SUMMARY
Animals have body parts made of similar cell types located at different axial positions (e.g. limbs). The identity and distinct morphology of each structure is often specified by the activity of different “master regulator” transcription factors. Although similarities in gene expression have been observed between body parts made of similar cell types, it is not known how regulatory information in the genome is differentially utilized to create morphologically diverse structures in development. Here, we use genome-wide open chromatin profiling to show that among the Drosophila appendages, the same DNA regulatory modules are accessible throughout the genome at a given stage of development, except at the loci encoding the master regulators themselves. In addition, while open chromatin profiles change over developmental time, these changes are coordinated between different appendages. We propose that master regulators create morphologically distinct structures by differentially influencing the function of the same set of DNA regulatory modules.
INTRODUCTION
Animals are comprised of a diversity of body parts, varied in form according to their function. Among species, changes in DNA sequence have been shown to underlie changes in morphology (Carroll, 2008; Wray, 2007). However, within a single animal, the same genome sequence gives rise to the full panoply of body parts through differential regulation of gene expression. During development, differences in body part identity are determined by the activity of master regulator transcription factors, often termed “selector” genes (Mann and Carroll, 2002). In Drosophila, the homeodomain transcription factor Distalless (Dll) (Gorfinkiel et al., 1997) and the zinc finger proteins Buttonhead and Sp1 (Estella and Mann, 2010) specify ventral appendage identities, including the legs. Dorsal appendage identities, such as the wing and haltere, are specified by Vestigial (Vg) and its TEA-domain DNA binding partner Scalloped (Sd) (Halder et al., 1998). Along the anterior-posterior axis, morphology of structures is diversified by other master regulator transcription factors such as the Hox proteins. For example, the Hox protein Ultrabithorax (Ubx) is responsible for specifying haltere identity over wing (Lewis, 1978). While many of the transcription factors that control growth and patterning during appendage development have been identified, little is known about how they access regulatory information in the genome to create different appendage morphologies. One possibility is that each master regulator, with its unique DNA binding specificity, accesses a unique set of cis-regulatory elements in the genome to differentially regulate gene expression between the appendages.
A major hurdle to understanding the mechanisms of developmental gene regulation is the identification of functional DNA regulatory elements in the genome. A variety of methods has been used to identify potential DNA regulatory elements with varying degrees of success, including prediction of transcription factor binding sites (Berman et al., 2002; Markstein et al., 2002; Rebeiz et al., 2002), DamID (van Steensel and Henikoff, 2000), chromatin immunoprecipitation (ChIP) (Fisher et al., 2012; Negre et al., 2011; Sandmann et al., 2007; Visel et al., 2013; Zinzen et al., 2009), STARR-seq (Arnold et al., 2013), and large-scale cloning efforts (Jory et al., 2012; Pfeiffer et al., 2008). Yet another approach to identify DNA regulatory elements is the identification of nucleosome-depleted or “open chromatin” sites. Methods such as DNase I hypersensitivity mapping (Dorschner et al., 2004) and FAIRE (Giresi et al., 2007; Nagy et al., 2003), provide a snapshot of genomic sites where nucleosomes have been depleted, often through competition with trans-acting factors. Nucleosome depletion identifies a variety of DNA regulatory elements, including those involved in DNA replication (MacAlpine et al., 2010), nuclear organization (Bartkuhn et al., 2009), and transcription (e.g. enhancers) (Song et al., 2011; Thomas et al., 2011). Thus, open chromatin profiling is an ideal method to compare how trans-acting factors read out the genome between different tissues, independently of the identity of those factors.
Here, we use development of the thoracic appendages in Drosophila to examine how a single genome sequence is utilized to give rise to morphologically diverse structures. We first demonstrate that open chromatin is an accurate and precise predictor of functional enhancer activity in developing embryos. Next, we ask how the genome is accessed in different appendages at two stages of their development. Although comprised of similar cell types, each appendage expresses a different combination of master regulator transcription factors that have different DNA binding domains, and therefore we hypothesized that in each appendage a significant subset of the enhancers used would be unique to that appendage. In contrast to our expectations, we find that the same set of enhancers is accessible in all three appendages, with the exception of enhancers that control expression of the master regulators themselves. We show that this shared set of appendage enhancers changes coordinately over developmental time. Finally, we provide functional evidence that the appendage master regulators differentially regulate the activity of the same enhancers to effect differences in gene expression between the appendages. Thus, morphologically distinct structures can be created using the same set of enhancers.
RESULTS
FAIRE identifies DNA bound by regulatory factors in developing animals
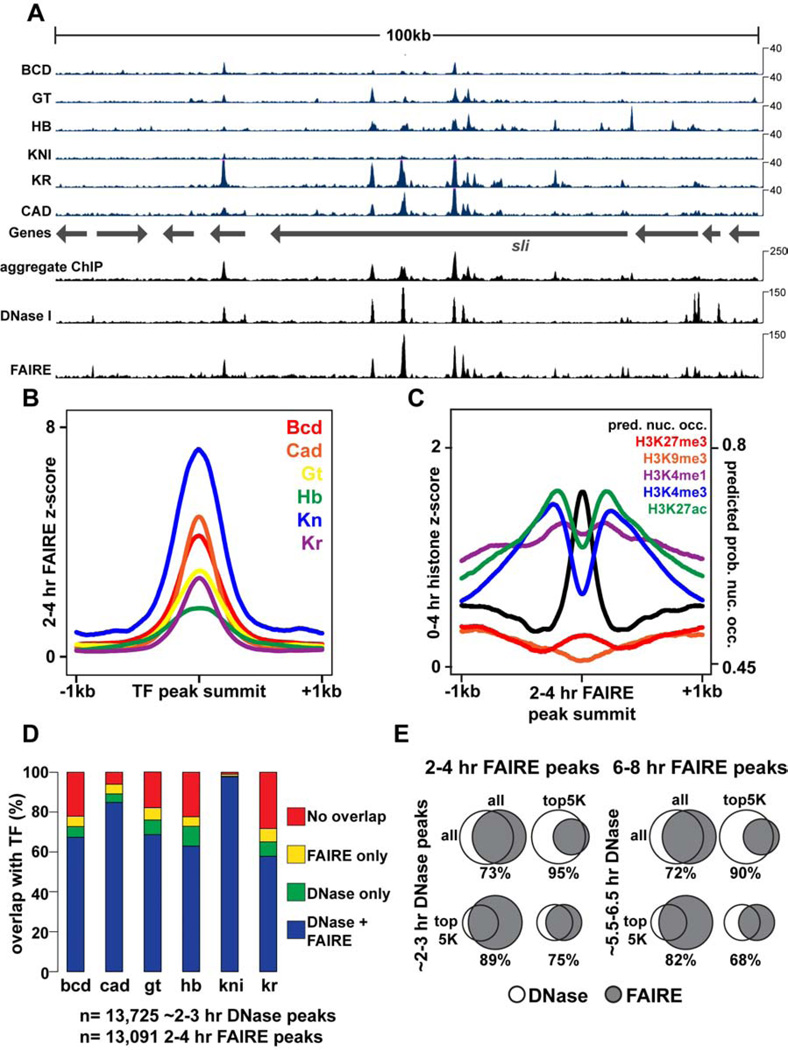
To identify genomic locations with gene regulatory activity, we performed Formaldehyde-Assisted Isolation of Regulatory Elements, which identifies nucleosome-depleted or “open” chromatin, followed by high-throughput sequencing (FAIRE-seq) (Giresi et al., 2007; Simon et al., 2012) and RNA-seq at three developmental timepoints in Drosophila embryos: 2–4 hr after egg laying (AEL) during initial establishment of the body axes and germ layers, 6–8 hr AEL during fine-scale cell fate specification through the action of local signaling pathways, and 16–18 hr AEL when many cells have terminally differentiated. Consistent with previous studies (Giresi et al., 2007; Song et al., 2011), we find FAIRE-enriched regions are bound by regulatory factors (Fig. 1, Fig. S1). FAIRE signal very closely resembles the aggregate transcription factor chromatin immunoprecipitation (ChIP) signal (Bradley et al., 2010) (Fig. 1A), supporting the well-established association between transcription factor binding and nucleosome depletion (Fig. 1B). Genomic locations with high FAIRE signal are evolutionarily conserved (Siepel et al., 2005) (Fig. S1) and are associated with high levels of “active” histone modifications (Fig. 1C, Fig. S1), including H3K4me1 and H3K27ac, marks associated with enhancer activity, and H3K4me3, a mark associated with active gene promoters. Correspondingly, high FAIRE signal is associated with low levels of “repressive” histone modifications such as H3K27me3 and H3K9me3 (Fig. 1C, Fig. S1). FAIRE data from embryos collected at 2–4 hr and 6–8 hr also closely match recent genome-wide DNase I hypersensitivity data from early Drosophila embryos (Thomas et al., 2011) (Fig. 1A, D, E). Thus, FAIRE identifies nucleosome-depleted regions during Drosophila development, which coincide with genomic sites bound by multiple regulatory factors. Both FAIRE-seq and RNA-seq experiments were highly reproducible (Fig. S1).
Figure 1. FAIRE identifies open chromatin bound by key developmental regulators.
