Abstract
miRNAs elicit gene silencing at the post-transcriptional level by several modes of action: translational repression, mRNA decay, and mRNA cleavage. Studies in animals have suggested that translational repression occurs at early steps of translation initiation, which can be followed by deadenylation and mRNA decay. Plant miRNAs were originally thought to solely participate in mRNA cleavage, but increasing evidence has indicated that they are also commonly involved in translational inhibition. Here we discuss recent findings on miRNA-mediated translational repression in plants. The identification of AMP1 in Arabidopsis as a protein required for the translational repression but not the mRNA cleavage activity of miRNAs links miRNA-based translational repression to the endoplasmic reticulum (ER). Future work is required to further elucidate the miRNA machinery on the ER.
Keywords: miRNA, argonaute, translational repression, mRNA cleavage, mRNA decay, endoplasmic reticulum (ER)
Introduction
Non-coding RNAs have been increasingly recognized as key players in gene and genome regulation that impacts development and diseases.1-3 microRNAs (miRNAs) are a class of non-coding RNAs of 20–24 nucleotides that serve as sequence-specific regulators of gene expression in diverse eukaryotes. The first miRNA that was discovered and characterized was Caenorhabditis elegans lin-4, which regulates developmental timing of the animal. Lin-4 was found to target the 3′ UTR of lin-14 and repress lin-14 translation.4-6 These elegant early studies uncovered translational repression as a mode of miRNA action in metazoans. To date, the mechanisms of miRNA biogenesis and mode of action have been extensively studied; and it is now known that animal miRNAs regulate target genes not only by repressing translation but also by RNA decay.7-9 In contrast to animal miRNAs, plant miRNAs were originally thought to only participate in mRNA cleavage.10,11 However, increasing evidence has shown that plant miRNAs are also commonly involved in translational repression.12 Now it is recognized that in either animals or plants, miRNAs elicit silencing through several modes of action: mRNA decay, mRNA cleavage, and translational repression. Most animal miRNAs reduce target mRNA levels through mRNA decay, which entails deadenylation and decapping followed by exonucleolytic degradation.9 In rare cases where an animal miRNA exhibits extensive complementarity to its target mRNA, the miRNA can induce target mRNA cleavage.13 Plant miRNAs have a high degree of sequence complementarity to their target mRNAs and direct the endonucleolytic cleavage of target mRNAs. Following this cleavage, the 3′ fragment is degraded by XRN4 (EXORIBONUCLEASE4);14,15 the 5′ fragment undergoes uridylation by an unknown enzyme followed by 3′ to 5′ exonucleolytic degradation,16 presumably by the exosome. Studies on miRNA biogenesis, miRNA-target recognition, or miRNA-mediated mRNA decay or cleavage have been comprehensively reviewed.7,8,12,17,18 In this Point of View, we focus on miRNA-mediated translational repression to highlight recent findings that connect this mode of action with the ER in plants.
miRNA-based translational repression in animals
The early observation that the lin-4 miRNA reduces LIN-14 protein levels without influencing lin-14 mRNA abundance in C. elegans established the role of this miRNA in translational repression.4-6 These studies in C. elegans and subsequent studies in cultured animal cells suggested that miRNAs interfere with polysomes that are engaged in translation elongation.4-6,19-21 However, many studies argued that miRNAs inhibit translation initiation.22-28 For example, m7GpppG-caped mRNAs but not artificial ApppG-capped mRNAs were found to be susceptible to miRNA-based translational inhibition.25 The identification of the initiation factor eIF4A2, which unwinds 5′ UTR secondary structures to allow the 40S ribosomal subunit to scan toward the start codon, as critical for miRNA-mediated translational repression is also consistent with miRNAs acting to prevent translation initiation.29 In recent years, several groups performed ribosome footprint profiling to assess whether miRNA affects translation initiation or elongation or causes ribosome drop-off.30-32 These studies did not observe a pattern of reduced ribosome density toward the 3′ ends of miRNA target transcripts, which would be predicted if miRNAs cause pre-mature ribosome drop-off or inhibition of translation elongation; instead, they found that miRNAs cause a decrease in ribosome occupancy over the length of target mRNAs, implying that miRNAs inhibit translation initiation.
