The HIV-1 Rev protein mediates export of unspliced and singly spliced transcripts by recruiting the export factor CRM1. This study examines the timing of Rev and CRM1 recruitment to transcripts targeted for export. The analyses reveal that Rev and CRM1 bind to the RNA with equivalent kinetics at the site of transcription.
Keywords: CRM1, HIV-1 Rev, nuclear export
Abstract
The HIV-1 Rev protein mediates export of unspliced and singly spliced viral transcripts by binding to the Rev response element (RRE) and recruiting the cellular export factor CRM1. Here, we investigated the recruitment of Rev to the transcription sites of HIV-1 reporters that splice either post- or cotranscriptionally. In both cases, we observed that Rev localized to the transcription sites of the reporters and recruited CRM1. Rev and CRM1 remained at the reporter transcription sites when cells were treated with the splicing inhibitor Spliceostatin A (SSA), showing that the proteins associate with RNA prior to or during early spliceosome assembly. Fluorescence recovery after photobleaching (FRAP) revealed that Rev and CRM1 have similar kinetics as the HIV-1 RNA, indicating that Rev, CRM1, and RRE-containing RNAs are released from the site of transcription in one single export complex. These results suggest that cotranscriptional formation of a stable export complex serves as a means to ensure efficient export of unspliced viral RNAs.
INTRODUCTION
The genome of HIV-1 consists of two copies of a single stranded ∼10-kb RNA molecule. Upon infection, the genomic HIV-1 RNAs are reverse-transcribed into DNA, which is integrated into the genome of the host cells. Transcription then produces one pre-mRNA molecule from which multiple mRNAs are generated by alternative splicing (Stoltzfus and Madsen 2006). The HIV-1 proteins env, vpu, vif, and vpr are translated from singly spliced HIV-1 mRNA; Tat, nef, and rev derive from doubly spliced mRNA; and the proteins gag and gag-pol are translated from unspliced mRNA (Meyer and Malim 1994; Cullen 1998). On one hand, splicing is an essential process that also stimulates nucleocytoplasmic transport (Katahira et al. 1999; Valencia et al. 2008). On the other hand, pre-mRNAs that have initiated spliceosomal assembly are retained in the nucleus until splicing is completed (Galy et al. 2004). Although nuclear retention of incompletely spliced mRNA can be beneficial for protein-coding genes, it poses a problem to retroviruses as unspliced genomic RNAs must reach the cytoplasm for gag and gag-pol translation as well as for viral assembly. HIV-1 has developed mechanisms to bypass nuclear retention by using an alternative export pathway mediated by the CRM1 protein (Fornerod et al. 1997; Fukuda et al. 1997). To couple unspliced HIV-1 RNA to the CRM1-dependent nucleocytoplasmic transport pathway, HIV-1 encodes an adaptor protein termed Rev. Rev recognizes the Rev response element (RRE) present in the unspliced and singly spliced HIV-1 RNA, oligomerizes upon binding, and recruits CRM1 (Malim et al. 1989; Neville et al. 1997; Askjaer et al. 1998; Daugherty et al. 2010). CRM1 finally mediates the translocation of HIV-1 RNAs across the nuclear pores. Rev further inhibits splicing, making Rev the key player in regulating export of unspliced HIV-1 RNA (Kjems et al. 1991).
Here, we investigated the spatial and temporal binding of Rev and CRM1 to the transcription sites of HIV-1 RNAs. We found that Rev associates with pre-mRNA at the transcription sites and recruits the export factor CRM1, demonstrating that Rev and CRM1 bind cotranscriptionally. Inhibition of splicing by spliceostatin A (SSA) does not abolish recruitment of Rev and CRM1, demonstrating that Rev binds prior to spliceosome activation to bypass nuclear retention mechanisms. Finally, FRAP analysis suggested that Rev, CRM1, and HIV-1 RNAs are released into the nucleoplasm as a single, stable complex.
