This paper reports a strategy for combining somatic mutation profiles of human tumors with gene networks to stratify tumors into biologically and clinically relevant subtypes. The method is applied to ovarian, uterine and lung cancers.
Supplementary information
The online version of this article (doi:10.1038/nmeth.2651) contains supplementary material, which is available to authorized users.
Subject terms: Cancer genetics, Machine learning, Systems biology, Genomic analysis
Abstract
Many forms of cancer have multiple subtypes with different causes and clinical outcomes. Somatic tumor genome sequences provide a rich new source of data for uncovering these subtypes but have proven difficult to compare, as two tumors rarely share the same mutations. Here we introduce network-based stratification (NBS), a method to integrate somatic tumor genomes with gene networks. This approach allows for stratification of cancer into informative subtypes by clustering together patients with mutations in similar network regions. We demonstrate NBS in ovarian, uterine and lung cancer cohorts from The Cancer Genome Atlas. For each tissue, NBS identifies subtypes that are predictive of clinical outcomes such as patient survival, response to therapy or tumor histology. We identify network regions characteristic of each subtype and show how mutation-derived subtypes can be used to train an mRNA expression signature, which provides similar information in the absence of DNA sequence.
Supplementary information
The online version of this article (doi:10.1038/nmeth.2651) contains supplementary material, which is available to authorized users.
Main
Cancer is a disease that is not only complex, i.e., driven by a combination of genes, but also wildly heterogeneous, in that gene combinations can vary greatly between patients. To gain a better understanding of these complexities, researchers involved in projects such as The Cancer Genome Atlas (TCGA) and the International Cancer Genome Consortium (ICGC) are systematically profiling thousands of tumors at multiple layers of genome-scale information, including mRNA and microRNA expression, DNA copy number and methylation, and DNA sequence1,2,3. There is now a strong need for informatics methods that can integrate and interpret genome-scale molecular information to provide insight into the molecular processes driving tumor progression. Such methods are also of pressing need in the clinic, where the impact of genome-scale tumor profiling has been limited by the inability to derive clinically relevant conclusions from the data4,5.
One of the fundamental goals of cancer informatics is tumor stratification, whereby a heterogeneous population of tumors is divided into clinically and biologically meaningful subtypes as determined by similarity of molecular profiles. Most prior attempts to stratify tumors with molecular profiles have used mRNA expression data2,6,7,8,9, resulting in the discovery of informative subtypes in diseases such as glioblastoma and breast cancer. On the other hand, in TCGA cohorts including colorectal adenocarcinoma and small-cell lung cancer, subtypes derived from expression profiles do not correlate with any clinical phenotype including patient survival and response to chemotherapy2,10. These results might be due to limitations of expression-based analysis11 such as issues with RNA sample quality, lack of reproducibility between biological replicates and ample opportunities for overfitting of data.
A promising new source of data for tumor stratification is the somatic mutation profile, in which high-throughput sequencing is used to compare the genome or exome of a patient's tumor to that of the germ line to identify mutations that have become enriched in the tumor cell population12. As this set of mutations is presumed to contain the causal drivers of tumor progression13, similarities and differences in mutations across patients could provide invaluable information for stratification. Although individual mutations in cancer genes have long been used to stratify patients14,15,16,17, stratification based on the entire mutation profile has been more challenging. Somatic mutations are fundamentally unlike other data types such as expression or methylation, in which nearly all genes or markers are assigned a quantitative value in every patient. Instead, somatic mutation profiles are extremely sparse, with typically fewer than 100 mutated bases in an entire exome (Supplementary Fig. 1). They are also remarkably heterogeneous, such that it is very common for clinically identical patients to share no more than a single mutation2,18,19.
Here we report that these problems can be largely overcome by integrating somatic mutation profiles with knowledge of the molecular network architecture of human cells. It is widely appreciated that cancer is a disease not of individual mutations, nor of genes, but of combinations of genes acting in molecular networks corresponding to hallmark processes such as cell proliferation and apoptosis20,21. We postulated that, although two tumors may not have any mutations in common, they may share the networks affected by these mutations (as per Waddington's original theory of 'genetic canalization'22). Although current cancer pathway maps are incomplete, much relevant information is available in public databases of human protein-protein, functional and pathway interactions. An increasing number of studies have successfully integrated these network databases with tumor molecular profiles to map the molecular pathways of cancer23,24,25,26,27. Here we focus on the orthogonal problem of using network knowledge to stratify a cohort into meaningful subsets. Using this knowledge, we were able to cluster somatic mutation profiles into robust tumor subtypes that are biologically informative and have a strong association to clinical outcomes such as patient survival time and emergence of drug resistance. As a proof of principle, we applied this method to stratify the somatic mutation profiles of three major cancers cataloged in TCGA: ovarian, uterine and lung adenocarcinoma.
Results
Overview of network-based stratification
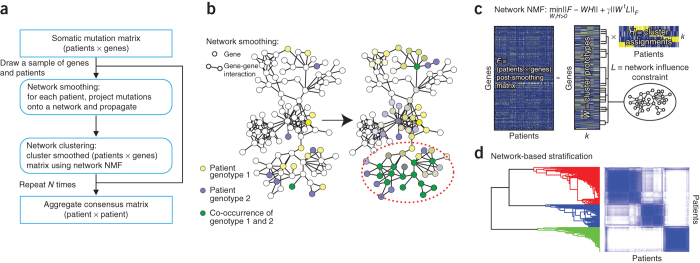
NBS combines genome-scale somatic mutation profiles with a gene interaction network to produce a robust subdivision of patients into subtypes (Fig. 1a). Briefly, somatic mutations for each patient are represented as a profile of binary (1, 0) states on genes, in which a '1' indicates a gene for which mutation (a single-nucleotide base change or the insertion or deletion of bases) has occurred in the tumor relative to germ line. For each patient, we project the mutation profile onto a human gene interaction network obtained from public databases28,29,30. Next we apply network propagation31 to spread the influence of each mutation over its network neighborhood (Fig. 1b). The resulting matrix of 'network-smoothed' patient profiles is clustered into a predefined number of subtypes (k = 2, 3, ... 12) via non-negative matrix factorization32 (NMF, Fig. 1c), an unsupervised technique. Finally, to promote robust cluster assignments, we use consensus clustering33, aggregating the results of 1,000 different subsamples from the entire data set into a single clustering result (Fig. 1d). For further details, see Online Methods. To evaluate the impact of different sources of network data, we used three interaction databases for this analysis: search tool for the retrieval of interacting genes (STRING)29, HumanNet28 or PathwayCommons30. Supplementary Table 1 summarizes the number of genes and interactions used in our analysis from each of these three networks. Our implementation of NBS is available as Supplementary Software; for updated versions, NBS may be downloaded from http://idekerlab.ucsd.edu/software/NBS/.
Figure 1. Overview of network-based stratification (NBS).