Apart from conflicting views on the steps of translation that miRNAs block, many studies also debated the roles of miRNA-mediated mRNA decay and translational repression in target regulation. In animals, miRNA-mediated mRNA degradation is not via endonucleolytic cleavage of targets, which occurs prevalently in plants, rather is via deadenylation followed by decapping and 5′-to-3′ mRNA degradation.33-39 Global analyses in mammalian cells, such as RNA-seq to determine steady-state transcript levels, quantitative proteomics to measure protein levels, and ribosome footprint profiling to uncover the status of translation of transcripts, found that the protein output could be largely explained by steady-state RNA levels,32,40-42 which led to the conclusion that mRNA decay is a major mode of action of mammalian miRNAs.42 However, using zebrafish embryos as well as Drosophila and human cultured cells, recent studies examined the two effects of miRNAs (mRNA decay and translational repression) with temporal resolution and revealed that miRNAs first cause translational repression, which may be followed by deadenylation and decay of the translationally inhibited mRNAs.30,43,44 As translational repression occurs prior to any appreciable RNA decay, translational repression is likely to be the primary function of animal miRNAs.
The two modes of miRNA-based target regulation, translational repression and RNA decay, appear to be mechanistically linked. Many factors involved in mRNA degradation were also implicated in translational control.45 A model for miRNA-mediated silencing that reconciles existing data involves the following scenarios: Argonaute (Ago) and associated GW182 (glycine-tryptophan protein of 182-kD) potentially interact with the m7G cap structure, thus interfering with some unknown early steps in translation initiation; meanwhile, GW182 recruits PABP (poly(A)-binding protein) which, in turn, directs mRNA deadenylation by PAN2-PAN3 (Pab1p-dependent poly(A) nuclease) and CAF1-CCR4-NOT1 deadenylase complexes; lastly, deadenylation may consolidate translational repression and lead to mRNA decay through recruitment of decapping factors DCP1/2.43,46 This model, however, does not convey the subcellular locations where these steps occur. In light of recent findings connecting mammalian RNAi and plant miRNA activities to the ER (see below), future investigations are necessary to place various events in miRNA-mediated gene silencing into a cell biology context.
miRNA-mediated translational repression in plants
In contrast to the situation in animals, where only a few miRNAs are highly complementary to their targets,47 in plants, miRNAs and their targets show a pattern of near complementarity, suggesting that plant miRNAs likely act through mRNA cleavage.10 The viewpoint was validated by studies of Arabidopsis miR171 that demonstrated that it cleaves Scarecrow-like mRNA targets.11
Subsequent studies, however, established translational repression as an important mode of action of plant miRNAs. Aukerman and Sakai showed that overexpression of Arabidopsis miR172 inhibits APETAL2 (AP2) protein accumulation without affecting AP2 RNA abundance.48 Our early work demonstrated that miR172 patterns floral organ and meristem development in Arabidopsis by repressing AP2 expression and showed that it causes disproportionate effects on AP2 expression at protein vs. mRNA levels.49 miR172 is evolutionarily conserved and also impacts floral and inflorescence development in rice.50,51 In rice, overexpression of miR172 phenocopies loss-of-function in its target gene SUPERNUMERARY BRACT (SNB) without affecting SNB transcript levels, suggesting that miR172 represses SNB through translation inhibition.50 It is worth noting that besides translational repression, miR172 is able to function via transcript cleavage. In both Arabidopsis and rice, 3′ fragments from miR172-guided cleavage of AP2 and other target transcripts can be detected with 5′ RACE-PCR.48,50,52 In fact, overexpression of miR172 leads to enhanced cleavage of AP2 mRNA.53 Despite the enhanced cleavage of AP2 mRNA, miR172 overexpression fails to affect the steady-state levels of the AP2 transcript,48,49 possibly due to feedback regulation that allows enhanced transcription of AP2.54 This highlights an advantage of translational repression over transcript cleavage—the former can effectively repress target gene expression despite feedback transcriptional regulation of the target genes.