All times below refer to hours After Egg Laying (AEL), which are estimated for data from other studies. DNase I data are from (Thomas et al., 2011). 2–3 hr ChIP data are from (Bradley et al., 2010). Transcription Factors (TFs): Bcd, Bicoid; Cad, Caudal; Gt, Giant; Hb, Hunchback; Kn, Knirps; Kr, Kruppel. (A) Browser representation of the slit locus. Below the genes track is ChIP signal (blue, Counts Per Million (CPM)) from 2–3 hr embryos, plotted for individual TFs. Above the genes track, from bottom to top, is the aggregate ChIP signal generated by summing the normalized signal from each individual TF, followed by 2–3 hr DNase I signal (CPM), and 2–4 hr FAIRE data (CPM). (B) 2–4 hr FAIRE signal at TF peaks from 2–3 hr embryos. (C) 0–4hr histone modification signals (Negre et al., 2011) and predicted probability of nucleosome occupancy based on DNA sequence (Kaplan et al., 2009) plotted for regions surrounding 2–4 hr AEL FAIRE peaks, centered on the maximum FAIRE signal for each peak. (D) Stacked bar charts showing overlap of ~2–3 hr DNase I and 2–4 hr FAIRE peaks with TF ChIP peaks from 2–3 hr embryos. (E) Venn diagrams depicting peak overlaps between ~2–3 hr DNase I peaks and 2–4 hr FAIRE peaks (left), and ~5.5–6.5 hr DNase I and 6–8 hr FAIRE peaks (right). See also Figure S1.
Open chromatin identifies enhancers and the timing of enhancer activity
A range of approaches has been used to identify functional DNA regulatory elements in the genome with varying degrees of success (Aerts et al., 2007). Since FAIRE identifies genomic regions that are bound by trans-acting proteins, it followed that FAIRE enrichment might be used as a predictor of enhancer activity at a given point in time. To test the sufficiency of individual FAIRE-enriched sites to control transcription, we cloned twenty-four different open chromatin regions for transgenic reporter assays (Table S1). To identify target regions for cloning, we used only FAIRE data, without consulting any other data sets (e.g. ChIP, evolutionary conservation). We chose previously uncharacterized regions that were differentially accessible across developmental stages or between tissues, and that are near developmentally important genes known to be expressed at these stages. We placed these selected regions upstream of a synthetic core promoter (Pfeiffer et al., 2010) to drive expression of the yeast transcription factor GAL4.
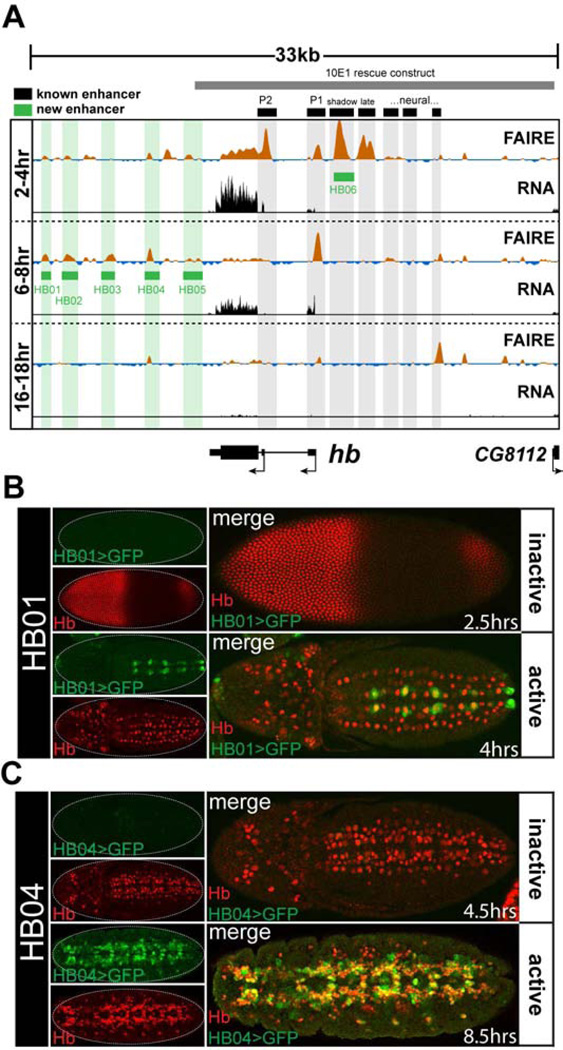
Despite extensive prior study of the loci selected for testing, we identified many previously undiscovered enhancers. Twenty-three of the twenty-four (96%) cloned regions recapitulated sharp, distinctive subsets of their gene’s expression pattern in transgenic reporter assays (Data File S1). For example, several enhancers were identified at the hunchback (hb) locus. Hb was first identified due to its function in anterior-posterior patterning of the blastoderm embryo (Nusslein-Volhard and Wieschaus, 1980). Consistent with that role, all hb enhancers previously known to control blastoderm expression (Gallo et al., 2011; Perry et al., 2011) coincide precisely with regions of open chromatin specifically at the 2–4 hr FAIRE timepoint (Fig. 2A black boxes). However, little is known about control of hb expression later in development when hb is required for proper development of the central nervous system (Hirono et al., 2012) and the tracheal system (Merabet et al., 2005).
Figure 2. FAIRE signal accurately predicts enhancer activity.
(A) FAIRE and RNA signals at the hunchback (hb) locus in embryos. Black boxes designate the locations of known enhancers: (left to right) P2 promoter, P1 promoter, blastoderm shadow enhancer, late blastoderm enhancer, and recently identified neural enhancers (Gallo et al., 2011; Hirono et al., 2012; Margolis et al., 1995; Perry et al., 2011). Green boxes designate enhancers that were identified and cloned in this study. The grey box indicates the boundaries of the 10E1 transgenic hb rescue construct (Margolis et al., 1995). (B, C) Confocal images of embryos from two transgenic lines (HB01, HB04) stained with antibodies for Hb (red) and GFP (green) protein. The estimated age of each embryo is indicated. The timing of chromatin opening coincides with timing of reporter activity. See also Table S1 and Data File S1.
We identified five hb enhancers in this study. The HB01 enhancer, which is accessible at the 6–8 hr timepoint (and to a lesser extent at the 2–4 hr timepoint), is active in a subset of Hb-positive neuroblasts in the ventral nerve cord beginning at 4hr AEL (Fig. 2B), whereas the enhancers HB04 and HB05, which are also accessible at 6–8 hr, are active in the Hb-positive progeny of these cells beginning around 5 hr AEL (Fig. 2C and Data File S1). Enhancers HB03 and HB04 recapitulate hb expression patterns in cells required for tracheal system development, in the mesoderm, and in the nervous system (Data File S1). The expression patterns of these enhancers show (1) that regulation of hb expression is divided between different enhancers for different lineages of hb-expressing cells, and (2) that there is a temporal division in the regulation of hb expression between different enhancers within hb-expressing cells of the developing nervous system. Interestingly, none of the 3’ hb enhancers we cloned are fully contained within the 10E1 hb construct (Fig. 2A grey box), which rescues hb function in blastoderm embryos, but is unable to provide appropriate hb function later in development, which leads to lethality (Margolis et al., 1995), This, along with our data from expression constructs, suggests that these newly-cloned enhancers are essential for regulating hb expression later in embryogenesis.
Finally, an important feature emerges from analysis of the newly-cloned enhancers: the timing of the appearance of open chromatin at enhancers coincides with the timing of their activity in vivo (Fig. 2, Table S1). Thus, FAIRE can identify not only the precise location in the genome of functional enhancers, but also the time at which these enhancers are active. Since FAIRE identifies any region of the genome that is depleted of nucleosomes, it is not expected that all FAIRE-enriched regions act as transcriptional enhancers. For example, many open chromatin regions identified by FAIRE correspond to Polycomb Response Elements (PREs) (Fig. 3C). Conversely, it is possible that regions of the genome that are not enriched by FAIRE act as transcriptional enhancers, or regulate gene expression through other mechanisms. Nevertheless, these reporter experiments demonstrate that FAIRE is an exceptionally accurate, sensitive, and precise predictor of gene regulatory activity.
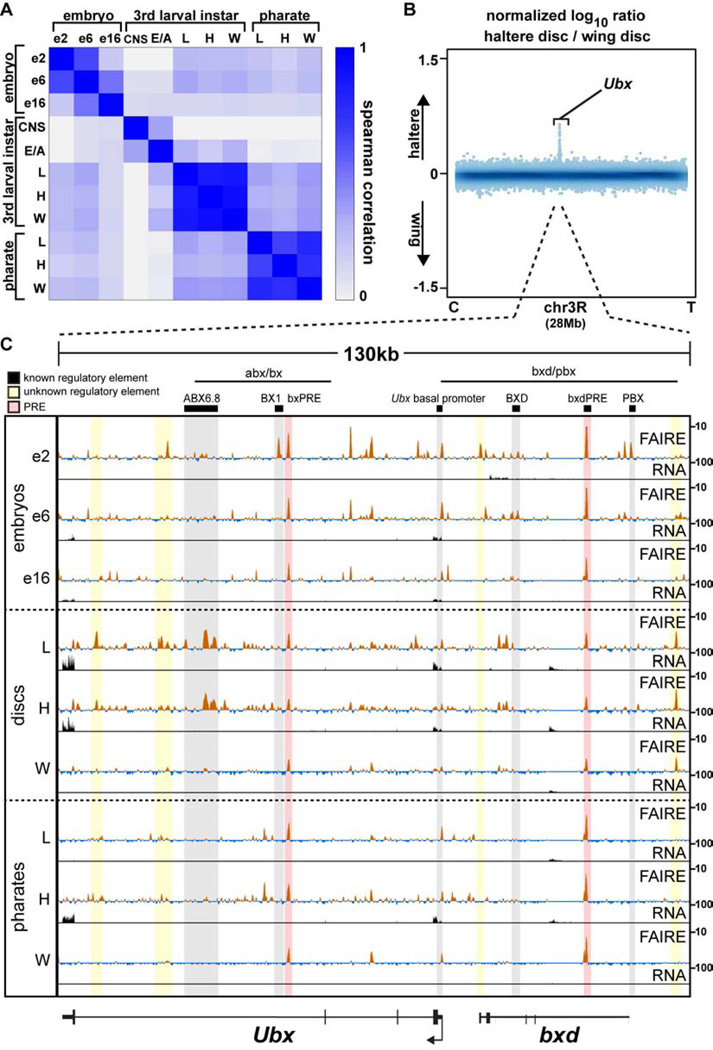
Figure 3. Appendage open chromatin profiles are very similar within a stage, except at master regulator loci.