(a) Flowchart of the approach. (b) Example illustrating smoothing of patient somatic mutation profiles over a molecular interaction network. Mutated genes are shown in yellow (patient 1) and blue (patient 2) in the context of a gene interaction network. Following smoothing, the mutational activity of a gene is a continuous value reflected in the intensity of yellow or blue; genes with high scores in both patients appear in green (dashed oval). (c) Clustering mutation profiles using non-negative matrix factorization (NMF) regularized by a network. The input data matrix (F) is decomposed into the product of two matrices: one of subtype prototypes (W) and the other of assignments of each mutation profile to the prototypes (H). The decomposition attempts to minimize the objective function shown, which includes a network influence constraint L on the subtype prototypes. k, predefined number of subtypes. (d) The final tumor subtypes are obtained from the consensus (majority) assignments of each tumor after 1,000 applications of the procedures in b and c to samples of the original data set. A darker blue color in the matrix coincides with higher co-clustering for pairs of patients.
Benchmarking and performance analysis
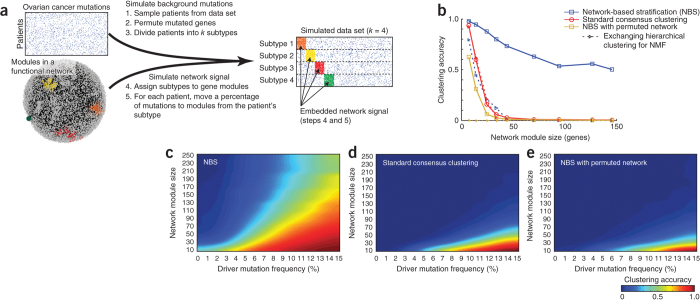
In an initial exploration of NBS, we simulated a somatic mutation data set using the structure of the TCGA ovarian tumor mutation data and the STRING gene interaction network (Fig. 2a). Mutation profiles were permuted, and patients were divided randomly and uniformly into a predefined number of subtypes (k = 4). Next we reassigned a fraction of mutations in each patient to fall within genes of a single 'network module' characteristic of that patient's subtype (the 'driver' mutation frequency f, varied from 0% to 15%); the remaining mutations were left to occur randomly. We selected the network modules randomly from the set of all network modules in STRING, defined as sets of densely interacting genes with size range s = 10–250 (see Online Methods for details and justification for the ranges of k, f and s). Although it is unknown whether these assumptions completely mirror the biology of cancer, they provide a reasonable model of a pathway-based genetic disease that is (i) driven by genetic circuits corresponding to a molecular network whose activity can be altered by mutations at multiple genes and (ii) characterized by many additional mutations that are noncausal 'passengers'.
Figure 2. Exploring performance of NBS through simulation.
(a) TCGA somatic mutations for ovarian cancer (top left) are combined with the STRING human protein interaction network (bottom left) to generate simulated mutation data sets embedded with known network structure (right). (b) Accuracy with which NBS clusters recover simulated subtype assignments, evaluated with and without network smoothing and using non-negative matrix factorization (NMF) versus hierarchical clustering. Accuracy is calculated as the Adjusted Rand Index57 of overlap between the clusters and correct subtype assignments, for which a score of 0 represents random overlap and 1 represents perfect overlap. Simulation was performed with a driver mutation frequency f = 7.5% with a single network module assigned to each subtype. (c) Accuracy landscape of NBS across varying driver mutation frequency and module size. (d) As in c, for a standard non-network–based clustering approach. (e) As in c, using a permuted network.
Using this simulation framework, we measured the ability of NBS to recover the correct subtype assignments in comparison to a standard consensus clustering approach not based on network knowledge (Online Methods). NBS showed a striking improvement in performance, especially for large network modules, as these can be associated with any of numerous different mutations across the patient population (Fig. 2b). As module size decreased, the chance of observing the same mutated gene in patients of the same subtype increased, and the standard clustering algorithm performed increasingly well. We found that the high performance of NBS depended not only on network smoothing but also on the NMF clustering approach; substitution of NMF with an alternative method such as hierarchical clustering resulted in relatively poor performance (Fig. 2b).
Next we investigated how NBS performance was affected as a function of mutation frequency (Fig. 2c). Standard consensus clustering was sufficient for stratification at high mutation frequencies and for small modules, for which there is substantial overlap in mutations among patients of the same subtype (Fig. 2d); however, NBS was able to accurately recover the correct subtypes for a much larger range of both variables. Applying NBS on a permuted network resulted in poor performance (Fig. 2e), which is on par with that observed with standard consensus clustering. These results were qualitatively similar when we used multiple network modules per patient (2–6) and/or a different network (Supplementary Fig. 2).
Network-based stratification of tumor mutations
We next sought to apply NBS to stratify patients profiled by TCGA full-exome sequencing for uterine, ovarian and lung cancers (see Online Methods for further details). In each of the three cancers, we observed that NBS resulted in robust subtype structure, whereas standard consensus clustering was unable to stratify the patient cohort (Fig. 3a for uterine cancer; Supplementary Figs. 3a and 4a for ovarian and lung cancers, respectively). Similar results were obtained when we used any of the three human networks (STRING, HumanNet and PathwayCommons).
Figure 3. NBS of somatic tumor mutations.
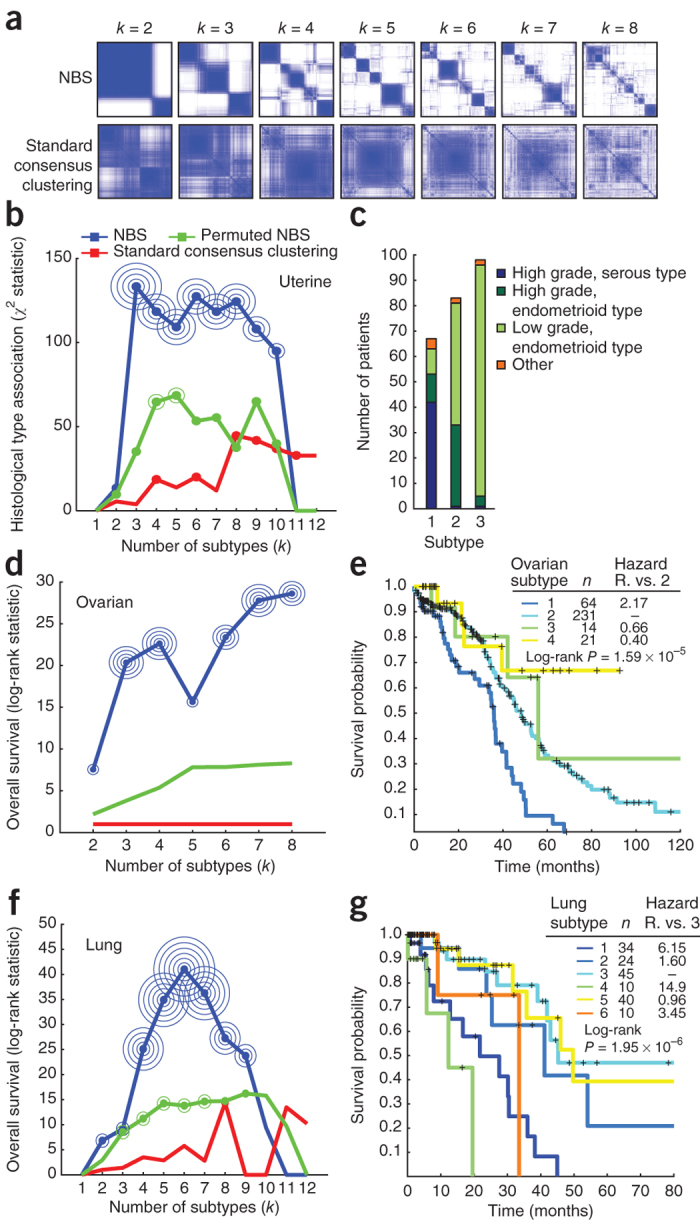
(a) Co-clustering matrices for uterine cancer patients, comparing NBS (STRING) (top) to standard consensus clustering (bottom). (b,c) Association of NBS subtypes with histology (b) and composition of NBS subtypes in terms of histological type and tumor grade (c) for uterine cancer. (d,e) Association of NBS subtypes (HumanNet) with patient survival time (d) and Kaplan-Meier survival plots for NBS subtypes (e) for ovarian cancer. (f,g) Association of NBS subtypes (HumanNet) with patient survival time (f) and Kaplan-Meier survival plots for NBS subtypes (g) for lung cancer. (b,d,f) P value of significance of 10−k is indicated by k concentric circles surrounding a data point (for example, three concentric circles indicate P < 0.001); in the case of uterine a significance of 10−5k is indicated by k concentric circles (for example, one circle indicates P < 10−5). Hazard R., hazard ratio, the ratio of fatalities between the two indicated subtypes over the studied time interval.