Studies on Arabidopsis miR156/157 also pinpointed translational repression as an important mode of action of this miRNA. miR156/157 targets the 3′ UTR of the SBP (SQUAMOSA promoter-binding protein) box gene SPL3 and inhibits SPL3 expression at the protein but not the RNA level.55 Recently, Yang et al. screened for Arabidopsis mutations that enhance the expression of miR156-regulated SPL3, and identified SUO as a component in miR156/157-mediated repression of SPL3.56 SUO does not affect the transcript levels of SPL3 or CSD2 (a target of miR39857), but reduces their protein levels in a miRNA-dependent manner, suggesting that SUO is essential for miRNA-mediated translational repression. SUO encodes a GW (glycine and tryptophan) repeat protein and further investigations are needed to determine if it is functionally related to GW182 found in animals.
The role of plant miRNAs in translational inhibition was strengthened by biochemical and genetic studies. Lanet et al. provided biochemical evidence that miRNAs and AGO1 are associated with polysomes.58 Brodersen et al. performed a genetic screen looking for Arabidopsis mutants unable to silence a constitutively expressed GFP mRNA that contains a miR171 target site downstream of the stop codon.59 They identified two microRNA biogenesis deficient (mbd) mutants (mbd1 and mbd2), and six miRNA action deficient (mad) mutants (mad1–6). Interestingly, mbd1,2 and mad1–4 have increased levels of GFP mRNA and protein compared with wild-type plants, whereas mad5 and mad6 have similar GFP mRNA levels but much higher GFP protein levels relative to wild-type plants, suggesting that MAD5 and MAD6 are required for miR171-mediated translational repression of the GFP reporter gene. Our recent work demonstrated that ALTERED MERISTEM PROGRAM1 (AMP1), which encodes an ER membrane protein, is required for miRNA-mediated translational repression.60 The fact that target genes of multiple miRNAs are de-repressed at the protein level in mad5, mad6, and amp1 mutants suggests that translational repression is a widespread activity of plant miRNAs. Although a disparity in plant miRNAs’ effects on the expression of their target genes at the protein vs. mRNA level has been attributed to their translational inhibition activity, such attribution was largely extrapolated from the translational repression activity of animal miRNAs and lacked experimental support until our studies on AMP1.60 Measurements of protein synthesis from the miR398 target gene CSD2 in wild-type and amp1 plants showed that the lower CSD2 protein levels in wild type as compared with amp1 were due to reduced protein synthesis. Similarly the lower protein levels from the miR165/166 target gene PHABULOSA in WT as compared with amp1 were also found to be due to reduced protein synthesis. These results demonstrate that plant miRNAs repress the translation of target mRNAs.
A membrane-small RNA connection in animals and plants
In an early attempt to isolate novel Golgi or ER proteins, Cikaluk et al. identified rat GERp95,61 which was later found to be an ortholog of mouse or human Ago2, a miRNA effector protein.62-64 GERp95 was found to be associated with the ER in many cell types and also with the Golgi in some cell types.61 Human Ago2, as well as human Dicer, was also found in the ER membrane fraction, although Dicer was mostly present in the cytosol.65 This raises the possibility that miRNA biogenesis and/or activity is associated with the endomembrane system. This viewpoint was reinforced by Gibbings et al.’s study that showed that human GW182 and Ago2 localize to the endosomes and multi-vesicular bodies (MVBs).66 They also showed that knockdown of ESCRT (endosomal sorting complex required for transport) components compromises miRNA activity. A parallel study identified Drosophila HSP4 (Hermansky-Pudlak syndrome 4), which is involved in endosome trafficking, as a critical factor in siRNA- and miRNA-mediated silencing.67 They showed that depletion of HSP4 or ESCRT factors that blocks MVB formation impairs miRNA silencing in both flies and human cell culture. These studies indicated that MVBs are the sites where siRNA-Ago2 or miRNA-Ago2 disassembly occurs, and the disassembly aids the recycling of Ago2 for another round of small RNA loading. Recently, Stalder et al. revealed that only a minor fraction of siRNAs or miRNAs has the potential to load into an RNA-induced silencing complex (RISC), and Ago2-bound siRNA or miRNA accumulates in the ER and Golgi fractions in HeLa cells.68 They demonstrated that Ago2, TRBP (transactivation-responsive RNA-binding protein), and a small amount of Dicer are associated with the rough ER (rER) membrane, suggesting that RISC loading occurs at rER. They further showed that Ago2 is anchored on the ER membrane through TRBP and PACT (protein activator of the interferon-induced protein kinase PKR), because depletion of TRBP or PACT disrupts the Ago2-membrane association. The products of siRNA-Ago2-mediated slicing of target mRNAs were also associated with the rER fractions, indicating that siRNA-Ago2 acts on the rER.