(A) Spearman correlation coefficients of FAIRE signal in 500-bp windows genome-wide for each pairwise comparison across all samples. (B) Log10 ratio (haltere/wing) of FAIRE signal from chromosome 3R (28Mb). Centromere (C), telomere (T), and the Ultrabithorax (Ubx) locus are indicated. (C) FAIRE (z score: −2 to 10) and RNA (FPKM: 0 to 100) signals at the Ubx and bithoraxoid (bxd) loci in embryos, imaginal discs, and pharate appendages. Horizontal black lines indicate the locations known Ubx regulatory regions (Simon et al., 1990). Black boxes designate the locations of known DNA regulatory elements: (left to right) ABX6.8 enhancer, BX1 enhancer, bxPRE, Ubx basal promoter, BXD enhancer, bxdPRE, PBX enhancer (Chan et al., 1994; Muller and Bienz, 1991; Pirrotta et al., 1995; Qian et al., 1991; Simon et al., 1990; Zhang et al., 1991). Shaded red rectangles indicate the locations of known PREs (Papp and Muller, 2006; Pirrotta et al., 1995). Shaded yellow rectangles indicate the locations of putative regulatory elements identified in this study. See also Figures S2, S3, S4.
Open chromatin profiles among leg, wing, and haltere imaginal discs are nearly identical at a given developmental stage
Similar to DNase I hypersensitivity patterns in embryos (Thomas et al., 2011), regulatory elements defined by FAIRE were highly dynamic from one embryonic stage to the next, with thousands of sites opening and closing between stages (Fig. S6). We next asked how information in the genome is utilized to generate morphologically diverse structures by mapping open chromatin during Drosophila appendage development. Insect appendages are thought to have evolutionary origins greater than 400 million years ago (Engel and Grimaldi, 2004; Garrouste et al., 2012), and they exhibit a stunning diversity of morphologies tailored to their functions (Grimaldi and Engel, 2005). The identity of each appendage is specified by a unique combination of master regulator transcription factors that differentially controls pattern formation, growth, and differentiation (Ashburner and Novitski, 1976; Estella and Mann, 2010; Gorfinkiel et al., 1997; Halder et al., 1998). Since the appendage master regulators possess different DNA-binding specificities, our hypothesis was that different transcriptional enhancers would be used to create each morphologically distinct appendage. To test this, we dissected the precursors of the thoracic appendages (called imaginal discs) from 3rd instar larvae (120 hAEL) and performed FAIRE. In sharp contrast to our findings from different stages of embryogenesis, and in refutation of our hypothesis, open chromatin profiles from the wing, haltere, and metathoracic (T3) leg imaginal discs were nearly identical to each other (Fig. 3A, Fig. S6). For example, the Spearman’s rank correlation coefficients of FAIRE signals between the thoracic appendage imaginal discs ranged between 0.85–0.90, whereas the same measures between different stages of embryogenesis ranged between 0.20–0.64. We describe these findings in more detail below.
Nearly all the differences in open chromatin between wing and haltere imaginal discs occur at the Ubx locus
Comparison of wing and haltere imaginal disc open chromatin profiles revealed an especially striking result. Among the most pronounced FAIRE peaks in wing and haltere discs across the entire genome (the top 20%, 3,525 peaks), only five sites are specifically open in haltere imaginal discs relative to wing imaginal discs (Fig. S2, Table S2). Four of these five regions are located within the Ubx locus (Fig. 3B, C, Fig. S3A). The function of Ubx in transforming wing identity into haltere is one of the best-characterized examples of transcription-factor dependent morphogenesis in development (Crickmore and Mann, 2008). Mutations in Ubx can lead to transformation of haltere into wing, resulting in a four-winged fly (Lewis, 1978). Although Ubx has been shown to regulate hundreds of target genes at specific stages of haltere development (Hersh et al., 2007; Pavlopoulos and Akam, 2011) (Fig. S4), the molecular mechanisms by which Ubx controls growth and patterning are largely unknown. Recent ChIP-chip experiments have identified putative Ubx binding sites in the developing haltere and T3 leg imaginal discs (Choo et al., 2011; Slattery et al., 2011a), but the pattern of Ubx binding suggests that only a subset of these sites are functional (Slattery et al., 2011a). Moreover, since Ubx is expressed in the haltere but not in the wing, these ChIP experiments cannot be used to compare how regulatory information is accessed in the haltere relative to the wing. We asked whether our FAIRE data could help to define functional Ubx binding events. We found that open chromatin sites bound by Ubx tend to be more conserved, and occur at Ubx-responsive genes (Fig. S3B – D). These data, combined with the data showing that only five sites are open in the haltere disc but not the wing disc, with four of these residing at the Ubx locus itself, means that Ubx binds to regulatory DNA in the haltere (where it is expressed) that is also accessible for use in the wing (where Ubx is not expressed), rather than to a set of enhancers that are specific to the haltere. Thus, these data suggest that morphologically distinct structures with a shared evolutionary origin can be made by acquiring transcription factor binding sites in existing enhancers, rather than by introducing a new set of enhancers de novo.
Differences in appendage open chromatin profiles are at loci encoding key developmental regulators
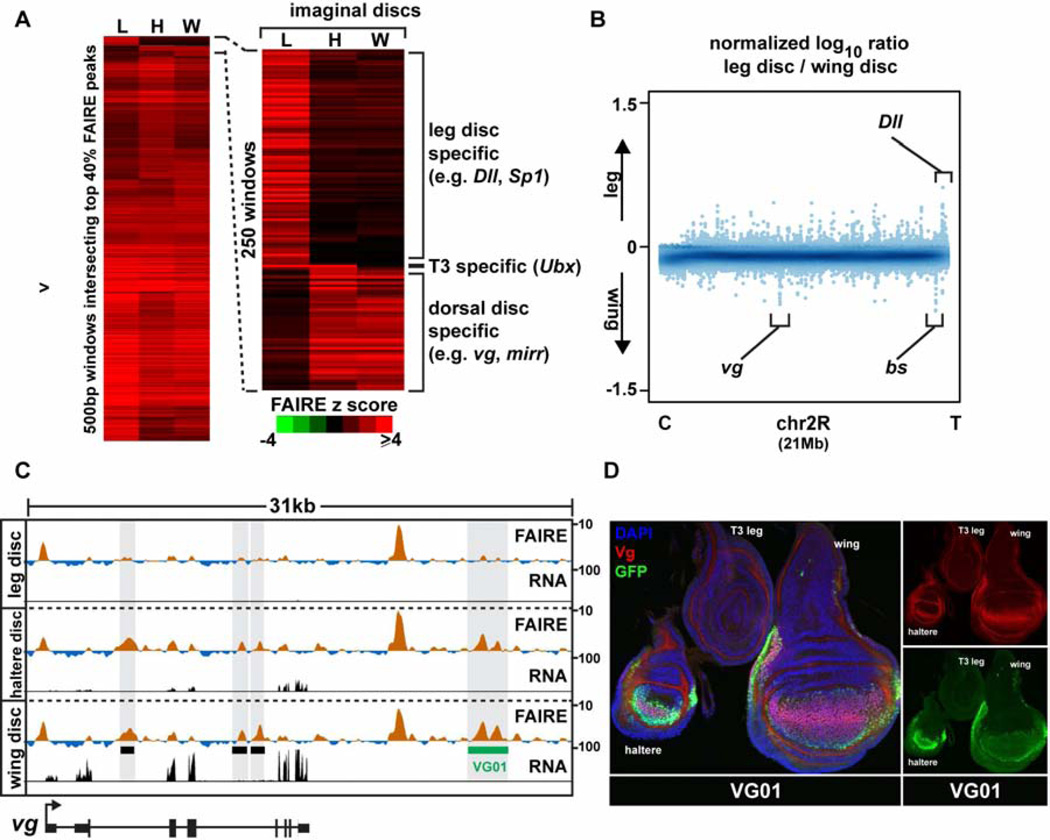
Given their diverse morphologies and transcription factor expression profiles (Fig. S5), we were surprised to find that wing and leg imaginal discs also share very similar open chromatin profiles. Of the most pronounced open chromatin regions (the top 20%, 3,525 peaks), only 110 were differentially open (Fig. S2, Table S2). We speculated that these few differences in open chromatin between wing and leg imaginal discs were important in determining morphological differences, as was the case with wing and haltere imaginal discs. Indeed, genes with open chromatin specific to the leg imaginal discs include Dll, and Sp1, the master regulators of leg development (Estella and Mann, 2010; Gorfinkiel et al., 1997) (Fig. 4A, B). Similarly, genes with open regions specific to the dorsal imaginal discs (wing and haltere) include vg and blistered, transcription factors required for development of these appendages (Kim et al., 1996; Montagne et al., 1996) (Fig. 4, Fig. S5). We tested whether these disc-specific open chromatin regions identified by FAIRE function as appendage-specific enhancers, and found that 6 of 7 accurately recapitulate gene expression in imaginal discs of late 3rd instar larvae (Table S1, Data File S1). Similar to our observations from the embryonic time course, the presence of disc-specific open chromatin correlated with disc-specific enhancer activity - the cloned imaginal disc enhancers are active only in the imaginal discs in which they are accessible. For example, the VG01 enhancer identified by this study, which is open specifically in wing and haltere imaginal discs, recapitulates vg expression specifically in wing and haltere imaginal discs and is not active in leg imaginal discs (Fig. 4D). Together, these data demonstrate that genomic regions accessible for use in thoracic appendage imaginal discs are nearly identical, except for at appendage master regulator gene loci.
