida, 2000), suggesting that abnormal expression of clock genes will affect Aqp5 expression, resulting in changes in salivary flow.
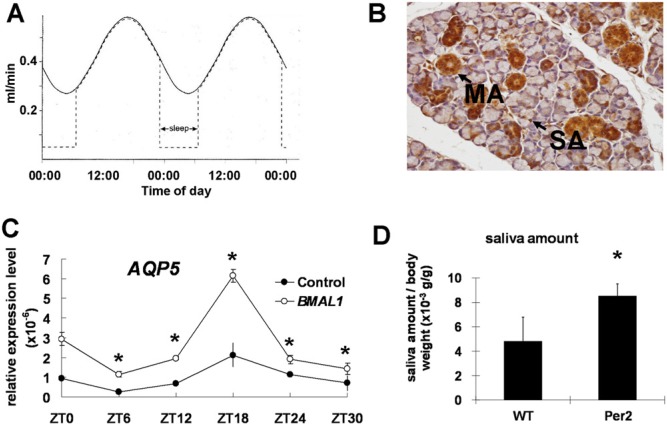
Figure 3.
The circadian clock of salivary glands regulates Aqp5 and cytokine expression and salivary flow levels. Circadian rhythms are detected in human unstimulated salivary flow (A). Immunohistochemistry results showed that the CLOCK protein is expressed in the nuclei of mucous acini (MA) and serous acini (SA), and in the epithelial cells of the ducts in mouse salivary glands. Expression was much weaker in duct cells. Scale bars = 20 μm. (B). AQP5 mRNA levels in HEK293 cells are up-regulated after transient transfection with a Bmal1 over-expression vector. All time interval calculations are based at the indicated zeitgeber (ZT; a zeitgeber is any external or environmental cue that entrains, or synchronizes, an organism’s biological rhythms) time; ZT 0 was considered to be 2 hrs after cell-cycle synchronization. The experiments were repeated 3 times, and 1 representative experiment is presented (*p < .05; compared with 0 hr). (C). The saliva level is higher in Per2 knock-out mice than in wild-type mice after simulation (D).
Saliva has numerous protective roles for oral tissue maintenance (Dowd, 1999). Adequate salivary flow and saliva content are directly related to health status. In contrast, reduced salivary flow rate is an indicator of xerostomia, which may be caused by SG disorders, such as Sjögren’s syndrome (SS), and chronic sialoadenitis. SS is an autoimmune disease with devastating oral health implications, including xerostomia (dry mouth due to hyposalivation) (Tzioufas et al., 2012). Patients with hyposalivation suffer from difficulties with swallowing and food mastication, dental caries, hampered speech, impaired taste, and nocturnal oral discomfort (Vissink et al., 2003). Of interest, analysis of our data in Per2 and Bmal1 knockout mice reveals that clock gene mutations affect saliva flow in salivary glands (Fig. 3). Preliminary evidence also suggests that circadian clock alterations are involved in the reduced saliva observed in persons with SS (unpublished observations). Further studies will help establish novel diagnostic tools and will provide a foundation for innovative treatment options for patients with SS and others.
Effects of the Circadian Clock in Human Diseases
Clock genes are key regulators of major cell functions, and research efforts are focusing on discovering how clock genes control cell proliferation during development and disease (Staels, 2006). More specifically, recent studies showed that alterations of the circadian clock are linked to disparate diseases, including cancer and rheumatoid arthritis (RA) (Shimba et al., 2011; Kouri et al., 2013). It remains unclear, however, whether perturbations of specific body clocks (e.g., sleep clock vs. feeding) preferentially predispose one to certain diseases. The direct links between body clocks and disease remain elusive. Regulation of chronic inflammation by the circadian clock may be one of the links between disease and normal body homeostasis (Gilbert and Slingerland, 2013; Fig. 4). More studies in these areas will benefit the fields of diagnosis and treatment of clock-related diseases.
Figure 4.