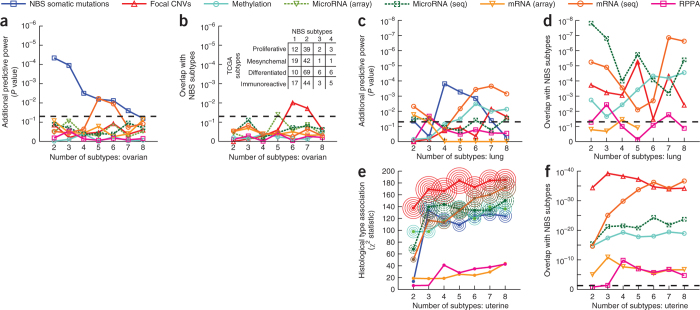
To determine the biological importance of the identified subtypes, we investigated whether they were predictive of observed clinical data. In uterine cancer, NBS subtypes (Supplementary Table 2) were closely associated with the recorded subtype on a histological basis (Fig. 3b,c and Supplementary Fig. 5). Survival analysis was not possible owing to low mortality rates for this cohort. In ovarian cancer, the identified subtypes (Supplementary Table 3) were significant predictors of patient survival time (log-rank P = 1.59 × 10−5; Fig. 3d,e and Supplementary Fig. 3b,c). Patients with the most aggressive ovarian tumor NBS subtype had a mean survival of approximately 32 months, compared to more than 80 months for those with the least aggressive NBS subtype (Supplementary Fig. 3d,e). Moreover, the NBS subtypes were predictive of survival independently of clinical covariates including tumor stage, age, mutation rate and residual tumor presence after surgery (Supplementary Fig. 6; likelihood ratio test, P = 3.75 × 10−5) and were also predictive of time to relapse after treatment with platinum chemotherapy ('platinum-free interval') (Supplementary Fig. 3f), as measured using a Kaplan-Meier analysis of platinum-free survival34. Finally, in lung cancer the identified NBS subtypes (Supplementary Table 4) were also significant predictors of patient survival (log-rank P = 1.95 × 10−6, Fig. 3f,g; median survival of 12 months versus approximately 50 months for the best-surviving subtype, Supplementary Fig. 4), with predictive value beyond known clinical covariates such as tumor stage, grade, mutation frequency, age at diagnosis and smoking status (likelihood ratio test, P = 3.3 × 10−4). Stratification using a network in which the mapping between mutated genes and the network was permuted, which disrupted the relationship between mutations and network structure, resulted in degraded predictive performance (Fig. 3b,d,f).
We compared these results to subtypes derived from other data types in the TCGA, including copy-number variation (CNV), methylation, mRNA expression, microRNA expression and protein profiles. For ovarian cancer, all other data types had inferior ability to predict survival beyond what could be predicted from clinical covariates (Fig. 4a) and led to different subtype assignments than NBS (Fig. 4b). In lung cancer, both NBS subtypes and those based on RNA-seq had good predictive power (Fig. 4c) and had some overlap in terms of patient assignments (Fig. 4d), whereas other data types were not predictive of survival. In uterine cancer, subtypes derived from all data types were highly predictive of histology (Fig. 4e; CNVs had highest predictive power overall) and also had very high overlap with NBS subtype assignments (Fig. 4f).
Figure 4. Predictive power and overlap of subtypes derived from different TCGA datasets.
(a) Predictive power in ovarian cancer. For each data type (line color), the power for predicting patient survival time beyond clinical indicators is shown as a function of number of subtypes. (b) Significance of overlap of ovarian cancer subtypes identified by each data type (line color) with subtypes identified by NBS. The table shows the number of patients shared between each NBS subtype and those defined by the TCGA using gene expression. (c) Predictive power in lung cancer, as for a. (d) Significance of overlap of lung cancer subtypes with NBS, as for b. (e) Association between uterine cancer subtype and tumor histology (y axis) as a function of the number of subtypes. P value of significance is indicated by concentric circles as in Figure 3. Colors are as in other panels, symbols have been omitted for clarity. (f) Significance of overlap of uterine cancer subtypes with NBS, as for b. Dashed horizontal lines indicate the P = 0.05 threshold of significance.
Distinct network modules associate with each tumor subtype
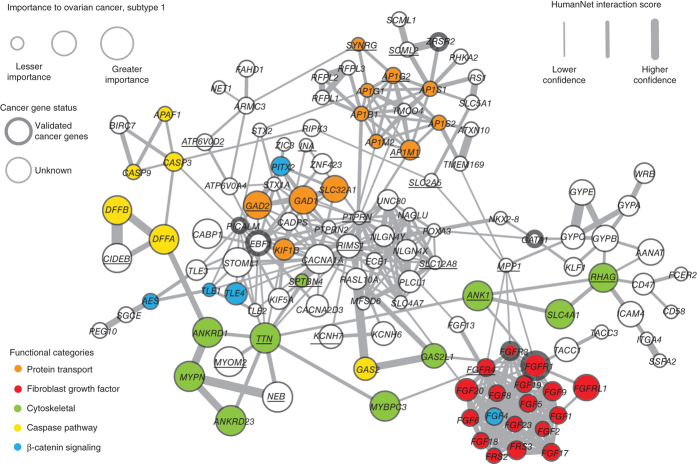
We next sought to identify the regions of the network that are most responsible for discriminating the somatic mutation profiles of tumors of different subtypes. Focusing on ovarian cancer as a proof of principle, for each subtype we identified genes for which the network-smoothed mutation state differs significantly for patients of that subtype versus the others (false discovery rate <0.05; Online Methods). This set of genes was projected onto the HumanNet network and visualized using Cytoscape35. The network for subtype 1 (Fig. 5), which had the worst overall survival and shortest platinum-free interval, contained over 20 genes in the fibroblast growth factor (FGF) signaling pathway, which has previously been implicated as a driver of tumor progression and associated with resistance to platinum and anti-VEGF therapy36. The network for subtype 2 was enriched in DNA damage–response genes including ATM, ATR, BRCA1, BRCA2, RAD51 and CHEK2 (Supplementary Fig. 7). Collectively these highlighted pathways are characteristic of a functional deficit in response to DNA damage, which has been referred to as 'BRCAness'7,37. Consistent with this finding, this subtype also included the vast majority of patients with BRCA1 and BRCA2 germ-line mutations (15 of 20 and 5 of 6 patients in the cohort, respectively). The network for subtype 3 was enriched for genes in the NF-κB pathway (Supplementary Fig. 8), whereas subtype 4 was enriched for genes involved in cholesterol transport and fat and glycogen metabolism (Supplementary Fig. 9). A similar analysis in uterine and lung cancers produced other subnetworks with unique characteristics, including enrichments for DNA-damage response, WNT signaling and histone modification (Supplementary Figs. 10,11,12,13,14,15,16). Thus, the NBS approach not only can stratify patients into clinically informative subtypes but may help identify the molecular network regions commonly mutated in each subtype.