Figure 4. Appendage open chromatin profiles differ primarily at loci of key developmental regulators.
(A) Hierarchical clustering of FAIRE signal from windows intersecting the top 7,000 imaginal disc peaks. Right, zoom-in of the most variable windows. (B) Log10 ratio (leg/wing) of FAIRE signal from chromosome 2R (21 Mb). Loci encoding key transcription factors are indicated. (C) Browser representation of the vg locus showing FAIRE (z score: −2 to 10) and RNA (FPKM: 0 to 100) signals in imaginal discs. Black boxes designate the locations of known enhancers: (left to right) boundary, vgAME, and quadrant enhancers (Kim et al., 1996; Stergachis et al., 2013; Williams et al., 1994). The green box designates the newly-cloned VG01 enhancer, which is active in the wing and haltere but not the leg. (D) Confocal images of imaginal discs from the VG01 transgenic line, stained with DAPI (blue), and antibodies for GFP (green), and Vg (red). The VG01 enhancer recapitulates vg expression in haltere and wing imaginal discs, and lack of expression in the leg disc. See also Figure S5, and Table S2.
Leg, haltere, and wing open chromatin profiles change coordinately over developmental time
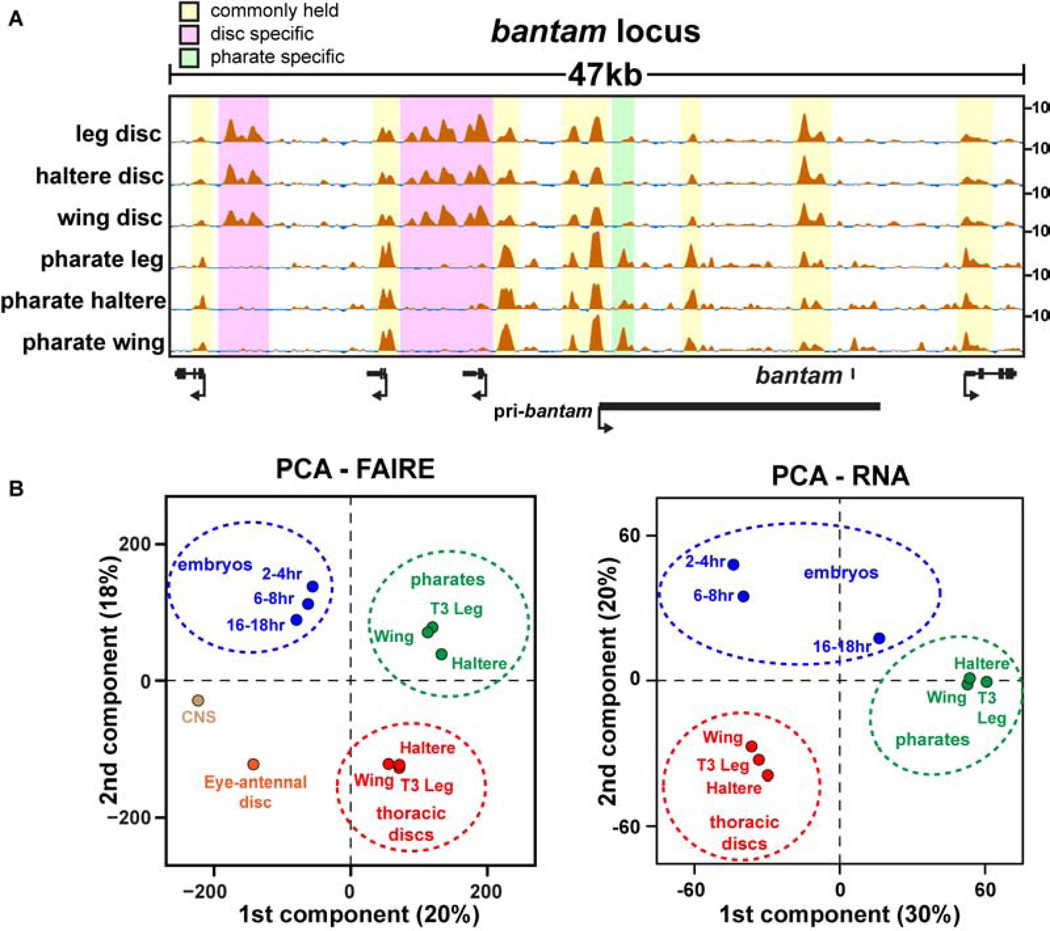
Although the fate of each disc is already determined by late third instar stages (Ashburner and Novitski, 1976), we thought that perhaps the similarity in thoracic imaginal disc open chromatin profiles might somehow be specific to this early stage of appendage formation. We therefore tested whether the terminally differentiated appendages that arose from these imaginal discs also share a similar open chromatin profile. We performed FAIRE on the fully-developed appendages of stage 13 and 14 pharate adults (~210hr). Like our observations in imaginal discs, the open chromatin profiles of the terminally differentiated appendages were strikingly similar to each other (Table S3). Spearman’s rank correlation coefficients between the pharate appendages ranged between 0.67–0.80 (Fig. 3A). Despite their similarity to each other, the open chromatin profiles in pharate appendages were markedly different from the open chromatin profiles in imaginal discs (Fig. 5A, Fig. S6). These data lead to the unexpected conclusion that open chromatin profiles of different appendages at the same developmental stage are more similar to each other than they are to their own lineage in subsequent stages (Fig. 5B). Thus, an imaginal wing disc is more similar to an imaginal leg disc than it is to its cellular progeny, the adult wing. This conclusion holds true regardless of whether FAIRE-seq or RNA-seq data are used in the analysis (Fig. 5B), or whether the data are pooled or analyzed as individual replicates (Fig. S7A, B). Although larval discs also give rise to body wall regions that are not present in pharate adult appendages, the many new open chromatin regions in the adult appendages support a large-scale change in open chromatin profiles over time.
Figure 5. Different appendages are more similar to each other at a given timepoint than they are to their own cellular progeny at a later timepoint.
(A) FAIRE signal surrounding the bantam locus from imaginal discs and pharate appendages. (B) Plots of PCA scores for the first two components from principal component analysis (PCA) of FAIRE and RNA signals. The percentage of the total variance represented by each component is shown in parentheses. See also Figure S6, and Table S2.
Different cell types have distinct chromatin profiles, but morphologically distinct tissues composed of similar cell types share open chromatin profiles at a given stage of development
Much like vertebrate limbs, the different Drosophila appendages are comprised of similar combinations of cell types (Klebes et al., 2002; Rodgers and Shearn, 1977; Taher et al., 2011). To test whether the similarities in thoracic imaginal disc open chromatin profiles also apply to body parts comprised of different combinations of cell types, we performed FAIRE on 3rd instar eye-antennal imaginal discs, which share developmental features of both dorsal and ventral appendages. The antenna is considered to be a ventral structure like the leg because mutations exist that transform antennal identity into leg (e.g. homothorax, antennapedia) (Casares and Mann, 1998). In contrast, the eye is considered to be a dorsal structure like the wing because mutations exist that transform eye tissue into wing (e.g. ophthalmoptera) (Morata and Lawrence, 1979). Therefore, since the wing and leg have very similar open chromatin profiles, one might expect the eye-antennal disc to have an open chromatin profile very similar to the wing and leg.