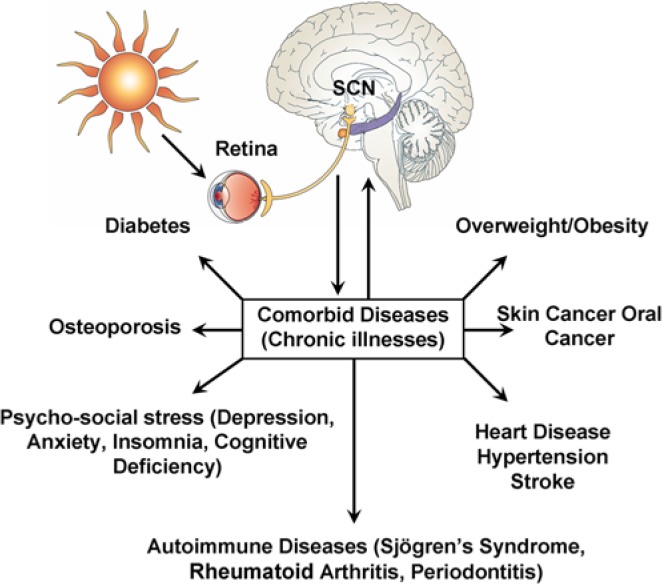
Emerging evidence strongly suggests that alterations of the circadian clock are directly or indirectly linked to an increasing list of chronic illnesses. This illustration summarizes the reported connections of the circadian clock with chronic diseases. Please note that patients can develop more than one of these diseases (co-morbidities). Modified from Kondratova and Kondratov, 2012.
Autoimmune Diseases
Accumulating evidence supports a complex relationship between the circadian rhythm and regulation of both the innate and adaptive immune systems. Cells including synovial fibroblasts, CD4+ T-cells, eosinophils, and macrophages are regulated by circadian oscillations and are therefore tightly controlled by clock gene transcription (Bollinger et al., 2011; Gibbs and Ray, 2013). As such, clock genes may have a role in the induction and elaboration of autoimmunity, in which normal mechanisms of self-tolerance and negative feedback are impaired. Like most autoimmune diseases, rheumatoid arthritis (RA), which causes both joint-based and systemic inflammatory disease, has clear genetic and environmental associations, as well as multiple pathways of immune dysregulation that are implicated in the disease (McInnes and Schett, 2011). Many of these pathways appear to be regulated by the circadian clock, including the pro-inflammatory cytokine IL-6 (Perry et al., 2009), regulation of the transcription of various peroxisome proliferator-activated receptor (PPAR) subtype genes (Kawai and Rosen, 2010), expression of tumor necrosis factor-α (TNF-α) (Kouri et al., 2013), and activity of the glucocorticoid receptor (Lamia et al., 2011). Therefore, it is reasonable to hypothesize that perturbations in clock gene expression may contribute to multiple downstream immunologic effects seen in a complex autoimmune disease such as RA. Indeed, induction of collagen-induced arthritis has been demonstrated to be more robust in a population of mice lacking 2 core clock genes (Cry1 and Cry2) compared with wild-type mice, in the absence of light cues (Hashiramoto et al., 2010).
Another autoimmune disease that often co-exists with RA is Sjögren’s syndrome (SS). Our group has recently shown that the mRNA and protein levels of clock genes are differentially expressed in the serous acini and duct cells of all major salivary glands in patients with primary Sjögren’s syndrome (PSS) (unpublished observations). SS can present either as a primary process or as a secondary phenomenon in patients with another underlying autoimmune disease, most commonly RA or systemic lupus erythematosus (SLE). SS is a chronic lymphoproliferative autoimmune disease associated with B- and T-lymphocytic infiltrate of exocrine glands, leading to xerostomia and keratoconjunctivitis sicca. Extra-glandular manifestations of this disease include central and peripheral nervous system disease, hematologic abnormalities, and cutaneous vasculitis, as well as a dramatically increased risk of malignant lymphoma and non-Hodgkin’s lymphoma (Voulgarelis and Moutsopoulos, 2008). In our recent study, expression levels of clock genes and aquaporin 5 (Aqp5) showed regular oscillatory patterns under both light/dark and complete dark conditions, suggesting that Aqp5 expression levels may be regulated by clock genes. Furthermore, RNA expression levels of clock genes correlated directly with salivary flow (Fig. 3) and are dramatically down-regulated in persons with SS (unpublished observations). In contrast, individuals with diminished salivary flow due to sialolithiasis showed clock genes levels similar to those of unaffected individuals (unpublished observations). This novel finding of down-regulation of clock gene expression and decreased salivary flow in persons with SS supports the hypothesis that altered clock gene mechanisms may have a role in SS, and indeed, may have a role in autoimmune diseases in general. Common features of autoimmune diseases such as RA, SS, and SLE include chronic fatigue, which can result from aberrant circadian oscillation, including the altered rhythms of circulating cortisol and melatonin, which have already been observed in patients with RA (Perry et al., 2009). Future study of the many effects of circadian oscillation on the immune system could have broad implications for the study and treatment of autoimmune diseases.