In plants, a link between endomembranes and miRNA activities is also emerging. Brodersen et al. cloned MAD3 and MAD4 in Arabidopsis, which are required for the target repression activities of miRNAs. These two genes participate in isoprenoid and lipid biosynthesis and are thus likely to help maintain normal membrane composition or states.69 They showed that AGO1 is present in both soluble and membrane fractions and it is a peripheral membrane protein, because AGO1 could be released from membranes upon washes with high-salt or alkaline solutions. This raised the question of which membrane AGO1 or possibly miRISC docks on. Our recent study showed that AGO1 is partially co-localized with the ER and interacts with an ER integral membrane protein AMP1, which is indispensable for miRNA-mediated translational repression60 (gray box in Fig. 1). Furthermore, we showed that AMP1 precludes the recruitment of miRNA target transcripts onto membrane-bound polysomes. This suggests that plant miRNAs inhibit the translation of their target mRNAs on the rER. These studies begin to build a link between miRNA activity and the ER membrane, and reveal a novel function of the ER. AMP1 is homologous to several mammalian proteins, some with N-acetylated-α-linked acidic dipeptidase activity; thus, whether its mammalian homologs have similar functions deserves further investigations.
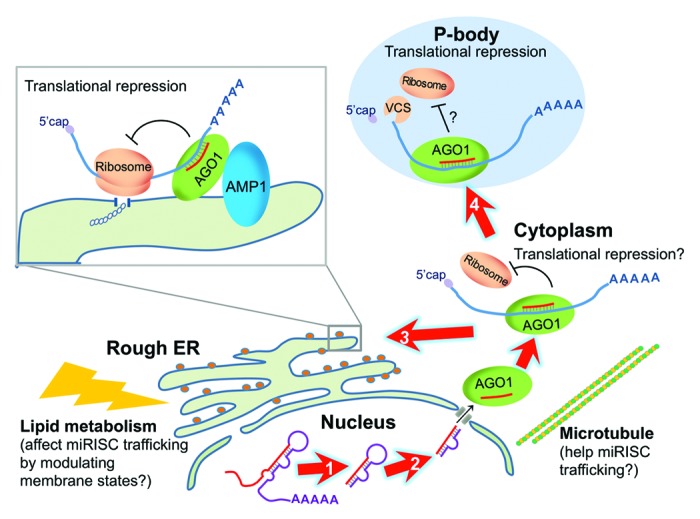
Figure 1. Subcellular locations where miRNA biogenesis or activity takes place in plants. A pri-miRNA is processed by a DCL1 complex into a pre-miRNA (arrow 1) and, subsequently, a miRNA:miRNA* duplex (arrow 2). The mature miRNA is loaded into AGO1 and represses target mRNAs through both mRNA cleavage (not diagramed) and translational repression. It is not known whether miRISC is capable of translational repression in the cytosol, but it is likely transported to the rough ER (arrow 3) or P-bodies (arrow 4), both of which are subcellular compartments where translational repression by miRNAs has been implicated to take place. The integral ER membrane protein AMP1 interacts with AGO1 and inhibits the loading of miRNA target mRNAs onto membrane-bound polysomes. How the P-body component and decapping factor VCS mediates the translational repression activity of plant miRNAs is currently unknown. Lipid metabolism (e.g., sterol synthesis), which influences membrane composition or states, also affects miRNA activity via an unknown mechanism. In addition, cytoskeleton (e.g., microtubules) dynamics may influence miRNA activities by affecting the subcellular trafficking of miRISCs.