RESULTS
Rev recruits CRM1 to the transcription sites of reporters that splice post-transcriptionally
To address how unspliced HIV-1 RNAs assemble with Rev and CRM1 into an export-competent complex in vivo, we first tested whether Rev and CRM1 colocalize with HIV-1 transcription sites. We used the well-established Exo1 cell line that expresses an HIV-1 reporter (termed pExo-MS2 × 24) (Fig. 1A), which was shown to splice post-transcriptionally (Boireau et al. 2007). This reporter features most HIV-1 elements, including the 5′ and 3′ long terminal repeats (LTR), the packaging sequence (ψ), the major splice donor (SD1), the RRE, and the splice acceptor (SA7) (Fig. 1A; Boireau et al. 2007). For detection of the HIV-1 RNA transcript, the reporter further contained 24 MS2-binding sites in the last exon to enable visualization of the RNA by either fluorescence in situ hybridization (FISH) or MS2-mCherry coexpression (Bertrand et al. 1998). In Exo1 cells, this reporter gene is inserted as a tandem array into the chromatin of U2OS cells, and its transcription site is visible as a bright nucleoplasmic spot upon induction with the transcription transactivator Tat (Fig. 1B).
FIGURE 1.
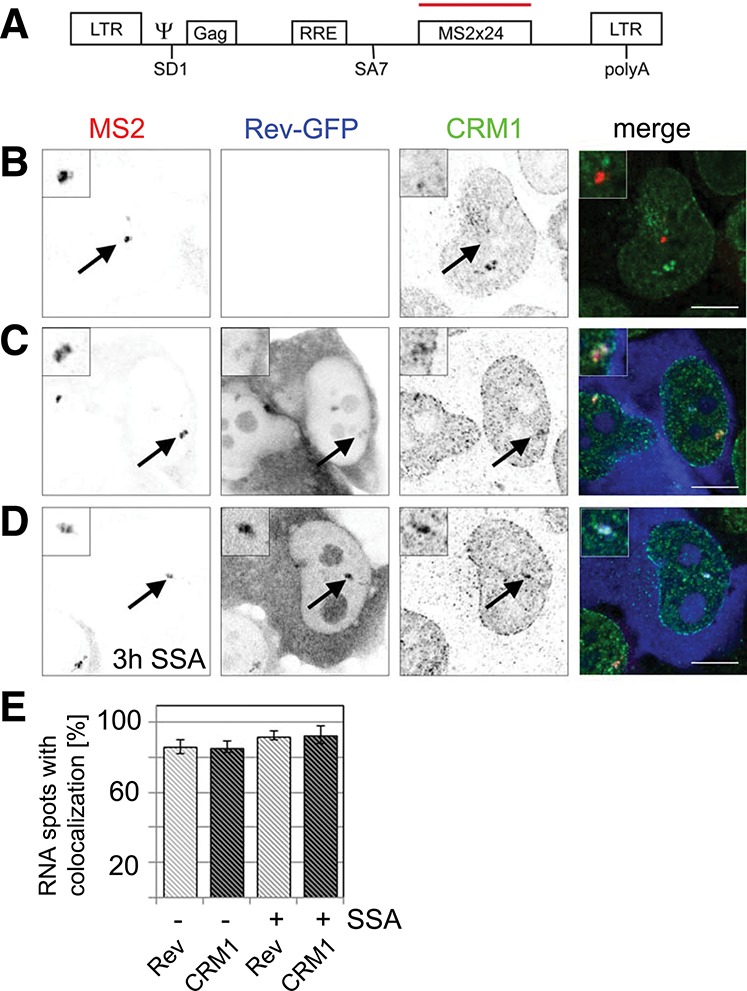
Rev and CRM1 accumulate at transcription sites of reporters that splice post-transcriptionally. (A) Scheme of the HIV-1 reporter. It contains the two HIV-1 LTRs, the packaging sequence Ψ, the splice donor SD1, the Rev-response element (RRE) within the intron, the splice acceptor SA7, and 24 MS2 repeats for detection in live and fixed cells. (B) Exo1 cells were transfected with Tat. HIV-1 RNAs were visualized by fluorescence in situ hybridization against the MS2 repeats and CRM1 by immunofluorescence. The overlay shows the RNA in red and CRM1 in green. (C) Exo1 cells were cotransfected with Tat and Rev-GFP. The RNA, Rev, and CRM1 were detected as in B. In the overlay, the RNA is depicted in red, CRM1 in green, and Rev-GFP in blue. (D) Same as C but cells were treated with spliceostatin A (SSA) for 3 h prior to fixation and immunolabeling. (E) Bar chart showing the percentage of transcription sites that colocalize with Rev-GFP (light gray) and CRM1 (dark gray) in absence and presence of SSA. Forty to 50 cells were counted in each condition. (Scale bar) 5 µm. Arrows indicate the position of the transcription sites.