Oral and Head and Neck Cancer
The circadian clock is a mechanism that controls most cellular processes, including cell proliferation (Huang et al., 2011). In industrialized societies, changes in lifestyles lead to frequent disruption of endogenous circadian rhythm in about 50% of the human population, which contributes to increased cancer development worldwide (Hrushesky et al., 2009). Recent literature suggests key roles of clock genes in cancer progression in humans. Expression of critical cancer-related genes, such as c-myc and p53, has been reported to follow a circadian rhythm in vivo (Fu et al., 2002; Hunt and Sassone-Corsi, 2007). Loss of function of circadian genes leads to deregulated DNA damage and neoplastic growth, resulting in oncogenic activation, uncontrolled cell proliferation, and increased tumor development in mice (Hunt and Sassone-Corsi, 2007). Among the clock genes, Bmal1 and Per2 have drawn particular attention, since they demonstrate functional characteristics as tumor suppressors. Mutations or alterations of the promoter methylation status of Per2 or Bmal1 are linked with tumor progression and poor clinical outcome in patients (Taniguchi et al., 2009; Xia et al., 2010). The role of clock genes in head and neck tumorigenesis remains unclear. This review provides some preliminary findings in the area of oral squamous cell carcinomas and salivary gland cancer.
Salivary gland (SG) cancers constitute 1% to 6% of head and neck malignancies in the United States and remains one of the less-characterized tumor types at the molecular level (Boukheris et al., 2009). Their behavior is variable, and currently, there are limited effective therapeutic options for this group of deadly cancers. Increasing our understanding of the molecular mechanisms governing SG tumorigenesis will allow for the development of better treatment options in the future. We have recently shown that the mRNA and protein levels of clock genes are differentially expressed in the serous acini and duct cells of all major salivary glands. Of interest, expression of clock genes and their targets solute carrier family 4, anion exchanger, member 2 (SLC4A2/AE2), and AQP5 is dramatically altered in human SG tumors (unpublished observations). Analysis of these data suggests that the SG circadian clock undergoes significant changes in human SG malignancies.
Very little is known with regard to the implications of clock genes in oral epithelium homeostasis and their disturbances in oral and head and neck carcinogenesis. Early studies have confirmed the presence of various clock genes in oral mucosa in healthy diurnally active volunteers and have assessed their 24-hour variations (Bjarnason et al., 2001). That study provided the first evidence in human oral mucosa of a rhythmic circadian expression profile of Per1, Cry1, and Bmal1 (early in the morning, in late afternoon, and at night, respectively), similar to that found in the SCN and the peripheral tissues in rodents (Bjarnason et al., 2001). Furthermore, the major peak in Per1 expression in oral mucosa coincided with a G1-phase marker (p53), while the peak for Bmal1 coincided with an M-phase marker (cyclin β1), suggesting a potential functional link between the circadian clock and the mammalian cell cycle, in support of a circadian coordination of cell-cycle events in oral mucosa (Bjarnason et al., 2001), similar to that in other human mucosa [e.g., the rectum (Buchi et al., 1991) and the gastrointestinal tract (GI) (Hoogerwerf, 2006)]. Interestingly, evidence suggests a wave of clock gene expression from the oral mucosa to the rectum and coherent cell proliferation, suggesting that findings from oral mucosa may extrapolate to the downstream gastrointestinal tissues (Pardini et al., 2005). Later studies have confirmed and extended the array of circadian gene profiles detected in normal oral mucosa (to 16 of the 20 clock genes) and identified new oral-mucosal CCGs such as the tumor suppressor gene SMAD5 [human mothers against decapentaplegic homolog 5, a critical component of the intracellular tumor growth factor (TGF)-beta pathway] and Nocturin (CCRN4L). SMAD5 expression parallels that of hPER2, is under the control of CLOCK/BMAL1, and shows regulation identical to that of hPER1 (Zieker et al., 2010). These findings suggest that detection of the asynchronous regulation of these genes would allow for a more accurate assessment of cancer risk or monitoring of tumor progression. Ulterior studies have confirmed p53 and cyclin B1 as direct targets of the circadian clock genes in humans (Hunt and Sassone-Corsi, 2007) as well as increased incidence of GI cancers in people with disrupted circadian rhythms (Schernhammer et al., 2003). The mechanisms involved in clock-related regulation of carcinogenesis remain elusive, but analysis of recent data suggests that circadian rhythms may control stem cell renewal and quiescence in epithelial tissues (Janich et al., 2011).