Figure 5. Network view of genes with high network-smoothed mutation scores in HumanNet ovarian cancer subtype 1 (relative to scores of other subtypes).
Subtype 1 had the lowest survival and highest platinum-resistance rates amongst the four recovered subtypes. Node size corresponds to smoothed mutation scores. Node color corresponds to a set of functional classes of interest recovered through manual examination of the resulting network with the aid of the GeneMania Cytoscape plug-in. Thickened node outlines indicate genes that are known cancer genes included in the COSMIC cancer-gene census. An underlined gene symbol in the network indicates that somatic mutations were found for that gene in the examined cohort.
Translation to predictive signatures
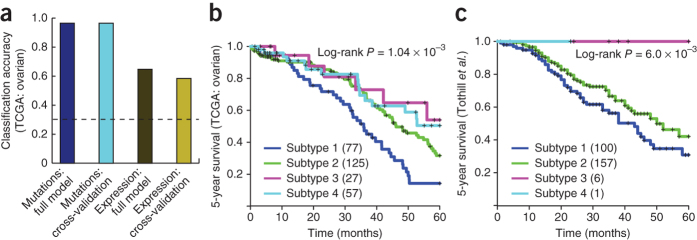
For NBS to be applicable to new patients not in the TCGA, it is necessary to complement it with a procedure for assigning a patient to one of the existing NBS subtypes. For this purpose, we explored the nearest shrunken centroid approach38, a standard method for sample classification that summarizes each subtype with a class 'centroid' and assigns new samples to the subtype with closest centroid. We found that this method was able to classify the network-smoothed mutation profile of an individual patient with over 95% accuracy (Fig. 6a; tenfold cross-validation).
Figure 6. From mutation-derived subtypes to expression signatures.
(a) Classification accuracy (fraction of correctly classified patients) when using a supervised learning method trained to learn a signature on the basis of either somatic mutation profiles or gene expression, showing training error and cross-validation error. Dashed line shows the accuracy for a random predictor. (b) Kaplan-Meier survival plots for the TCGA ovarian cancer patients using a classifier trained on subtypes from NBS of mutation data in TCGA. (c) Results of the same classifier applied to serous ovarian cancer samples from an independent data set (Tothill et al.40).
However, mRNA expression data are presently much more widely available than are full genome or exome sequences: there are numerous existing cohorts of cancer patients that have been profiled in mRNA expression but not in somatic mutations7,39,40,41,42. We therefore sought to test whether, having used NBS to define subtypes within TCGA somatic mutation data, we could assign a new patient to these subtypes using an expression signature. To explore this idea, we used the mRNA expression profiles available for the TCGA ovarian tumor cohort to learn an expression signature for each subtype defined earlier by NBS, again using the nearest shrunken centroid approach38. We found that expression performed as an adequate surrogate for mutation profile, albeit at a reduced accuracy (Fig. 6a; >95% for mutations, ∼60% for expression and ∼30% at random). This expression signature was nonetheless able to recover stratification predictive of survival (Fig. 6b).
We examined the predictive value of this gene expression signature in two independent studies of serous ovarian tumors by Tothill et al.40 and Bonome et al.42 as well as in a meta-analysis including over 1,000 patients, which subsumes Tothill, Bonome and TCGA samples that included expression profiles but lacked somatic mutation profiles41 (Fig. 6c and Supplementary Fig. 17) and incorporates an unknown number of nonserous ovarian cancer samples. Using the expression signature we had learned from NBS analysis of TCGA data, all patients could be assigned to one of the four NBS subtypes. In the Tothill data set, the subtype assignments were found to be significantly predictive of patient survival and platinum drug resistance (log-rank P = 6.1 × 10−3 and 1.65 × 10−6 respectively; Fig. 6c and Supplementary Fig. 17), following the same trends observed in the original TCGA cohort. In the Bonome and the meta-analysis data sets, the recovered subtypes were again significantly associated with patient survival (log-rank P = 1.40 × 10−3 and 1.22 × 10−4, respectively; Supplementary Fig. 17). We note that the proportions of the recovered subtypes in each of the three independent expression cohorts appeared to be different (Supplementary Table 2), a phenomenon possibly due to different criteria for inclusion in each study (for example: the TCGA ovarian cohort is primarily composed of high-grade, late-stage patients) or possibly differences due to population substructure. As a final control, we performed clustering of the Tothill expression profiles independent of NBS subtypes; this resulted in a different set of subtypes that associated with survival to a more limited extent (P = 0.01, Supplementary Fig. 18). These results show that tumor subtypes defined by NBS can be identified in independent data sets when gene expression is used as a surrogate biomarker.
Effects of different classes of mutation on stratification
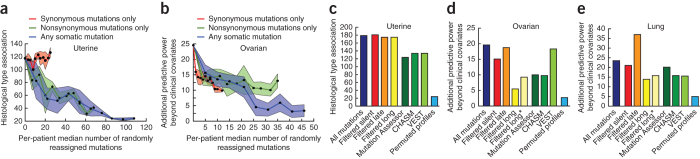
We studied the impacts of different classes of somatic mutation on the NBS approach. We first tested the effect on NBS of disrupting synonymous mutations by reassigning them to new randomly chosen gene locations. For uterine and lung cancers (Fig. 7a and Supplementary Fig. 19, respectively), disruption of synonymous mutations had little effect on NBS performance. In sharp contrast, disruption of nonsynonymous mutations or of all mutations greatly affected stratification performance. Interestingly, in the ovarian cancer cohort (Fig. 7b), disruption of either synonymous or nonsynonymous mutations was detrimental to performance.
Figure 7. Effects of different types of mutations on stratification.