Microtubule, P-body, and lipids are linked to miRNA activities, including translational inhibition
As mentioned earlier, Arabidopsis mad5 and mad6 mutants are defective in miRNA-based translational repression.59 MAD5 encodes the microtubule-severing enzyme KATANIN. Interestingly, a link between miRNA activities and microtubules was also observed in animals. For example, genome-wide screens in C. elegans identified numerous cytoskeleton components linked to miRNA function.70 In sea urchin, SEAWI, a homolog of PIWI/AGO, is a component of a microtubule-associated ribonucleoprotein complex.71 Thus, miRNA-based translational repression or miRNA activities in general may rely on cytoskeleton dynamics in diverse species.
miRNA activities have also been associated with mRNA-processing bodies (P-bodies). P-bodies were originally named by Sheth and Parker, who found that mRNA decapping and 5′ to 3′ degradation occur in discrete cytoplasmic foci (thereafter termed as P-bodies) in yeast.72 Later studies revealed that Ago2/miRISC localizes to P-bodies.73,74 Although miRNA localization is clearly linked to P-bodies, Chu and Rana suggested that miRNA-mediated translational repression in cultured human cells does not require P-bodies, as disruption of P-bodies by depleting Lsm1 has no effect on RNAi activity and does not affect the interaction of Ago2 with RCK/p54, a DEAD box helicase essential for miRNA-mediated translational repression.75 They proposed that transfer of miRISC to P-bodies is not a cause, but rather a consequence of translational repression. In contrast, studies in plants implicate P-bodies in miRNA-mediated translational repression. In Arabidopsis, VCS, Decapping 1 (DCP1), and DCP2 form a decapping complex localized to P-bodies.76 Brodersen et al. observed that in Arabidopsis, vcs-1 and vcs-7 mutants have increased protein accumulation without changes at mRNA levels for a few miRNA target genes, suggesting that the decapping factor VCS is required for miRNA-based translational repression59 (Fig. 1). Another P-body component, DCP5, was found to be required for translational repression of certain cellular mRNAs,77 but whether it acts in miRNA-mediated translational repression is unknown. Although P-bodies may have different roles in miRNA activities between animals and plants, P-bodies may represent an important location during miRISC’s intracellular sorting in both animals and plants.
Lastly, miRNA activities have also been linked to cellular lipid metabolism. Arabidopsis MAD3 was recently found to encode 3-hydroxy-3-methylglutaryl CoA (HMG-CoA) reductase.69 Interestingly, in C. elegans, HMG-CoA synthase is also required for miRNA activity.78 Both HMG-CoA reductase and HMG-CoA synthase are enzymes in the mevalonate pathway of lipid metabolism. The end product of the mevalonate pathway is isoprenoids, which feed into a variety of biosynthetic pathways including those of sterols, cholesterol, dolichol, heme, etc. Arabidopsis MAD4 encodes sterol C-8 isomerase; this suggests that sterols, as an output of the isoprenoid pathway, contribute to miRNA activities in plants. In C. elegans, the dolichol pathway for protein N-linked glycosylation, also an output of the isoprenoid pathway, is needed for miRNA activity.78 Because N-glycosylation assists protein folding in the ER, and sterols are membrane components, these findings in Arabidopsis and C. elegans bolster the connection between miRNAs and the endomembrane system. But how the isoprenoid pathway impacts miRNA activities remains to be uncovered.
Conclusions and Future Perspectives
miRNAs elicit silencing by several distinct but related modes of regulation: translational repression, mRNA cleavage, and mRNA decay. The current, simplified view on miRNA-mediated translational inhibition and mRNA decay in animals is as follows: translational repression occurs at an early step of translation initiation, followed by deadenylation, and extensive mRNA decay. But so far, studies on the mechanisms of action of siRNAs or miRNAs in animals have largely been conducted without reference to the subcellular locations of the small RNAs or the proteins involved. Given the emerging link between the activities of small RNAs to membrane vesicles or the ER, it is essential that mechanistic studies in the future take into consideration the involvement of membranes.