Another retrospective study has investigated the expression profiles of 9 circadian clock genes in a cohort of 40 patients with head and neck squamous cell carcinomas (HNSCC) comparatively with their non-cancerous counterpart tissues (Hsu et al., 2012). The expression of hPER1, hPER2, hPER3, hCRY1, hCRY2, hCKIϵ, and hBMAL1 showed significant down-regulation in the tumoral tissues. Down-regulated hPER3, hCRY2, and hBMAL1 expression was correlated with more advanced cancer stages, while down-regulated hPER3 correlated with larger tumor size, deeper tumor invasion, and poor survival (Hsu et al., 2012). These results indicate a possible association of circadian clock genes, especially PER3, with the pathogenesis of HNSCC. However, the direct links between aberrant circadian clock gene expression and human malignancies, including oral and head and neck carcinomas, remain largely unknown.
Of interest, recent clinical studies indicate that perioperative stress in patients with esophageal and early gastric cancer has a considerable effect on the circadian expression of clock genes (Azama et al., 2007). Several factors have been incriminated as able to desynchronize the peripheral and central clocks, such as: types of surgical methods, operative duration, volume of blood loss, volume of infusion, time-imposed restricted feeding and parenteral nutrition, duration of hospitalization, and post-operative changes or complications and other associated treatments (glucocorticoid injections, anxiolytic drugs, etc.) (Azama et al., 2007). Recent investigations have also tried to determine the predictors of the poor sleep quality in patients with head and neck cancer (Shuman et al., 2010) and how healthy behaviors (a proper daily life routine, including physical activity, diet, sleep, etc.) influence inflammatory mediators and thus survival in these patients (Duffy et al., 2013). This emerging clinical evidence shows promise for chronobiology-based individualized treatment strategies in patients with HNSCC (Ohdo, 2010).
The Role of Epigenetics Modifications in Clock Gene Dysregulation and Related Diseases
It has been shown that polymorphisms and mutations of clock genes are directly linked to disease predisposition and to disease pathogenesis for a great number of diseases. There is also growing evidence that epigenetics may play an essential role in the control of the circadian clock (Zhu et al., 2011). The term ‘epigenetics’, literally meaning “beyond genetics”, refers to heritable changes and is generally considered to encompass several modes of regulation that do not include changes to the actual DNA sequence. These modes of regulation include the DNA methylation state at cytosine-guanine (CpG) sites and a multitude of chemical modifications to histone tails (including acetylation, methylation, ubiquitination, and phosphorylation) across the genome. More recently, the term epigenetics has included the various levels of small non-coding RNAs (e.g., microRNAs) in a cell (Girardot et al., 2012). All of these epigenetic mechanisms play a regulatory role in gene expression, thus affecting phenotypes at both cellular and organismal levels, and all are associated with diseases such as oral cancers (Gasche and Goel, 2012). Increased DNA methylation in CpG dense regions termed ‘CpG islands’, which often reside in gene promoter regions, is associated with transcriptional inhibition. Conversely, a lack of DNA methylation at CG sites outside of genic regions is associated with transcriptional activation.