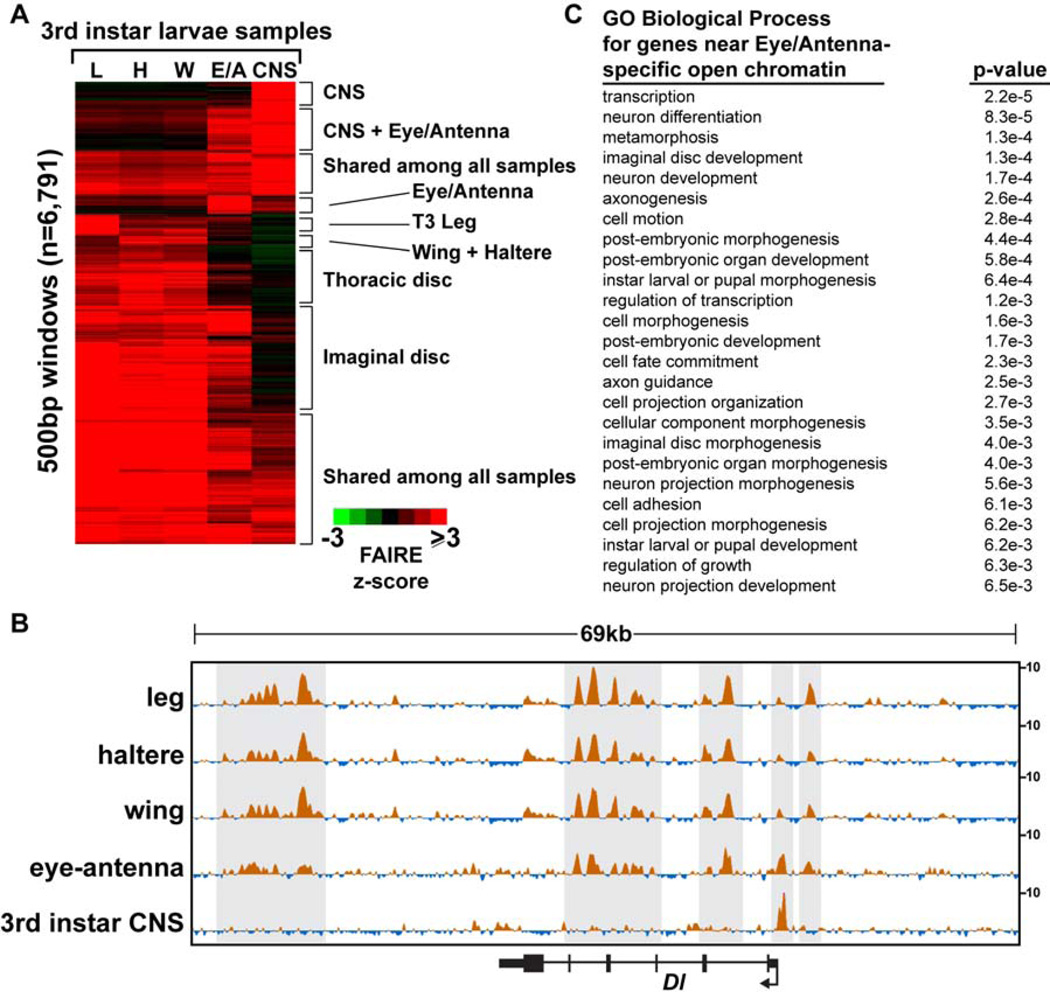
The open chromatin profile of the eye-antennal disc is indeed very similar to those of the thoracic imaginal discs (Fig. 3A, Fig. 6A,C). For example, many open chromatin regions are held in common between the eye-antennal disc and the thoracic imaginal discs at the Delta (Dl) locus (Fig. 6B). These similarities in open chromatin occur despite differences in Dl expression in these tissues. For example, Dl is transcribed in photoreceptors and cells within the morphogenetic furrow of the eye (Parks et al., 1995), whereas it is expressed in rings near the presumptive joints of leg imaginal discs (Bishop et al., 1999), and in stripes near the presumptive veins of wing imaginal discs (de Celis et al., 1997). While there are many similarities in the open chromatin profiles between these imaginal discs, the eye-antennal disc open chromatin profile also deviates from the thoracic disc open chromatin profiles at many locations in the genome (Fig. 6A, B). Many of these differences are found at genes that function in neural cells, particularly regions that are open in the eye-antennal disc but are closed in the thoracic discs (Fig. 6C). This is consistent with the known presence of neural cells in the eye half of the disc. To test this hypothesis, we compared the open chromatin profiles of the eye-antennal disc and the thoracic discs to those of the central nervous system of the same larval stage (late 3rd instar CNS). These data demonstrate that the open chromatin profile from the eye-antennal disc can be reconstructed nearly completely from the profiles of the thoracic discs plus the CNS (Fig. 6A, B). Thus, not all cells at a given developmental stage share the same open chromatin profiles. Instead, open chromatin profiles are likely shared by cells with similar identities. We have not yet explored the spatial heterogeneity of the open chromatin profiles within a given body part.
Figure 6. Eye-antennal open chromatin profiles share features with appendage and CNS open chromatin profiles.
(A) Hierarchical clustering of FAIRE signal in windows intersecting the union set of top 5K FAIRE peaks from third instar larval samples. The eye-antennal signal can be reconstructed nearly completely from the profiles of the thoracic discs plus the CNS. (B) Browser representations of the Delta (Dl) locus, a gene with known roles in 3rd instar imaginal discs and CNS (see text). Note the eye-antennal signal shares features with both the thoracic discs and the CNS. (C) Gene ontology terms of the genes nearest to peaks that are present in eyeantennal discs but not present in the thoracic imaginal discs. Genes with neural cell functions are enriched. The Bonferroni corrected p-value is shown.
Appendage master regulator transcription factors differentially interpret the same enhancers
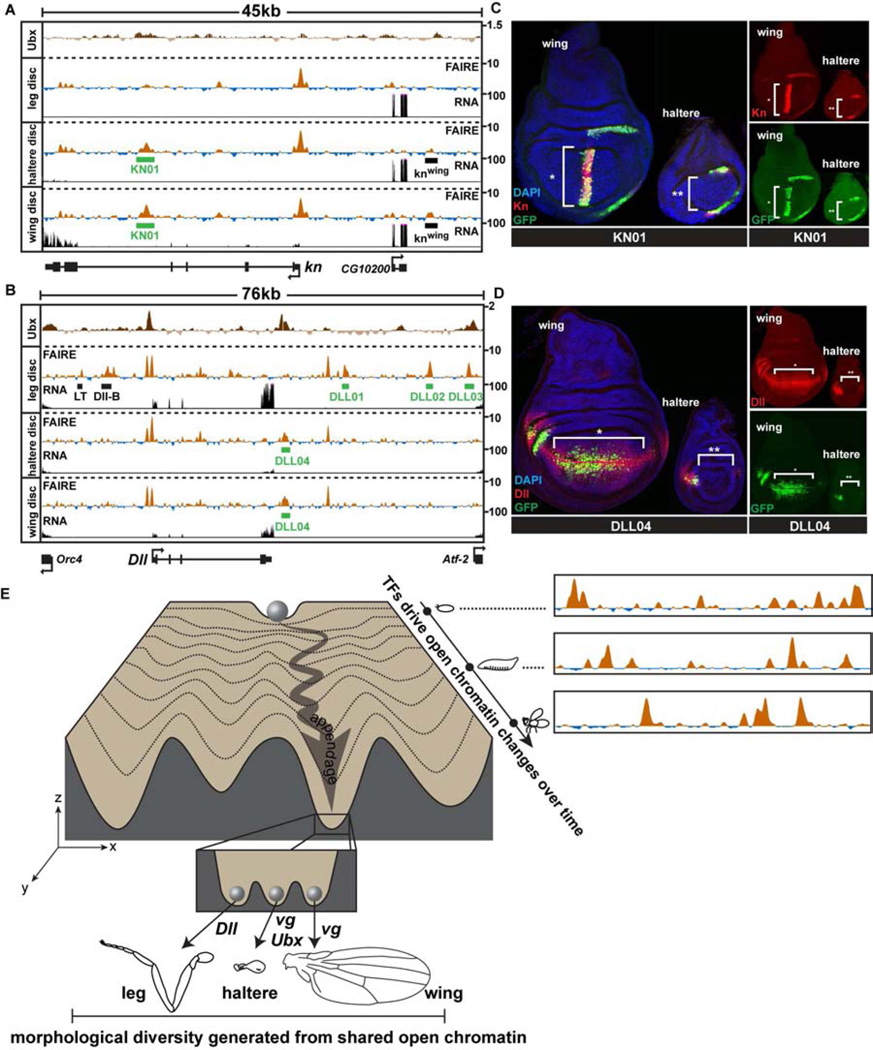
If the same set of enhancers is accessible between the developing appendages, how do master regulators such as Ubx produce differential gene expression? The knot (kn) gene is a known Ubx target that encodes a transcription factor required for cell fates between L3 and L4 wing veins (Vervoort et al., 1999). In wing imaginal discs, kn is expressed at high levels in a wide stripe of cells near the anterior-posterior boundary of the wing pouch, and at lower levels in the wing hinge (Vervoort et al., 1999) (Fig. 7A). In the haltere disc, kn is also expressed at low levels in the presumptive hinge region (Fig. 7B), but due to repression by Ubx, kn is not expressed in the pouch (Hersh and Carroll, 2005). Despite this difference in expression, the wing and haltere open chromatin profiles at the kn locus are identical (Fig. 7A). For example, a previously characterized enhancer that recapitulates kn expression specifically in the wing pouch (Hersh and Carroll, 2005) is open in both wing and haltere discs (Fig. 7A, knwing). We cloned a separate open chromatin region from the 4th kn intron that is highly accessible in both wing and haltere discs (KN01). Remarkably, the KN01 enhancer has strikingly different patterns of activity in the wing and haltere (Fig. 7B). In the wing, the KN01 enhancer is active in the pouch and hinge, whereas in the haltere, it is active only in the hinge.
Figure 7. Transcription factors differentially regulate the activity of the same enhancers in different appendages.
(A) FAIRE (z score), RNA (CPM), and Ubx ChIP (log2 ratio) (Choo et al., 2011; Slattery et al., 2011a) signal at the knot (kn) and (B) Distalless (Dll) loci in imaginal discs, with locations of enhancers KN01 and DLL01-04 (green boxes) identified in this study, plotted as in Figure 3C. (C) Confocal images showing reporter activity of KN01 and (D) DLL04 in wing and haltere imaginal discs. Discs were stained for DAPI (blue), and antibodies to GFP (green), and Kn (C) or Dll (D) (red). (E) A conceptual model of the appendage shared open chromatin profiles, depicted within the framework of Waddington’s epigenetic landscape (Waddington, 1957). A range of open chromatin profiles exists within the fly (x-axis) at any single stage of development. These profiles are dynamic over time (y-axis), and differ by varying degrees between tissues (z-axis). Each valley (z-axis) represents the shared open chromatin profile of a developing anatomical structure or tissue (e.g. appendage). The inset depicts the specific group of selector genes expressed in each developing appendage acting upon the same set of open chromatin regions to create morphologically diverse tissues. See also Figure S7.