(a,b) Effects of permuting a progressively larger fraction of mutations per patient for different types of somatic mutation, for the uterine (a) and ovarian (b) tumor cohorts. Lines show the median performance, and colored regions represent the median absolute deviation. (c–e) Different types of filters were applied as a preprocessing step before NBS was run on the uterine (c), ovarian (d) and lung (e) cohorts. In blue is the full data set; in red we filter all synonymous mutations; in orange and yellow we filter the top 2% late-to-replicate and long genes, respectively (long*: top 2% long genes, with any COSMIC cancer gene census genes included in the analysis). In green are three types of filters based on predictors of the functional effect of mutation; in light blue is the performance we observed after permuting all mutations within each patient separately as a control. (a–e) For uterine cancer, we report the median χ2 statistic; for ovarian and lung cancer, we report the median likelihood difference of a full model to a base model including just clinical covariates (age, grade, stage, mutation rate and residual tumor after surgery).
We also studied the effect of removing mutations judged to be nonfunctional in cancer by methods such as MutationAssessor43, cancer-specific high-throughput annotation of somatic mutations (CHASM)13 and the variant effect scoring tool (VEST)44, which use features such as sequence conservation and protein structural information to assess the likely impact of mutations. Filtering mutations with these tools resulted in decreased association of NBS subtypes with patient survival in all three cancers (Fig. 7c–e, with the possible exception of VEST for ovarian tumors: Fig. 7d). Finally, we studied the effect of removing genes with long sequences or late cell-cycle replication times: both of these characteristics have been postulated to accrue high numbers of mutations that may be unrelated to tumor progression45. We found that removal of long genes substantially degraded the ability to identify ovarian and lung subtypes predictive of survival (Fig. 7d,e). However, removal of late-replicating genes had little effect and, in the case of the lung tumor cohort, actually increased predictive power (Fig. 7e).
Discussion
Here we have reported the discovery that, through the use of prior knowledge captured in molecular networks, a set of tumor mutation profiles can be stratified into subtypes that are both biologically and clinically informative. These subtypes are distinct from those recovered through stratification of other types of data and are independent of other clinical markers known to be associated with survival. We can identify network modules characteristic of each subtype, which may provide new insight into the biological mechanisms driving tumor progression. To our knowledge, this is the first time that somatic mutation profiles have been used to stratify patients in an unsupervised fashion.
One might consider at least three potential reasons for the good performance of NBS. First, somatic mutations represent a digital signal in that a given gene can be considered either mutated or not, whereas most other data layers are analog signals representing measurements of continuous values. In general, digital systems have improved accuracy and reproducibility and are more robust to noise46. Second, somatic mutation profiles are differential measurements between tumor and normal tissue, whereas expression and other 'omics profiles are absolute measurements in each patient. The differential analysis filters out mutations or variants present in the patient's germ line, leaving only tumor-specific changes. In contrast, it has been difficult to identify a true 'baseline' gene expression state for a tissue, as these measurements are dynamic and highly context specific. Finally, the somatic mutation profile captures the causal genetic events underlying tumor progression, whereas mRNA or protein expression profiles are a functional readout of the current cell state and are influenced by external factors that may be unrelated to tumor biology.
The network modules we identified as characteristic for each tumor subtype provide new insights into the biology of cancer and raise many new questions. One particularly promising finding was the prominence of the FGF pathway in ovarian tumor subtype 1 (Fig. 5). This pathway has been implicated in tumor proliferation and angiogenesis, and many inhibitors for this pathway are in clinical development47. Specifically, it has been shown that increased expression of FGF1 is associated with poor survival in ovarian cancer48, and inhibition of FGFR1 and FGFR2 increases sensitivity to cisplatin in ovarian cancer cell lines36. An intriguing question for future work is whether subtype 1 patients are particularly responsive to therapy directed at network-identified targets, such as treatment with inhibitors of FGFR1.
Another interesting observation is that several network modules are enriched for long genes. For example, for ovarian tumor subtype 2, a total of 12 of 176 genes in the module are in the top 2% by length (P = 2.3 × 10−4). One prominent example is TTN, the longest known coding gene. Although prominent 'gold-standard' catalogs of cancer genes—such as the Catalogue of Somatic Mutations in Cancer (COSMIC) cancer gene census49 and the list of Vogelstein et al.50—are also enriched for long genes (for example, 17 of 125 in the Vogelstein list, P = 5.11 × 10−10), there remains some controversy about the roles these genes may play in cancer. On the one hand, it is possible that long genes are highly mutated not because they are drivers of cancer but simply owing to chance because they are a bigger 'target' to hit. On the other hand, there is no definitive evidence that mutations in long genes are not functional or do not contribute to tumor progression. Our analysis provides some evidence that these long genes should not be ignored. In the molecular network, long mutated genes were highly interconnected to other functionally related genes of all lengths, which are also found to be mutated in patients of that subtype. For example, the network region for ovarian tumor subtype 1 (Fig. 5) showed TTN interconnected to genes such as NEB, ANK1 and MYOM2, all of which are also mutated in patients of this subtype. These genes encode components of the cytoskeleton thought to have both structural and signaling roles51. Although TTN is a long gene and thus might accrue mutations by chance, it is striking that other members of the same protein interaction neighborhood are also found to be mutated in tumors of the same subtype. Using permutation analysis, we estimated that the chance of TTN having an immediate network neighborhood with this same number of mutations is roughly P < 0.0001. Thus, one possibility is that the TTN and other cytoskeletal components are required for platinum-induced, P53-independent apoptosis, and that mutation in either structural or signaling proteins in this pathway leads to platinum resistance. In support of this theory is prior work demonstrating that cell shape is associated with chemotherapy response in ovarian cancer52.
Another interesting observation is that synonymous mutations, though dispensable for stratification of uterine and lung tumors, appear to have some predictive power in stratification of ovarian tumors. In support of this finding, a number of high-profile studies have suggested that synonymous mutations may indeed play a causal role in cancer progression53,54,55,56. Further study is needed to understand whether ovarian cancer is indeed the outlier in this respect and whether and how synonymous mutations truly function in this disease.
Finally, we see many opportunities to improve upon the basic concept of NBS in future work. First, integrating multiple layers of information beyond somatic mutations (for example: CNVs, epigenome, transcriptome, etc.) into a composite stratification method might further expand our ability to identify subtypes with clinically relevant differences. Second, although we have shown the utility of three sources of gene-gene interactions, there are other types of networks worth exploring, such as those involved in signaling, metabolism or transcription. Although this study focused on uterine, ovarian and lung cancers, the NBS method is broadly applicable to any cohort of cancer patients for which somatic mutations are known. Finally, analyzing NBS subtypes across all cancers simultaneously (i.e., a pan-cancer analysis) will offer the intriguing opportunity to explore whether the genes and networks underlying the progression of a tumor are more informative of clinical outcome than its tissue of origin.
Methods
Expanded overview of network-based stratification.