Although mRNA cleavage was initially thought to be the major mode of action of plant miRNAs, studies in the past 10 y led to the unequivocal conclusion that translational inhibition is an important mode of action of plant miRNAs. Despite this conceptual advance, knowledge on the scale of miRNA-mediated translational repression in plants is still lacking, as only a handful of miRNAs have been shown to act via translational repression. We anticipate global analyses such as ribosome footprint profiling, mRNA-seq, and quantitative proteomics to be conducted to reveal miRNA-based gene regulation (by either translational repression or mRNA cleavage) in a genome-wide scale in plants. Such global analyses will facilitate the unraveling of miRNA-regulated gene/protein networks in plants.
Our recent studies on AMP1 in Arabidopsis reveal the ER as a pivotal site of miRNA-based translational repression and prompt future studies that examine the cell biology of RNAi. Although accumulative evidence has suggested that miRISC undergoes trafficking via ER/Golgi, P-bodies, MVB, and lysosome/exosome, a clear picture of miRISC’s intracellular dynamics remains to be unveiled. Many questions remain to be addressed. Are the different modes of miRNA action physically separated in the cell? Where does miRNA-guided cleavage occur? How does cytoskeleton dynamics affect miRNA activities? How do lipids influence miRNA activities? What is the miRNA machinery on the ER that is responsible for translational repression? Answers to these questions will place RNA silencing in the context of subcellular structures and may be essential to solving various controversies currently existing in the field and fully understanding the molecular mechanisms of RNA silencing.
Acknowledgments
Research in the Chen lab is supported by a grant from NIH (GM61146) and by Howard Hughes Medical Institute and Gordon and Betty Moore Foundation (through Grant GBMF3046). XM is supported by a postdoctoral fellowship from Shenzhen University and by a grant from National Natural Science Foundation of China (Grant No.31210103901) to X Cao and BM.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/rnabiology/article/26313
References
- 1.Gong Z, Zhang S, Zhang W, Huang H, Li Q, Deng H, Ma J, Zhou M, Xiang J, Wu M, et al. Long non-coding RNAs in cancer. Sci China Life Sci. 2012;55:1120–4. doi: 10.1007/s11427-012-4413-9. [DOI] [PubMed] [Google Scholar]
- 2.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng W, Feng Y. MicroRNAs in neural cell development and brain diseases. Sci China Life Sci. 2011;54:1103–12. doi: 10.1007/s11427-011-4249-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-Y. [DOI] [PubMed] [Google Scholar]
- 5.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 6.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 9.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12:99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 10.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–20. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 11.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 12.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 14.Souret FF, Kastenmayer JP, Green PJ. AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell. 2004;15:173–83. doi: 10.1016/j.molcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 15.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26:941–6. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 16.Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 17.Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013;25:2383–99. doi: 10.1105/tpc.113.113159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu G. Plant microRNAs and development. J Genet Genomics. 2013;40:217–30. doi: 10.1016/j.jgg.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–14. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 20.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–7. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 21.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 22.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 23.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–60. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 25.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–8. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–7. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 28.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–62. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meijer HA, Kong YW, Lu WT, Wilczynska A, Spriggs RV, Robinson SW, Godfrey JD, Willis AE, Bushell M. Translational repression and eIF4A2 activity are critical for microRNA-mediated gene regulation. Science. 2013;340:82–5. doi: 10.1126/science.1231197. [DOI] [PubMed] [Google Scholar]