A similarly noteworthy result was obtained with an enhancer we identified in this study from the Dll gene that is highly open in both wing and haltere discs (Fig. 7C, DLL04). Although Dll specifies leg identity, it is also required for development of cells near the margin of the wing, where Dll is expressed in late 3rd instar larvae (Gorfinkiel et al., 1997) (Fig. 7D). In the haltere, Ubx represses Dll expression in the center of the disc (Fig. S7C), such that Dll is expressed only at the extreme anterior aspect of the pouch (Fig. 7D); in contrast, Ubx does not repress Dll in the T3 leg disc despite Ubx expression because Dll is controlled by a different set of regulatory elements in leg discs (Estella et al., 2008; McKay et al., 2009) (Fig. S7C, D, E, Data File S1). Similar to our findings from the kn gene, the activity of the DLL04 enhancer in halteres is markedly different from its activity in wings, despite equivalent open chromatin profiles in both tissues (Fig. 7D). Importantly, ChIP data show that both KN01 and DLL04 are specifically bound by Ubx in vivo (Choo et al., 2011; Slattery et al., 2011a). These results provide functional evidence that Ubx controls haltere morphogenesis by modulating the activity of the set of enhancers utilized in the wing, rather than by creating a haltere-specific set of enhancers.
DISCUSSION
We address a longstanding question in developmental biology: How does a single genome give rise to a diversity of structures? Our results indicate that the unique combination of transcription factors expressed in each thoracic appendage acts upon a shared set of enhancers to create different morphological outputs, rather than operating on a set of enhancers that is specific to each tissue (Fig. 7E). This conclusion is based upon the surprising observation that the open chromatin profiles of the developing appendages are nearly identical at a given developmental stage. Therefore, rather than each master regulator operating on a set of enhancers that is specific to each tissue, the master regulators instead have access to the same set of enhancers in different tissues, which they differentially regulate. We also find that tissues composed of similar combinations of cell types have very similar open chromatin profiles, suggesting that a limited number of distinct open chromatin profiles may exist at a given stage of development, dependent on cell type identity.
Considerations regarding the sensitivity of FAIRE and the spatial heterogeneity of open chromatin profiles within a given body part
We dissected different tissues from developing flies to compare their open chromatin profiles. These tissues are composed of different cell types, each with its own gene expression profile. Our FAIRE data thus represent the average signal across all cells present in a sample. However, data from embryos and imaginal discs indicate that FAIRE is a very sensitive detector of functional DNA regulatory elements. For example, the Dll01 enhancer is active in 2–4 neurons of the leg imaginal disc, yet the FAIRE signal at Dll01 is as strong as the Dll04 enhancer, which is active in hundreds of cells of the wing pouch (Figure 7B, D, Data File S1). Thus, FAIRE may detect nearly all of the DNA regulatory elements that are in use among the cells of an imaginal disc. Our study does not rule out the existence of DNA regulatory elements that are not marked by open chromatin or otherwise not detected by FAIRE.
Despite this sensitivity, our approach does not identify which cells within the tissue have a particular open chromatin profile. For a given locus, it is possible that all cells in the tissue share a single open chromatin profile, or that the FAIRE signal originates from only a subset of cells in which a given enhancer is active. Our comparisons between eye-antennal discs, larval CNS, and thoracic discs (Fig. 3A, Fig. 6) suggest that the latter scenario is most likely, with open chromatin profiles among cells within a tissue shared by cells with similar identities at a given developmental stage.
Differential regulation of a shared set of enhancers as a mechanism of generating morphological diversity
Our observation that halteres and wings share open chromatin profiles demonstrates that Hox proteins like Ubx can differentially interpret the DNA sequence within the same subset of enhancers to modify one structure into another. This is consistent with the idea that morphological differences are largely dependent on the precise location, duration, and magnitude of expression of similar genes (Crickmore and Mann, 2006; Weatherbee et al., 1998), and it is further supported by the similarity in gene expression profiles observed between Drosophila appendages (Klebes et al., 2002) (Fig. S4), and observed between vertebrate limbs (Taher et al., 2011). However, it was not known that such dramatic differences in morphology could be achieved by using the same subset of DNA regulatory modules in different tissues genome-wide. Our findings provide a molecular framework to support the hypothesis that Hox factors function as “versatile generalists”, rather than stable binary switches (Akam, 1998). The similarity in open chromatin profiles between wings and legs suggests that this framework also extends to other classes of master regulators beyond the Hox genes. We also note that, like the Drosophila appendages, vertebrate limbs are composed of similar combinations of cell types that differ in their pattern of organization. Moreover, the Drosophila appendage master regulators share a common evolutionary origin with the master regulators of vertebrate limb development (Mann and Carroll, 2002), suggesting that the concept of shared open chromatin profiles may also apply to human development.
Our data suggest that open chromatin profiles vary both over time for a given lineage and between cell types at a given stage of development. Given the dramatic differences in the FAIRE landscape observed during embryogenesis, and between the CNS and the appendage imaginal discs during larval stages, it appears as though the alteration of the chromatin landscape is especially important for specifying different cell types from a single genome. After cell-type specification, open chromatin profiles in the appendages continued to change as they proceeded toward terminal differentiation, suggesting that stage-specific functions require significant opening of new sites or the closing of existing sites. These findings contrast with those investigating hormone-induced changes in chromatin accessibility (John et al., 2011), in which the majority of open chromatin sites did not change after hormone treatment, including sites of de novo hormone-receptor binding. Thus, it may be that genome-wide remodeling of chromatin accessibility is reserved for the longer time-scales and eventual permanence of developmental processes rather than the shorter time-scales of environmental responses.
What determines the appendage open chromatin profiles?
Different combinations of “master regulator” transcription factors, often termed selector genes, are expressed in the developing appendages. Selectors are thought to specify the identity of distinct regions of developing animals by regulating the expression of transcription factors, signaling pathway components, and other genes that act as effectors of identity (Mann and Carroll, 2002). One property attributed to selectors to explain their unique power to specify identity during development is the ability to act as pioneer transcription factors (Budry et al., 2012; Fakhouri et al., 2010). In such models, selectors are the first factors to bind target genes; once bound, selectors then create a permissive chromatin environment for other transcription factors to bind. Our finding that the same set of enhancers are accessible for use in all three appendages, with the exception of the enhancers that control expression of the selector genes themselves and other primary determinants of appendage identity, suggests that the selectors expressed in each appendage do not absolutely control the chromatin accessibility profile; otherwise, the haltere chromatin profile (for example) would differ from that of the wing due to the expression of Ubx.
What then determines the appendage open chromatin profiles? Since open chromatin is likely a consequence of transcription factor binding, two non-exclusive models are possible. First, different combinations of transcription factors could specify the same open chromatin profiles. In this scenario, each appendage’s selectors would bind to the same enhancers across the genome. For example, the wing selector Vg, with its DNA binding partner Sd, would bind the same enhancers in the wing as Dll and Sp1 bind in the leg. In the second model, transcription factors other than the selectors could specify the appendage open chromatin profiles. Selector genes are a small fraction of the total number of transcription factors expressed in the appendages (Fig. S5). Many of the non-selector transcription factors are expressed at similar levels in each appendage, and thermodynamic models would predict them to bind the same enhancers (Biggin, 2011). This model could also help to explain how the appendage open chromatin profiles coordinately change over developmental time despite the steady expression of the appendage selector genes during this same period. It is possible that stage-specific transcription factors determine which enhancers are accessible at a given stage of development. This would help to explain the temporal specificity of target genes observed for selectors such as Ubx (Pavlopoulos and Akam, 2011). Recent work supports the role of hormone-dependent transcription factors in specifying the temporal identity of target genes in the developing appendages (Mou et al., 2012). Further experiments, including ChIP of the selectors from each of the appendages, will be required to determine the extent to which either of these models is correct.
What determines the differential activity of enhancers in different appendages?
We show that binding of Ubx results in differential activity of enhancers in the haltere imaginal disc relative to the wing, despite equivalent accessibility of the enhancers in both discs, indicating that master regulators control morphogenesis by differentially regulating the activity of the same set of enhancers. It is likely that functional specificity of enhancers is achieved through multiple mechanisms. These include differential recruitment of co-activators and co-repressors, modulation of binding specificity through interactions with co-factors (Slattery et al., 2011b), differential utilization of binding sites within a single enhancer (Bradley et al., 2010), or regulation of binding dynamics through an altered chromatin context (Lickwar et al., 2012). This last mechanism would allow for epigenetic modifications early in development to impact subsequent gene regulatory events. For example, the activity of Ubx enhancers in the early embryo (Fig. 3C) may control recruitment of Trithorax or Polycomb complexes to the PREs within the Ubx locus, which then maintain Ubx in the ON or OFF state at subsequent stages of development (Papp and Muller, 2006; Pirrotta et al., 1995). Consistent with this model, Ubx enhancers active in the early embryo are only accessible in our 2–4 hr timepoint, whereas the accessibility of Ubx PREs varies little across developmental time or between tissues at a given developmental stage.