Methylation and/or acetylation of histone tails leads to chromatin remodeling, which can mediate transcriptional response of CCGs to circadian signals such as light or nutrients (Sahar and Sassone-Corsi, 2013). For example, the histone deacetylase HDAC3 displays a rhythmic binding pattern which, when disrupted, resulted in hepatic steatosis in mice (Feng et al., 2011). The histone methyltransferase MLL3 was identified as being clock-regulated and that, in turn, it regulates many CCGs in the liver (Valekunja et al., 2013). Given the role that CLOCK and related genes play in regulating metabolism via histone modifications, it would not be surprising if circadian dysregulation of chromatin acetylation and/or methylation were involved in oral diseases.
We have recently found evidence that circadian genes are also involved in dysregulation of DNA methylation in oral diseases. In particular, preliminary analysis of publicly available data from The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov/) suggests dysregulation of the genes Per2 and Per3 via DNA methylation. Three sites near Per2 were observed to be hypermethylated in head and neck cancers (n = 277), compared with matched normal controls, and to be significantly more hypermethylated in tumors negative for Human Papillomavirus (HPV) (19 % to 25% more methylated than normal, on average) than in those that were HPV-positive (11 % to 15% more methylated than normal, on average). Correspondingly, expression of Per2 was ~1.5-fold higher in HPV-positive tumors than in those that were HPV-negative. Based on the same data, Per3 was also observed to be hypermethylated in head and neck cancer, but mainly in the HPV-negative tumors compared with normal (5 sites ranging from 11% to 23% more methylated, on average), with a resulting 2.3-fold decrease in expression in HPV-negative compared with HPV-positive tumors. It will be interesting to determine whether these differences correlate with disease-free survival or other prognostic indicators. In addition, we have shown that variability in DNA methylation exists within subtypes of HNSCC and is influenced by environmental factors such as diet (Sartor et al., 2011; Colacino et al., 2012). Future research is needed to show the extent to which circadian rhythm genes are involved in epigenetic dysregulation in oral diseases.
Conclusions
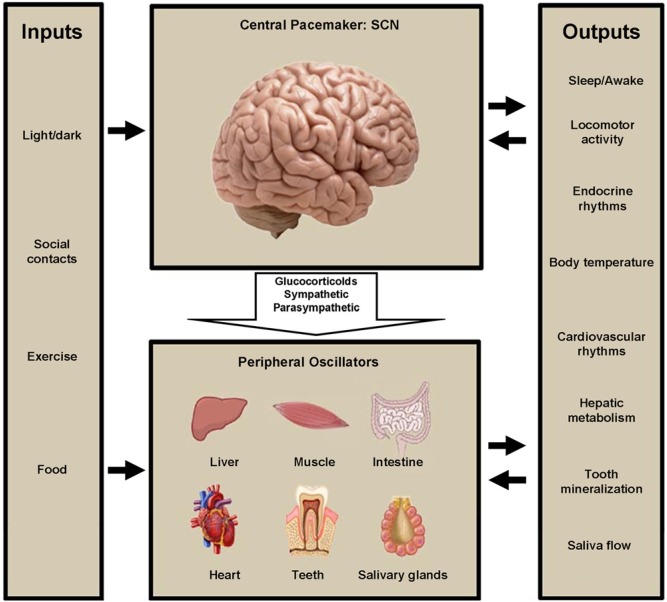
Elucidation of the circadian clock mechanisms guarantees a better understanding of oral tissue formation. It also opens a fascinating new area of research linking oral and systemic health. The list of the inputs that maintain the circadian rhythms in our bodies is increasing (Fig. 5). It is unknown what regulates circadian oral clocks. Additional to the previously known circadian clock outputs, we can now postulate that mineralized tissue formation and saliva production and content may be valid outputs of peripheral circadian clocks (Fig. 5). It is unclear at this point how the different outputs influence the general communication between the central and peripheral clocks regulating the whole body as a system.
Figure 5.
This illustration summarizes the list of the known inputs and outputs of the circadian clock system. It also shows the most well-characterized interactions between the central and peripheral clocks. We have included tooth mineralization and saliva production as novel outputs in this list. Modified after Garaulet and Madrid, 2009.
Dissecting the molecular circadian clock mechanisms involved in oral and systemic diseases will also open new areas of research contributing to the diagnosis and treatment of several clock-related diseases (CRD). Cross-disciplinary research in well-defined populations is necessary to further enhance knowledge and develop clinical strategies to coordinate therapy and risk assessments of CRD diseases. This information will help care-providers identify the best time to treat certain diseases in each of their patients, and understand how the diseases may be influenced by the patients’ immune systems’ daily peaks and related body functions.