The technique of network-based stratification (NBS) combines genome-scale somatic mutation profiles with a gene interaction network to produce a robust subdivision of patients into subtypes (Fig. 1a). Briefly, somatic mutations for each patient are represented as a profile of binary (1, 0) states on genes, in which a '1' indicates a gene for which mutation has occurred in the tumor relative to germ line (i.e., a single-nucleotide base change or the insertion or deletion of bases). For each patient independently we project the mutation profiles onto a human gene interaction network obtained from public databases28,29,30. Next, the technique of network propagation31 is applied to spread the influence of each mutation profile over its network neighborhood (Fig. 1b). The result is a 'network-smoothed' profile in which the state of each gene is no longer binary but reflects its network proximity to the mutated genes in that patient along a continuous range [0, 1]. Following this 'network smoothing', patient profiles are clustered into a predefined number of subtypes (k = 2, 3, ... 12) using the unsupervised technique of non-negative matrix factorization32 (NMF; Fig. 1c). For NBS we use a variant of NMF that encourages the selection of gene sets supporting each subtype according to high network connectivity (NetNMF)58. Finally, to promote robust cluster assignments, we use the technique of consensus clustering33, in which the above procedure is repeated for 1,000 different subsamples in which subsets of 80% of patients and genes are drawn randomly without replacement from the entire data set. The results of all 1,000 runs are aggregated into a (patient × patient) co-occurrence matrix, which summarizes the frequency with which each pair of patients has cosegregated into the same cluster. This co-occurrence matrix is then clustered a second time to recover a final stratification of the patients into clusters/subtypes (Fig. 1d). Our implementation of the NBS method is available for download as a Matlab package from http://idekerlab.ucsd.edu/software/NBS/ or as Supplementary Software. The former should be used for obtaining the most up-to-date versions.
Processing of patient mutation profiles.
High-grade serous ovarian cancer, uterine endometrial carcinoma and lung adenocarcinoma somatic mutation data were downloaded from the TCGA data portal on 8 August 2012, 1 January 2013 and 1 January 2013, respectively. Only mutation data generated using the Illumina GAIIx platform were retained for subsequent analysis, and patients with fewer than 10 mutations were discarded. This left 356 patients with mutations in 9,850 genes for the TCGA ovarian cohort, 248 patients with mutations in 17,968 genes for the TCGA uterine endometrial cohort and 381 patients with mutations in 15,967 genes in the TCGA lung adenocarcinoma cohort. Patient mutation profiles were constructed as binary vectors such that a bit is set if the gene corresponding to that position in the vector harbors a mutation in that patient. Additional details on processing and organization of the data are available in a previous TCGA publication2.
Sources of molecular network data.
Patient mutation profiles were mapped onto gene interaction networks from three sources: STRING v.9 (ref. 29), HumanNet v.1 (ref. 28) and PathwayCommons30 (Supplementary Table 1). STRING integrates protein-protein interactions from literature curation, computationally predicted interactions, and interactions transferred from model organisms based on orthology. HumanNet uses a naïve Bayes approach to weight different types of evidence together into a single interaction score focusing on data collected in humans, yeast, worms and flies. PathwayCommons aggregates interactions from several pathway and interaction databases, focused primarily on physical protein-protein interactions (PPIs) and functional relationships between genes in canonical regulatory, signaling and metabolic pathways (including hallmark pathways of cancer). Supplementary Table 1 summarizes the number of genes and interactions used in our analysis from each of these three networks.
All network sources comprise a combination of interaction types, including direct protein-protein interactions between a pair of gene products and indirect genetic interactions representing regulatory relationships between pairs of genes (for example, coexpression or TF activation). The PathwayCommons network was filtered to remove any nonhuman genes and interactions, and all remaining interactions were used for subsequent analysis. Only the most confident 10% of interactions for both the STRING and HumanNet networks were used for this work, ordered according to the quantitative interaction score provided as part of both networks. This threshold was chosen using an independent ROC analysis with respect to a set of Gene Ontology–derived gold standards (data not shown). After filtering of edges, all networks were used as unweighted, undirected networks.
Network smoothing.
After mapping a patient mutation profile onto a molecular network, network propagation31 is applied to 'smooth' the mutation signal across the network. Network propagation uses a process that simulates a random walk on a network (with restarts) according to the function
F0 is a patient-by-gene matrix, and A is a degree-normalized adjacency matrix of the gene interaction network, created by multiplying the adjacency matrix by a diagonal matrix with the inverse of its row (or column) sums on the diagonal. α is a tuning parameter governing the distance that a mutation signal is allowed to diffuse through the network during propagation. The optimal value of α is network dependent (0.7, 0.5 and 0.7, for HumanNet, PathwayCommons and STRING, respectively), but the specific value seems to have only a minor effect on the results of NBS over a sizable range (for example, 0.5–0.8). The propagation function is run iteratively with t = [0, 1, 2, ...] until Ft+1 converges (the matrix norm of Ft+1 – Ft < 1 × 10−6). Following propagation, the rows of the resultant matrix Ft are quantile normalized to ensure that the smoothed mutation profile for each patient follows the same distribution.
Network-regularized NMF.
Network-regularized NMF is an extension that constrains NMF to respect the structure of an underlying gene interaction network. This is accomplished by minimizing the following objective function using an iterative method32,58,59:
W and H form a decomposition of the patient × gene matrix F (resulting from network smoothing as described above) such that W is a collection of basis vectors, or 'metagenes', and H is the basis vector loadings. The trace(WtKW) function constrains the basis vectors in W to respect local network neighborhoods. The term K is and adjacency matrix of a nearest neighbors network derived from the graph Laplacian of an influence distance matrix23 that is derived from the original network. The degree to which local network topology versus global network topology constrains W is determined by the number of nearest neighbors. We experimented with neighbor counts ranging from 5 to 50 to include in the nearest network, and we observed only small changes in outcome (data not shown). For the work presented in this manuscript, the 11 most influential neighbors of each gene in the network as determined by network influence distance were used.
Consensus clustering.
Clustering was performed with a standard consensus clustering framework, discussed in detail by Monti et al.33 and used in previous TCGA publications2,18,60. Briefly, we used network-regularized NMF (see above) to derive a stratification of the input cohort. In order to ensure robust clustering, network-regularized NMF was performed 1,000 times on subsamples of the data set. In each subsample, we sampled 80% of the patients and 80% of the mutated genes at random without replacement. The set of clustering outcomes for the 1,000 samples was then transformed into a co-clustering matrix. This matrix records the frequency with which each patient pair was observed to have membership in the same subtype over all clustering iterations in which both patients of the pair were sampled. The result is a similarity matrix of patients, which we then used to stratify the patients by applying either average linkage hierarchical clustering or a second symmetric NMF step. Patients showing poor cluster association to a single subtype were excluded from further analysis.
Simulation of somatic mutation cohorts.
We used simulations to determine the ability of NBS to recover subtypes from somatic mutation profiles. In order to quantify the performance of NBS, we needed a cohort with specified subtypes as a 'ground truth' reference and to be able to control the properties of the simulated signal determining the different subtypes. We simulated a somatic mutation cohort as follows. Patient mutation profiles were sampled with replacement from the TCGA ovarian data set. For each patient, the mutation profile was permuted, whereas the per-patient mutation frequency was kept invariant; this resulted in a background mutation matrix with no subtype signal. For simulation of an underlying network structure for NBS to detect, a network-based signal was added to the patient-by-mutation matrix as follows. First, we established a set of network communities (i.e., connected components enriched for edges shared within community members) in the input network (STRING, HumanNet or PathwayCommons) using the network community detection algorithm QCut61. Next, we divided the patient cohort randomly into four equal-sized subtypes (four was selected as reasonable owing to the four expression-based subtypes that have been identified for glioblastoma, ovarian and breast cancers2,18,60,62). Each subtype was assigned a small number (for example, 1–6) of network modules that together had a combined size s ranging from 10 to 250 genes. These network modules represent 'driver' subnetworks characterizing the subtype. For each patient, we reassigned a fraction of the patient's mutations f to genes covered by the driver modules for that patient's subtype. This procedure resulted in a patient × gene mutation matrix with underlying network structure while maintaining the per-patient mutation frequency.