Evolutionary significance
Our results also have implications for the evolution of morphological diversity. Halteres and wings are considered to have a common evolutionary origin, but the relationship between insect wings and legs is unresolved (Averof and Cohen, 1997; Jockusch and Ober, 2004). Our observation that wings and legs share open chromatin profiles supports the hypothesis that wings and legs also share a common evolutionary origin in flies. Since legs appear in the fossil record before wings, the similarity in their open chromatin profiles suggests that the existing leg cis-regulatory network was co-opted for use in creation of dorsal appendages during insect evolution.
EXPERIMENTAL PROCEDURES
RNA and FAIRE sample collections
Drosophila strains were grown and collected as previously reported (Agelopoulos et al., 2012; Estella et al., 2008). RNA-seq and FAIRE-seq experiments were performed essentially as described (Simon et al., 2012). See Supplementary Experimental Procedures for details.
Sequence data analysis
FAIRE-seq data were processed essentially as previously described (Simon et al., 2012). FAIRE signal was converted to z-scores: genomic DNA signal (normalized to read depth) was subtracted from FAIRE signal (normalized to read depth) at each base, and z-scores were generated at each base by calculating the mean and standard deviation of the FAIRE base coverage signal for individual chromosome arms, subtracting the mean signal from the signal at each base on the given chromosome arm, and dividing by the standard deviation. FAIRE and DNaseI peaks were called with MACS2 (Zhang et al., 2008). Hierarchical clustering and principal component analysis was performed with Cluster 3.0 (de Hoon et al., 2004). RNA-seq data were aligned to the reference genome (dm3) using TopHat (version 1.1.4), and assembled into transcripts with Cufflinks (Trapnell et al., 2009) (version 0.9.3). Differential gene expression calls were made with Cuffdiff (version 0.9.3), as outlined in Fig. S4. The UCSC genome browser was used to visualize data (Kent et al., 2002) (http://genome.ucsc.edu). See Supplementary Experimental Procedures for details. Data has been deposited in the Gene Expression Omnibus under accession numbers GSE38727. Included in the dataset are raw sequencing reads, processed FAIRE signal tracks, FAIRE peaks calls, and RNA-seq FPKM values.
Defining regions of differential open chromatin in appendages
For the analysis shown in Figures S2, S5, and Tables S2 and S3, we focused on the most pronounced open chromatin regions because we hypothesized that these would be more likely to be associated with regulatory activity. We reasoned that DNA regulatory modules that are most likely to have mutually exclusive activity between appendages would exhibit large-scale differences in the degree to which they are open. Therefore, we defined a peak as differentially open if it was within the top 20% of FAIRE peaks (ranked by their MACS q-values) from the first sample, and did not intersect with a peak in the top 60% from the second sample. The number of FAIRE peaks in each of the two datasets being compared was kept equal for each comparison. See Fig. S2 for details.
For details on data processing, enhancer cloning, and immunofluorescence experiments, see Supplemental Experimental Procedures
Supplementary Material
Highlights.
Open chromatin accurately predicts enhancer activity in developing animals
Drosophila appendages use the same set of enhancers at a given developmental stage
Appendage open chromatin profiles change coordinately over developmental time
Master regulators differentially influence the same enhancers among appendages
ACKNOWLEDGEMENTS
We thank J. Pearson, C. Doe, R. Mann, M. Croatzier, G. Struhl, and A. Laughon for sharing reagents; P. Giresi, J. Simon, and S. Pott for technical assistance; B. Duronio, M. Peifer, G. Matera, and V. Bautch for providing the infrastructure for Drosophila work, and for comments on the manuscript. This work was supported in part by NIH grant F32GM090759. D.J.M and J.D.L designed the study. D.J.M. performed the experiments. D.J.M. and J.D.L. performed data analysis and wrote the paper.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
REFERENCES
- Aerts S, van Helden J, Sand O, Hassan BA. Fine-tuning enhancer models to predict transcriptional targets across multiple genomes. PLoS ONE. 2007;2:e1115. doi: 10.1371/journal.pone.0001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agelopoulos M, McKay DJ, Mann RS. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012;1:350–359. doi: 10.1016/j.celrep.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akam M. Hox genes: from master genes to micromanagers. Curr Biol. 1998;8:R676–R678. doi: 10.1016/s0960-9822(98)70433-6. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Stelzer C, Boryn LM, Rath M, Stark A. Genome-wide quantitative enhancer activity maps identified by STARR-seq. Science. 2013;339:1074–1077. doi: 10.1126/science.1232542. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Novitski E. The Genetics and biology of Drosophila. London: New York, Academic Press; 1976. [Google Scholar]
- Averof M, Cohen SM. Evolutionary origin of insect wings from ancestral gills. Nature. 1997;385:627–630. doi: 10.1038/385627a0. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman BP, Nibu Y, Pfeiffer BD, Tomancak P, Celniker SE, Levine M, Rubin GM, Eisen MB. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc Natl Acad Sci U S A. 2002;99:757–762. doi: 10.1073/pnas.231608898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin MD. Animal transcription networks as highly connected, quantitative continua. Dev Cell. 2011;21:611–626. doi: 10.1016/j.devcel.2011.09.008. [DOI] [PubMed] [Google Scholar]
- Bishop SA, Klein T, Arias AM, Couso JP. Composite signalling from Serrate and Delta establishes leg segments in Drosophila through Notch. Development. 1999;126:2993–3003. doi: 10.1242/dev.126.13.2993. [DOI] [PubMed] [Google Scholar]
- Bradley RK, Li XY, Trapnell C, Davidson S, Pachter L, Chu HC, Tonkin LA, Biggin MD, Eisen MB. Binding site turnover produces pervasive quantitative changes in transcription factor binding between closely related Drosophila species. PLoS Biol. 2010;8:e1000343. doi: 10.1371/journal.pbio.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budry L, Balsalobre A, Gauthier Y, Khetchoumian K, L'Honore A, Vallette S, Brue T, Figarella-Branger D, Meij B, Drouin J. The selector gene Pax7 dictates alternate pituitary cell fates through its pioneer action on chromatin remodeling. Genes Dev. 2012;26:2299–2310. doi: 10.1101/gad.200436.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Casares F, Mann RS. Control of antennal versus leg development in Drosophila. Nature. 1998;392:723–726. doi: 10.1038/33706. [DOI] [PubMed] [Google Scholar]
- Chan CS, Rastelli L, Pirrotta V. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 1994;13:2553–2564. doi: 10.1002/j.1460-2075.1994.tb06545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo SW, White R, Russell S. Genome-wide analysis of the binding of the Hox protein Ultrabithorax and the Hox cofactor Homothorax in Drosophila. PLoS ONE. 2011;6:e14778. doi: 10.1371/journal.pone.0014778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP, Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore MA, Mann RS. Hox control of organ size by regulation of morphogen production and mobility. Science. 2006;313:63–68. doi: 10.1126/science.1128650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crickmore MA, Mann RS. The control of size in animals: insights from selector genes. Bioessays. 2008;30:843–853. doi: 10.1002/bies.20806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Bray S, Garcia-Bellido A. Notch signalling regulates veinlet expression and establishes boundaries between veins and interveins in the Drosophila wing. Development. 1997;124:1919–1928. doi: 10.1242/dev.124.10.1919. [DOI] [PubMed] [Google Scholar]
- de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Dorschner MO, Hawrylycz M, Humbert R, Wallace JC, Shafer A, Kawamoto J, Mack J, Hall R, Goldy J, Sabo PJ, et al. High-throughput localization of functional elements by quantitative chromatin profiling. Nat Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- Engel MS, Grimaldi DA. New light shed on the oldest insect. Nature. 2004;427:627–630. doi: 10.1038/nature02291. [DOI] [PubMed] [Google Scholar]
- Estella C, Mann RS. Non-redundant selector and growth-promoting functions of two sister genes, buttonhead and Sp1, in Drosophila leg development. PLoS Genet. 2010;6:e1001001. doi: 10.1371/journal.pgen.1001001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estella C, McKay DJ, Mann RS. Molecular integration of wingless, decapentaplegic, and autoregulatory inputs into Distalless during Drosophila leg development. Dev Cell. 2008;14:86–96. doi: 10.1016/j.devcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri TH, Stevenson J, Chisholm AD, Mango SE. Dynamic chromatin organization during foregut development mediated by the organ selector gene PHA-4/FoxA. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher WW, Li JJ, Hammonds AS, Brown JB, Pfeiffer BD, Weiszmann R, MacArthur S, Thomas S, Stamatoyannopoulos JA, Eisen MB, et al. DNA regions bound at low occupancy by transcription factors do not drive patterned reporter gene expression in Drosophila. Proc Natl Acad Sci U S A. 2012;109:21330–21335. doi: 10.1073/pnas.1209589110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo SM, Gerrard DT, Miner D, Simich M, Des Soye B, Bergman CM, Halfon MS. REDfly v3.0: toward a comprehensive database of transcriptional regulatory elements in Drosophila. Nucleic Acids Res. 2011;39:D118–D123. doi: 10.1093/nar/gkq999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrouste R, Clement G, Nel P, Engel MS, Grandcolas P, D'Haese C, Lagebro L, Denayer J, Gueriau P, Lafaite P, et al. A complete insect from the Late Devonian period. Nature. 2012;488:82–85. doi: 10.1038/nature11281. [DOI] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Morata G, Guerrero I. The homeobox gene Distal-less induces ventral appendage development in Drosophila. Genes Dev. 1997;11:2259–2271. doi: 10.1101/gad.11.17.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi DA, Engel MS. Evolution of the insects. Cambridge U.K.: New York, Cambridge University Press; 2005. [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh BM, Carroll SB. Direct regulation of knot gene expression by Ultrabithorax and the evolution of cis-regulatory elements in Drosophila. Development. 2005;132:1567–1577. doi: 10.1242/dev.01737. [DOI] [PubMed] [Google Scholar]