Footnotes
P.P. is supported by NIH grant DE018878 and the University of Michigan (OVPR funds, M-cube) funds. W.M. is supported by K12HD001438 from NIH/ORWH. E.C.S. is supported by K01ES019909. M.A.S. is supported by NCI grant # R01CA158286-01A1. S.P. is supported by the UM Department of Otolaryngology – Head and Neck Surgery.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Athanassiou-Papaefthymiou M, Kim D, Harbron L, Papagerakis S, Schnell S, Harada H, et al. (2011). Molecular and circadian controls of ameloblasts. Eur J Oral Sci 119(Suppl 1):35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azama T, Yano M, Oishi K, Kadota K, Hyun K, Tokura H, et al. (2007). Altered expression profiles of clock genes hPer1 and hPer2 in peripheral blood mononuclear cells of cancer patients undergoing surgery. Life Sci 80:1100-1108. [DOI] [PubMed] [Google Scholar]
- Bass J, Takahashi JS. (2010). Circadian integration of metabolism and energetics. Science 330:1349-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjarnason GA, Jordan RC, Wood PA, Li Q, Lincoln DW, Sothern RB, et al. (2001). Circadian expression of clock genes in human oral mucosa and skin: association with specific cell-cycle phases. Am J Pathol 158:1793-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger T, Leutz A, Leliavski A, Skrum L, Kovac J, Bonacina L, et al. (2011). Circadian clocks in mouse and human CD4+ T cells. PLoS One 6:e29801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukheris H, Curtis RE, Land CE, Dores GM. (2009). Incidence of carcinoma of the major salivary glands according to the WHO classification, 1992 to 2006: a population-based study in the United States. Cancer Epidemiol Biomarkers Prev 18:2899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchi KN, Moore JG, Hrushesky WJ, Sothern RB, Rubin NH. (1991). Circadian rhythm of cellular proliferation in the human rectal mucosa. Gastroenterology 101:410-415. [DOI] [PubMed] [Google Scholar]
- Colacino JA, Arthur AE, Dolinoy DC, Sartor MA, Duffy SA, Chepeha DB, et al. (2012). Pretreatment dietary intake is associated with tumor suppressor DNA methylation in head and neck squamous cell carcinomas. Epigenetics 7:883-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C. (1972). Circadian rhythms in human salivary flow rate and composition. J Physiol 220:529-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. (2010). The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol 72:517-549. [DOI] [PubMed] [Google Scholar]
- Dowd FJ. (1999). Saliva and dental caries. Dent Clin North Am 43:579-597. [PubMed] [Google Scholar]
- Duffy SA, Teknos T, Taylor JM, Fowler KE, Islam M, Wolf GT, et al. (2013). Health behaviors predict higher interleukin-6 levels among patients newly diagnosed with head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 22:374-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. (2011). A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331:1315-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Pelicano H, Liu J, Huang P, Lee C. (2002). The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111:41-50. [DOI] [PubMed] [Google Scholar]
- Garaulet M, Madrid JA. (2009). Chronobiology, genetics and metabolic syndrome. Curr Opin Lipidol 20:127-134. [DOI] [PubMed] [Google Scholar]
- Gasche JA, Goel A. (2012). Epigenetic mechanisms in oral carcinogenesis. Future Oncol 8:1407-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JE, Ray DW. (2013). The role of the circadian clock in rheumatoid arthritis. Arthritis Res Ther 15:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert CA, Slingerland JM. (2013). Cytokines, obesity, and cancer: new insights on mechanisms linking obesity to cancer risk and progression. Annu Rev Med 64:45-57. [DOI] [PubMed] [Google Scholar]
- Girardot M, Cavaille J, Feil R. (2012). Small regulatory RNAs controlled by genomic imprinting and their contribution to human disease. Epigenetics 7:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashiramoto A, Yamane T, Tsumiyama K, Yoshida K, Komai K, Yamada H, et al. (2010). Mammalian clock gene Cryptochrome regulates arthritis via proinflammatory cytokine TNF-alpha. J Immunol 184:1560-1565. [DOI] [PubMed] [Google Scholar]