A plausible range for the number of driver mutation in a tumor was recently proposed to be between 2 and 8 driver mutations50. We note that in our simulation framework, a 4% mutation rate corresponds to between 1 and 9 mutations with a median of 3, which is on par with the aforementioned estimate. In order to estimate the appropriate size of cancer pathways (s), we examined the known cancer pathways in the NCI-Nature pathway interaction database63. We observe that pathways in the database are of varying sizes, 2–139 genes, with a median size of 34, and over 23% of pathways include over 50 genes.
Identifying differentially mutated subnetworks.
After applying NBS, we identified genes that were enriched for mutation in each of the subtypes relative to the whole cohort. To do this we applied the significance analysis of microarrays (SAM) method64 on the network-smoothed mutation profiles. This is a nonparametric method developed for discovering differentially expressed genes in microarray experiments. We used a rank-based Wilcoxon-type statistic and compared each subtype against the remaining cohort. Significance was assessed using the SAM permutation scheme with 1,000 permutations. The resulting set of genes for each subtype was overlaid on the network used for network smoothing.
Survival analysis.
Survival analysis was performed using the R “survival” package. We fit a Cox-proportional hazards model65 to determine the relationship between the NBS-assigned subtypes and patient survival. A likelihood-ratio test and associated P value is calculated by comparing the full model, which includes subtypes and clinical covariates, against a baseline model that includes covariates only. Clinical covariates available in TCGA and included in the model were age, grade, stage, residual surgical resection and mutation rate, as well as cigarette smoking status for the lung cancer cohort.
Comparing predictive power and overlap with TCGA subtypes.
Added predictive power is estimated using a likelihood-ratio test comparing the Cox proportional hazards model given subtypes and clinical covariates (age, stage, grade, mutation frequency and residual tumor presence after surgery) compared to a covariate-only model. Significance of overlap is assessed using a Pearson's χ2 test of independence between NBS subtypes with a specific network and number of subtypes (ovarian, HumanNet, four subtypes; lung, HumanNet, six subtypes; uterine, STRING, three subtypes) and the different data types with varying number of subtypes reported in the TCGA and subtyped using consensus-clustering NMF. TCGA subtypes were downloaded from the Firehose run from 25 May 2012 (http://gdac.broadinstitute.org/runs/analyses__2012_05_25/reports/cancer/OV/).
Shrunken-centroid prediction on expression profiles.
We used shrunken centroids to derive an expression signature equivalent to the somatic mutation-based NBS subtypes. Expression data were provided by Győrffy et al.41, who aggregated several expression data sets as part of a meta-analysis of ovarian cancer. In this analysis, all data were regularized using quantile and MAS5 normalization. We performed this analysis on the Tothill et al.40 (ovarian serous samples only), Bonome et al.42 and TCGA data sets, as well as across the full meta-analysis cohort. We used the “pamr” R package with default parameters to train a shrunken-centroid model38 on mRNA expression levels for all genes in the TCGA ovarian data set with subtype assignment as the class label. The trained model was next used to predict subtype labels on the held-out Tothill et al. and Bonome et al. data or the full meta-analysis expression cohort (excluding any TCGA samples included in the training set).
We include a table of the class centroids for each of the three TCGA somatic mutation cohorts and the four expression cohorts of ovarian cancer included in this study (Supplementary Table 5).
Missense-mutation scoring.
Missense mutations were scored using three methods: CHASM13, VEST44 and MutationAssessor43. CHASM and VEST use supervised machine learning to score mutations. The CHASM training set is composed of a positive class of driver mutations from the COSMIC database and a negative class of synthetic passenger mutations simulated according to the mutation spectrum observed in the tumor type under study. The VEST training set comprises a positive class of disease mutations from the Human Gene Mutation Database66 and a negative class of variants detected in the ESP6500 (http://evs.gs.washington.edu/EVS/) cohort with an allele frequency of >1%. MutationAssessor uses patterns of conservation from protein alignments of large numbers of homologous sequences to assess the functional impact of missense mutations. CHASM and VEST scores were obtained from the CRAVAT webserver44 (http://www.cravat.us/). MutationAssessor precomputed mutation scores were downloaded from http://mutationassessor.org/. After using each method to score all mutations across all patients, we picked a permissive threshold for retaining mutations to use for NBS (retaining the top 75% of mutations as scored by CHASM and VEST and using MutationAssessor with the “low threshold” setting).
Replication timing.
RepliSeq67 data for the GM12878 cell line were downloaded from the ENCODE project website (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeUwRepliSeq/, downloaded March 2013). Summed normalized tag densities were used as a proxy for replication time (higher counts indicating that a transcript was replicated earlier in the cell cycle). Normalized tag densities for RefSeq protein coding regions were retrieved using bigWigAverageOverBed68 with RefSeq gene sequence features in .gff3 format downloaded from http://www.yandell-lab.org/software/VAAST/data/hg19/Features/refGene_hg19.gff3. Tag densities were averaged for each transcript, and the longest transcript was selected to represent each gene.
Supplementary information
Supplementary Figures 1–19 and Supplementary Table 1 (PDF 2745 kb)
Class assignment and summary of class assignment and clinical data in uterine cancer (XLSX 6253 kb)
Class assignment summary of and clinical data in ovarian cancer (XLSX 1791 kb)
Class assignment and summary of clinical data in lung cancer (XLSX 675 kb)
Class centroid vectors for reported subtypes (XLSX 3105 kb)
Network-based stratification (ZIP 1433 kb)
Acknowledgements
We would like to thank all members of the Ideker lab, specifically J. Dutkowski, R. Srivas, G. Bean, M. Yu and M. Choueiri, for many fruitful discussions during various stages of this project. We also thank G. Hofree for her input, patience and support. J.P.S. is supported in part by grants from the Marsha Rivkin Center for Ovarian Cancer Research and the Conquer Cancer Foundation of the American Society of Clinical Oncology. This work was supported by US National Institutes of Health grants P41 GM103504 and P50 GM085764.
Author Contributions
M.H. and T.I. conceived of and designed the approach. M.H. performed the data analysis, implemented the method and performed all computational experiments. J.P.S. assisted in characterizing clinical aspects of the cohorts and interpreting the biological implication of the resulting subtypes. H.C. assisted in analyzing the contributions of different mutation types and the functional scoring of mutations. A.G. compared resulting subtypes to other data layers. M.H., J.P.S. and T.I. wrote the manuscript. All authors approved the final version of this manuscript.
Competing interests
The authors declare no competing financial interests.