- Hersh BM, Nelson CE, Stoll SJ, Norton JE, Albert TJ, Carroll SB. The UBX-regulated network in the haltere imaginal disc of D. melanogaster. Dev Biol. 2007;302:717–727. doi: 10.1016/j.ydbio.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono K, Margolis JS, Posakony JW, Doe CQ. Identification of hunchback cis-regulatory DNA conferring temporal expression in neuroblasts and neurons. Gene Expr Patterns. 2012;12:11–17. doi: 10.1016/j.gep.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Pearson JC, Crews ST. Diverse modes of Drosophila tracheal fusion cell transcriptional regulation. Mech Dev. 2010;127:265–280. doi: 10.1016/j.mod.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch EL, Ober KA. Hypothesis testing in evolutionary developmental biology: a case study from insect wings. J Hered. 2004;95:382–396. doi: 10.1093/jhered/esh064. [DOI] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jory A, Estella C, Giorgianni MW, Slattery M, Laverty TR, Rubin GM, Mann RS. A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of Drosophila melanogaster. Cell Rep. 2012;2:1014–1024. doi: 10.1016/j.celrep.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sebring A, Esch JJ, Kraus ME, Vorwerk K, Magee J, Carroll SB. Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature. 1996;382:133–138. doi: 10.1038/382133a0. [DOI] [PubMed] [Google Scholar]
- Klebes A, Biehs B, Cifuentes F, Kornberg TB. Expression profiling of Drosophila imaginal discs. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-8-research0038. RESEARCH0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Lickwar CR, Mueller F, Hanlon SE, McNally JG, Lieb JD. Genome-wide protein-DNA binding dynamics suggest a molecular clutch for transcription factor function. Nature. 2012;484:251–255. doi: 10.1038/nature10985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAlpine HK, Gordan R, Powell SK, Hartemink AJ, MacAlpine DM. Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res. 2010;20:201–211. doi: 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Carroll SB. Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev. 2002;12:592–600. doi: 10.1016/s0959-437x(02)00344-1. [DOI] [PubMed] [Google Scholar]
- Margolis JS, Borowsky ML, Steingrimsson E, Shim CW, Lengyel JA, Posakony JW. Posterior stripe expression of hunchback is driven from two promoters by a common enhancer element. Development. 1995;121:3067–3077. doi: 10.1242/dev.121.9.3067. [DOI] [PubMed] [Google Scholar]
- Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci U S A. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DJ, Estella C, Mann RS. The origins of the Drosophila leg revealed by the cis-regulatory architecture of the Distalless gene. Development. 2009;136:61–71. doi: 10.1242/dev.029975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabet S, Ebner A, Affolter M. The Drosophila Extradenticle and Homothorax selector proteins control branchless/FGF expression in mesodermal bridge-cells. EMBO Rep. 2005;6:762–768. doi: 10.1038/sj.embor.7400462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagne J, Groppe J, Guillemin K, Krasnow MA, Gehring WJ, Affolter M. The Drosophila Serum Response Factor gene is required for the formation of intervein tissue of the wing and is allelic to blistered. Development. 1996;122:2589–2597. doi: 10.1242/dev.122.9.2589. [DOI] [PubMed] [Google Scholar]
- Morata G, Lawrence PA. Development of the eye-antenna imaginal disc of Drosophila. Dev Biol. 1979;70:355–371. doi: 10.1016/0012-1606(79)90033-2. [DOI] [PubMed] [Google Scholar]
- Mou X, Duncan DM, Baehrecke EH, Duncan I. Control of target gene specificity during metamorphosis by the steroid response gene E93. Proc Natl Acad Sci U S A. 2012;109:2949–2954. doi: 10.1073/pnas.1117559109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Bienz M. Long range repression conferring boundaries of Ultrabithorax expression in the Drosophila embryo. EMBO J. 1991;10:3147–3155. doi: 10.1002/j.1460-2075.1991.tb04876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci U S A. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471:527–531. doi: 10.1038/nature09990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Papp B, Muller J. Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins. Genes Dev. 2006;20:2041–2054. doi: 10.1101/gad.388706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Turner FR, Muskavitch MA. Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech Dev. 1995;50:201–216. doi: 10.1016/0925-4773(94)00336-l. [DOI] [PubMed] [Google Scholar]
- Pavlopoulos A, Akam M. Hox gene Ultrabithorax regulates distinct sets of target genes at successive stages of Drosophila haltere morphogenesis. Proc Natl Acad Sci U S A. 2011;108:2855–2860. doi: 10.1073/pnas.1015077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MW, Boettiger AN, Levine M. Multiple enhancers ensure precision of gap gene-expression patterns in the Drosophila embryo. Proc Natl Acad Sci U S A. 2011;108:13570–13575. doi: 10.1073/pnas.1109873108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc Natl Acad Sci U S A. 2008;105:9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Ngo TT, Hibbard KL, Murphy C, Jenett A, Truman JW, Rubin GM. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V, Chan CS, McCabe D, Qian S. Distinct parasegmental and imaginal enhancers and the establishment of the expression pattern of the Ubx gene. Genetics. 1995;141:1439–1450. doi: 10.1093/genetics/141.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian S, Capovilla M, Pirrotta V. The bx region enhancer, a distant cis-control element of the Drosophila Ubx gene and its regulation by hunchback and other segmentation genes. EMBO J. 1991;10:1415–1425. doi: 10.1002/j.1460-2075.1991.tb07662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeiz M, Reeves NL, Posakony JW. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc Natl Acad Sci U S A. 2002;99:9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers ME, Shearn A. Patterns of protein synthesis in imaginal discs of Drosophila melanogaster. Cell. 1977;12:915–921. doi: 10.1016/0092-8674(77)90155-6. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–449. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Peifer M, Bender W, O'Connor M. Regulatory elements of the bithorax complex that control expression along the anterior-posterior axis. EMBO J. 1990;9:3945–3956. doi: 10.1002/j.1460-2075.1990.tb07615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JM, Giresi PG, Davis IJ, Lieb JD. Using formaldehyde-assisted isolation of regulatory elements (FAIRE) to isolate active regulatory DNA. Nat Protoc. 2012;7:256–267. doi: 10.1038/nprot.2011.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Ma L, Negre N, White KP, Mann RS. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS ONE. 2011a;6:e14686. doi: 10.1371/journal.pone.0014686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Riley T, Liu P, Abe N, Gomez-Alcala P, Dror I, Zhou T, Rohs R, Honig B, Bussemaker HJ, et al. Cofactor binding evokes latent differences in DNA binding specificity between Hox proteins. Cell. 2011b;147:1270–1282. doi: 10.1016/j.cell.2011.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Zhang Z, Grasfeder LL, Boyle AP, Giresi PG, Lee BK, Sheffield NC, Graf S, Huss M, Keefe D, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–1767. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergachis AB, Neph S, Reynolds A, Humbert R, Miller B, Paige SL, Vernot B, Cheng JB, Thurman RE, Sandstrom R, et al. Developmental fate and cellular maturity encoded in human regulatory DNA landscapes. Cell. 2013;154:888–903. doi: 10.1016/j.cell.2013.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher L, Collette NM, Murugesh D, Maxwell E, Ovcharenko I, Loots GG. Global gene expression analysis of murine limb development. PLoS ONE. 2011;6:e28358. doi: 10.1371/journal.pone.0028358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome Biol. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Henikoff S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat Biotechnol. 2000;18:424–428. doi: 10.1038/74487. [DOI] [PubMed] [Google Scholar]
- Vervoort M, Crozatier M, Valle D, Vincent A. The COE transcription factor Collier is a mediator of short-range Hedgehog-induced patterning of the Drosophila wing. Curr Biol. 1999;9:632–639. doi: 10.1016/s0960-9822(99)80285-1. [DOI] [PubMed] [Google Scholar]
- Visel A, Taher L, Girgis H, May D, Golonzhka O, Hoch RV, McKinsey GL, Pattabiraman K, Silberberg SN, Blow MJ, et al. A high-resolution enhancer atlas of the developing telencephalon. Cell. 2013;152:895–908. doi: 10.1016/j.cell.2012.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. London: Allen & Unwin; 1957. [Google Scholar]
- Weatherbee SD, Halder G, Kim J, Hudson A, Carroll S. Ultrabithorax regulates genes at several levels of the wing-patterning hierarchy to shape the development of the Drosophila haltere. Genes Dev. 1998;12:1474–1482. doi: 10.1101/gad.12.10.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Paddock SW, Vorwerk K, Carroll SB. Organization of wing formation and induction of a wing-patterning gene at the dorsal/ventral compartment boundary. Nature. 1994;368:299–305. doi: 10.1038/368299a0. [DOI] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Zhang CC, Muller J, Hoch M, Jackle H, Bienz M. Target sequences for hunchback in a control region conferring Ultrabithorax expression boundaries. Development. 1991;113:1171–1179. doi: 10.1242/dev.113.4.1171. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE. Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature. 2009;462:65–70. doi: 10.1038/nature08531. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.