- Hoogerwerf WA. (2006). Biologic clocks and the gut. Curr Gastroenterol Rep 8:353-359. [DOI] [PubMed] [Google Scholar]
- Hrushesky WJ, Grutsch J, Wood P, Yang X, Oh EY, Ansell C, et al. (2009). Circadian clock manipulation for cancer prevention and control and the relief of cancer symptoms. Integr Cancer Ther 8:387-397. [DOI] [PubMed] [Google Scholar]
- Hsu CM, Lin SF, Lu CT, Lin PM, Yang MY. (2012). Altered expression of circadian clock genes in head and neck squamous cell carcinoma. Tumour Biol 33:149-155. [DOI] [PubMed] [Google Scholar]
- Huang W, Ramsey KM, Marcheva B, Bass J. (2011). Circadian rhythms, sleep, and metabolism. The J Clin Invest 121:2133-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt T, Sassone-Corsi P. (2007). Riding tandem: circadian clocks and the cell cycle. Cell 129:461-464. [DOI] [PubMed] [Google Scholar]
- Ishikawa Y, Ishida H. (2000). Aquaporin water channel in salivary glands. Jpn J Pharmacol 83:95-101. [DOI] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos-Suarez A, Batlle E, Ripperger J, Albrecht U, et al. (2011). The circadian molecular clock creates epidermal stem cell heterogeneity. Nature 480:209-214. [DOI] [PubMed] [Google Scholar]
- Kawai M, Rosen CJ. (2010). PPARgamma: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol 6:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JK, Forger DB. (2012). A mechanism for robust circadian timekeeping via stoichiometric balance. Mol Syst Biol 8:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratova AA, Kondratov RV. (2012). The circadian clock and pathology of the ageing brain. Nat Rev Neurosci 13:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouri VP, Olkkonen J, Kaivosoja E, Ainola M, Juhila J, Hovatta I, et al. (2013). Circadian timekeeping is disturbed in rheumatoid arthritis at molecular level. PLoS One 8:e54049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, et al. (2011). Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nature 480:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HM, Chen R, Kim H, Etchegaray JP, Weaver DR, Lee C. (2011). The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc Natl Acad Sci USA 108:16451-16456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Kuwahara S, Ito H, Tanaka K, Hayakawa T, Seki M. (2008). Expression and localization of aquaporins in the kidney of the musk shrew (Suncus murinus). J Histochem Cytochem 56:67-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes IB, Schett G. (2011). The pathogenesis of rheumatoid arthritis. N Engl J Med 365:2205-2219. [DOI] [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. (2012). Central and peripheral circadian clocks in mammals. Annu Rev Neurosci 35:445-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak B, Tyson JJ. (2008). Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9:981-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohdo S. (2010). Chronotherapeutic strategy: rhythm monitoring, manipulation and disruption. Adv Drug Deliv Rev 62:859-875. [DOI] [PubMed] [Google Scholar]
- Pardini L, Kaeffer B, Trubuil A, Bourreille A, Galmiche JP. (2005). Human intestinal circadian clock: expression of clock genes in colonocytes lining the crypt. Chronobiol Int 22:951-961. [DOI] [PubMed] [Google Scholar]
- Perry MG, Kirwan JR, Jessop DS, Hunt LP. (2009). Overnight variations in cortisol, interleukin 6, tumour necrosis factor alpha and other cytokines in people with rheumatoid arthritis. Ann Rheum Dis 68:63-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, et al. (2002). The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110:251-260. [DOI] [PubMed] [Google Scholar]