References
- 1.The International Cancer Genome Consortium. International network of cancer genome projects. Nature464, 993–996 (2010). [DOI] [PMC free article] [PubMed]
- 2.The Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature474, 609–615 (2011). [DOI] [PMC free article] [PubMed]
- 3.The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature497, 67–73 (2013). [DOI] [PMC free article] [PubMed]
- 4.Brunham LR, Hayden MR. Whole-genome sequencing: the new standard of care? Science. 2012;336:1112–1113. doi: 10.1126/science.1220967. [DOI] [PubMed] [Google Scholar]
- 5.Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat. Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 6.Konstantinopoulos PA, Spentzos D, Cannistra SA. Gene-expression profiling in epithelial ovarian cancer. Nat. Clin. Pract. Oncol. 2008;5:577–587. doi: 10.1038/ncponc1178. [DOI] [PubMed] [Google Scholar]
- 7.Konstantinopoulos PA, et al. Gene expression profile of BRCAness that correlates with responsiveness to chemotherapy and with outcome in patients with epithelial ovarian cancer. J. Clin. Oncol. 2010;28:3555–3561. doi: 10.1200/JCO.2009.27.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–1823. doi: 10.1016/S0140-6736(11)61539-0. [DOI] [PubMed] [Google Scholar]
- 9.Esteva FJ, et al. Prognostic role of a multigene reverse transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin. Cancer Res. 2005;11:3315–3319. doi: 10.1158/1078-0432.CCR-04-1707. [DOI] [PubMed] [Google Scholar]
- 10.The Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature487, 330–337 (2012). [DOI] [PMC free article] [PubMed]
- 11.Raspe E, Decraene C, Berx G. Gene expression profiling to dissect the complexity of cancer biology: pitfalls and promise. Semin. Cancer Biol. 2012;22:250–260. doi: 10.1016/j.semcancer.2012.02.011. [DOI] [PubMed] [Google Scholar]
- 12.Mardis ER. Genome sequencing and cancer. Curr. Opin. Genet. Dev. 2012;22:245–250. doi: 10.1016/j.gde.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carter H, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang K, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011;43:1219–1223. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 16.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013;45:478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Allegra CJ, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J. Clin. Oncol. 2009;27:2091–2096. doi: 10.1200/JCO.2009.21.9170. [DOI] [PubMed] [Google Scholar]
- 18.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature490, 61–70 (2012). [DOI] [PMC free article] [PubMed]
- 19.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreeger PK, Lauffenburger DA. Cancer systems biology: a network modeling perspective. Carcinogenesis. 2010;31:2–8. doi: 10.1093/carcin/bgp261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/150563a0. [DOI] [PubMed] [Google Scholar]
- 23.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J. Comput. Biol. 2011;18:507–522. doi: 10.1089/cmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 24.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dutkowski J, Ideker T. Protein networks as logic functions in development and cancer. PLoS Comput. Biol. 2011;7:e1002180. doi: 10.1371/journal.pcbi.1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. doi: 10.1101/gr.118992.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cerami EG, et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2011;39:D685–D690. doi: 10.1093/nar/gkq1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vanunu O, Magger O, Ruppin E, Shlomi T, Sharan R. Associating genes and protein complexes with disease via network propagation. PLoS Comput. Biol. 2010;6:e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–791. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 33.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: a resampling-based method for class discovery and visualization of gene expression microarray data. Mach. Learn. 2003;52:91–118. doi: 10.1023/A:1023949509487. [DOI] [Google Scholar]
- 34.Yang D, et al. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. J. Am. Med. Assoc. 2011;306:1557–1565. doi: 10.1001/jama.2011.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cole C, et al. Inhibition of FGFR2 and FGFR1 increases cisplatin sensitivity in ovarian cancer. Cancer Biol. Ther. 2010;10:495–504. doi: 10.4161/cbt.10.5.12585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wysham WZ, et al. BRCAness profile of sporadic ovarian cancer predicts disease recurrence. PLoS ONE. 2012;7:e30042. doi: 10.1371/journal.pone.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. USA. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le Page C, et al. Gene expression profiling of primary cultures of ovarian epithelial cells identifies novel molecular classifiers of ovarian cancer. Br. J. Cancer. 2006;94:436–445. doi: 10.1038/sj.bjc.6602933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tothill RW, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 41.Győrffy B, Lánczky A, Szállási Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 42.Bonome T, et al. A gene signature predicting for survival in suboptimally debulked patients with ovarian cancer. Cancer Res. 2008;68:5478–5486. doi: 10.1158/0008-5472.CAN-07-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carter H, Douville C, Stenson PD, Cooper DN, Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14 (suppl. 3):s3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat. Genet. 2009;41:393–395. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabiner, L.R. & Gold, B. Theory and Application of Digital Signal Processing (Prentice Hall, 1975).
- 47.Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat. Rev. Cancer. 2010;10:116–129. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 48.Birrer MJ, et al. Whole genome oligonucleotide-based array comparative genomic hybridization analysis identified fibroblast growth factor 1 as a prognostic marker for advanced-stage serous ovarian adenocarcinomas. J. Clin. Oncol. 2007;25:2281–2287. doi: 10.1200/JCO.2006.09.0795. [DOI] [PubMed] [Google Scholar]
- 49.Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic 'network of networks'. Nat. Rev. Mol. Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, et al. Integrated analysis of gene expression and tumor nuclear image profiles associated with chemotherapy response in serous ovarian carcinoma. PLoS ONE. 2012;7:e36383. doi: 10.1371/journal.pone.0036383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strauss BS. Role in tumorigenesis of silent mutations in the TP53 gene. Mutat. Res. 2000;457:93–104. doi: 10.1016/S0027-5107(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 54.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 55.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 56.Salzman DW, Weidhaas JB. miRNAs in the spotlight: making 'silent' mutations speak up. Nat. Med. 2011;17:934–935. doi: 10.1038/nm0811-934. [DOI] [PubMed] [Google Scholar]
- 57.Rand WM. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 1971;66:846–850. doi: 10.1080/01621459.1971.10482356. [DOI] [Google Scholar]
- 58.Cai, D., He, X., Wu, X. & Han, J. Non-negative matrix factorization on manifold. in 8th IEEE Int. Conf. Data Mining 63–72 (IEEE, 2008).
- 59.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. USA. 2004;101:4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruan J, Zhang W. Identifying network communities with a high resolution. Phys. Rev. E. 2008;77:016104. doi: 10.1103/PhysRevE.77.016104. [DOI] [PubMed] [Google Scholar]
- 62.Verhaak RG, et al. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J. Clin. Invest. 2013;123:517–525. doi: 10.1172/JCI65833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaefer CF, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen PK, Gill RD. Cox's regression model for counting processes: a large sample study. Ann. Stat. 1982;10:1100–1120. doi: 10.1214/aos/1176345976. [DOI] [Google Scholar]
- 66.Stenson PD, et al. The Human Gene Mutation Database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr. Protoc. Bioinformatics. 2012;39:1.13. doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 67.Hansen RS, et al. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc. Natl. Acad. Sci. USA. 2010;107:139–144. doi: 10.1073/pnas.0912402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–19 and Supplementary Table 1 (PDF 2745 kb)
Class assignment and summary of class assignment and clinical data in uterine cancer (XLSX 6253 kb)
Class assignment summary of and clinical data in ovarian cancer (XLSX 1791 kb)
Class assignment and summary of clinical data in lung cancer (XLSX 675 kb)
Class centroid vectors for reported subtypes (XLSX 3105 kb)
Network-based stratification (ZIP 1433 kb)