- 30.Bazzini AA, Lee MT, Giraldez AJ. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science. 2012;336:233–7. doi: 10.1126/science.1215704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadler M, Artiles K, Pak J, Fire A. Contributions of mRNA abundance, ribosome loading, and post- or peri-translational effects to temporal repression of C. elegans heterochronic miRNA targets. Genome Res. 2012;22:2418–26. doi: 10.1101/gr.136515.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–7. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–9. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 37.Eulalio A, Rehwinkel J, Stricker M, Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M, Izaurralde E. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 2007;21:2558–70. doi: 10.1101/gad.443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piao X, Zhang X, Wu L, Belasco JG. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–94. doi: 10.1128/MCB.01481-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 42.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Béthune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep. 2012;13:716–23. doi: 10.1038/embor.2012.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–86. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Djuranovic S, Nahvi A, Green R. A parsimonious model for gene regulation by miRNAs. Science. 2011;331:550–3. doi: 10.1126/science.1191138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin C, Nam JW, Farh KKH, Chiang HR, Shkumatava A, Bartel DP. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell. 2010;38:789–802. doi: 10.1016/j.molcel.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu QH, Upadhyaya NM, Gubler F, Helliwell CA. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa) BMC Plant Biol. 2009;9:149. doi: 10.1186/1471-2229-9-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee DY, An G. Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J. 2012;69:445–61. doi: 10.1111/j.1365-313X.2011.04804.x. [DOI] [PubMed] [Google Scholar]
- 52.Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA unction. Dev Cell. 2003;4:205–17. doi: 10.1016/S1534-5807(03)00025-X. [DOI] [PubMed] [Google Scholar]
- 53.Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D. Specific effects of microRNAs on the plant transcriptome. Dev Cell. 2005;8:517–27. doi: 10.1016/j.devcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 54.Yant L, Mathieu J, Dinh TT, Ott F, Lanz C, Wollmann H, Chen X, Schmid M. Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell. 2010;22:2156–70. doi: 10.1105/tpc.110.075606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gandikota M, Birkenbihl RP, Höhmann S, Cardon GH, Saedler H, Huijser P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007;49:683–93. doi: 10.1111/j.1365-313X.2006.02983.x. [DOI] [PubMed] [Google Scholar]
- 56.Yang L, Wu G, Poethig RS. Mutations in the GW-repeat protein SUO reveal a developmental function for microRNA-mediated translational repression in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:315–20. doi: 10.1073/pnas.1114673109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sunkar R, Kapoor A, Zhu JK. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell. 2006;18:2051–65. doi: 10.1105/tpc.106.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lanet E, Delannoy E, Sormani R, Floris M, Brodersen P, Crété P, Voinnet O, Robaglia C. Biochemical evidence for translational repression by Arabidopsis microRNAs. Plant Cell. 2009;21:1762–8. doi: 10.1105/tpc.108.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–90. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Liu L, Zhuang X, Yu Y, Liu X, Cui X, Ji L, Pan Z, Cao X, Mo B, et al. MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell. 2013;153:562–74. doi: 10.1016/j.cell.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell. 1999;10:3357–72. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–9. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, Ma E, Mane S, Hannon GJ, Lawson ND, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–8. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–8. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tahbaz N, Kolb FA, Zhang H, Jaronczyk K, Filipowicz W, Hobman TC. Characterization of the interactions between mammalian PAZ PIWI domain proteins and Dicer. EMBO Rep. 2004;5:189–94. doi: 10.1038/sj.embor.7400070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–9. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 67.Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim H, Cho IS, et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–6. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stalder L, Heusermann W, Sokol L, Trojer D, Wirz J, Hean J, Fritzsche A, Aeschimann F, Pfanzagl V, Basselet P, et al. The rough endoplasmatic reticulum is a central nucleation site of siRNA-mediated RNA silencing. EMBO J. 2013;32:1115–27. doi: 10.1038/emboj.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brodersen P, Sakvarelidze-Achard L, Schaller H, Khafif M, Schott G, Bendahmane A, Voinnet O. Isoprenoid biosynthesis is required for miRNA function and affects membrane association of ARGONAUTE 1 in Arabidopsis. Proc Natl Acad Sci U S A. 2012;109:1778–83. doi: 10.1073/pnas.1112500109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–22. doi: 10.1016/j.cub.2007.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, DI Carlo M, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–56. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–23. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–6. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 75.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu J, Yang JY, Niu QW, Chua NH. Arabidopsis DCP2, DCP1, and VARICOSE form a decapping complex required for postembryonic development. Plant Cell. 2006;18:3386–98. doi: 10.1105/tpc.106.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21:3270–9. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi Z, Ruvkun G. The mevalonate pathway regulates microRNA activity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2012;109:4568–73. doi: 10.1073/pnas.1202421109. [DOI] [PMC free article] [PubMed] [Google Scholar]