- Raspe E, Mautino G, Duval C, Fontaine C, Duez H, Barbier O, et al. (2002). Transcriptional regulation of human Rev-erbalpha gene expression by the orphan nuclear receptor retinoic acid-related orphan receptor alpha. J Biol Chem 277:49275-49281. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. (2013). The epigenetic language of circadian clocks. Handb Exp Pharmacol 217:29-44. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Dolinoy DC, Jones TR, Colacino JA, Prince ME, Carey TE, et al. (2011). Genome-wide methylation and expression differences in HPV(+) and HPV(-) squamous cell carcinoma cell lines are consistent with divergent mechanisms of carcinogenesis. Epigenetics 6:777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schernhammer ES, Laden F, Speizer FE, Willett WC, Hunter DJ, Kawachi I, et al. (2003). Night-shift work and risk of colorectal cancer in the nurses’ health study. J Natl Cancer Inst 95:825-828. [DOI] [PubMed] [Google Scholar]
- Schmutz I, Ripperger JA, Baeriswyl-Aebischer S, Albrecht U. (2010). The mammalian clock component PERIOD2 coordinates circadian output by interaction with nuclear receptors. Genes Dev 24:345-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, et al. (2011). Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One 6:e25231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman AG, Entezami P, Chernin AS, Wallace NE, Taylor JM, Hogikyan ND. (2010). Demographics and efficacy of head and neck cancer screening. Otolaryngol Head Neck Surg 143:353-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer JP, Papagerakis P, Smith CE, Fisher DC, Rountrey AN, Zheng L, et al. (2010). Regulation of dental enamel shape and hardness. J Dent Res 89:1024-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B. (2006). When the Clock stops ticking, metabolic syndrome explodes. Nat Med 12:54-55. [DOI] [PubMed] [Google Scholar]
- Stow LR, Gumz ML. (2011). The circadian clock in the kidney. J Am Soc Nephrol 22:598-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Fernandez AF, Setien F, Ropero S, Ballestar E, Villanueva A, et al. (2009). Epigenetic inactivation of the circadian clock gene BMAL1 in hematologic malignancies. Cancer Res 69:8447-8454. [DOI] [PubMed] [Google Scholar]
- Tzioufas AG, Tatouli IP, Moutsopoulos HM. (2012). Autoantibodies in Sjögren’s syndrome: clinical presentation and regulatory mechanisms. Presse Med 41(9 Pt 2):e451-e460. [DOI] [PubMed] [Google Scholar]
- Valekunja UK, Edgar RS, Oklejewicz M, van der Horst GT, O’Neill JS, Tamanini F, et al. (2013). Histone methyltransferase MLL3 contributes to genome-scale circadian transcription. Proc Natl Acad Sci USA 110:1554-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissink A, Jansma J, Spijkervet FK, Burlage FR, Coppes RP. (2003). Oral sequelae of head and neck radiotherapy. Crit Rev Oral Biol Med 14:199-212. [DOI] [PubMed] [Google Scholar]
- Voulgarelis M, Moutsopoulos HM. (2008). Mucosa-associated lymphoid tissue lymphoma in Sjögren’s syndrome: risks, management, and prognosis. Rheum Dis Clin North Am 34:921-933, viii. [DOI] [PubMed] [Google Scholar]
- Weaver DR. (1998). The suprachiasmatic nucleus: a 25-year retrospective. J Biol Rhythms 13:100-112. [DOI] [PubMed] [Google Scholar]
- Xia HC, Niu ZF, Ma H, Cao SZ, Hao SC, Liu ZT, et al. (2010). Deregulated expression of the Per1 and Per2 in human gliomas. Can J Neurol Sci 37:365-370. [DOI] [PubMed] [Google Scholar]
- Zheng L, Papagerakis S, Schnell SD, Hoogerwerf WA, Papagerakis P. (2011). Expression of clock proteins in developing tooth. Gene Expr Patterns 11:202-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Seon YJ, McHugh J, Papagerakis S, Papagerakis P. (2012). Clock genes show circadian rhythms in salivary glands. J Dent Res 91:783-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Seon YJ, Mourao MA, Schnell S, Kim D, Harada H, et al. (2013). Circadian rhythms regulate amelogenesis. Bone 55:158-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Stevens RG, Hoffman AE, Tjonneland A, Vogel UB, Zheng T, et al. (2011). Epigenetic impact of long-term shiftwork: pilot evidence from circadian genes and whole-genome methylation analysis. Chronobiol Int 28:852-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zieker D, Jenne I, Koenigsrainer I, Zdichavsky M, Nieselt K, Buck K, et al. (2010). Circadian expression of clock- and tumor suppressor genes in human oral mucosa. Cell Physiol Biochem 26:155-166. [DOI] [PubMed] [Google Scholar]