To address the spatial binding of CRM1 to HIV-1 RNA, Exo1 cells were first transfected with Tat, and the HIV-1 RNAs were detected by FISH against the MS2 repeats; whereas endogenous CRM1 protein was labeled by immunofluorescence. CRM1 distributed in a typical punctuate pattern throughout the nucleoplasm and delineated the nuclear periphery but did not accumulate at the transcription site (Fig. 1B). Exo1 cells were then cotransfected with Tat and Rev-GFP. The latter localized to the cytoplasm and nucleoli but also accumulated to the HIV-1 transcription site (Fig. 1C). In the presence of Rev-GFP, CRM1 also localized to the HIV-1 transcription site, demonstrating that Rev recruits CRM1 to nascent RNAs (Fig. 1C). This recruitment was specific because we could not see recruitment of Rev by reporters lacking the RRE (see below). We counted the number of transcription sites that colocalized with Rev-GFP and CRM1, and found that for both proteins, 85% of the spots colocalized (Fig. 1E).
We next sought to address the role of the spliceosome assembly process in the recruitment of Rev and CRM1, and we repeated this experiment in the presence of SSA (Kaida et al. 2007). This compound inhibits spliceosome activation by stalling the spliceosome at an A-like complex before cleavage of the 5′ splice site. Competitive RT-PCR experiments confirmed that splicing of the HIV-1 construct was inhibited by SSA (Supplemental Fig. 1). Treatment of cells with SSA did not abolish the localization of Rev-GFP to the transcription site, nor that of CRM1, confirming that they were recruited at an early step of spliceosome assembly (Fig. 1D). Instead, splicing inhibition led to a slight increase in the number of transcription sites that colocalize with Rev-GFP and CRM1 to >90% (Fig. 1E).
Rev recruits CRM1 to cotranscriptionally spliced reporter RNAs
Next, we wanted to test whether cotranscriptional splicing prevented recruitment of Rev and CRM1 to RRE-containing RNAs. To this end, we cloned the RRE into the intron of chimeric HIV-1/MINX reporters (MINX_RREin) (Fig. 2A). The MINX reporter, which derives from the major late transcript of adenovirus-2, contains strong splicing signals and splices entirely cotranscriptionally (Zillmann et al. 1988; Schmidt et al. 2011). The MINX reporter was put under the control of the HIV-1 LTR and placed upstream of the LacZ gene, whereas the 3′ end processing was controlled by the 3′ polyA signal of the bovine growth hormone (bGH). As a control, we used the HIV-1/MINX reporter that did not contain an RRE, termed MINX_ΔRRE. We were unable to establish stable clones expressing MINX_RREin, and U2OS cells were thus transiently transfected with the MINX reporters, Tat and Rev-GFP (Fig. 2B). The resulting RNAs were detected by FISH using probes hybridizing to the last exon (LacZ sequence), and CRM1 was labeled by immunofluorescence. In cells expressing MINX_ΔRRE, the RNA FISH revealed several distinct dots within the nucleoplasm and a strong cytoplasmic staining. Rev and CRM1 did not colocalize with the nuclear MINX RNA, in agreement with the lack of Rev binding sites within this RNA (Fig. 2B). In contrast, the MINX_RREin reporter recruited both Rev-GFP and CRM1 into distinct dots in the nucleus (Fig. 2B). Counting these dots revealed that ∼75% and 50% colocalized with Rev-GFP and CRM1, respectively (Fig. 2D). We next tested whether inhibition of splicing with SSA would affect the recruitment of Rev and CRM1 to the MINX_RREin RNAs. Upon treatment with SSA, the MINX_ΔRRE RNA accumulated in nuclear speckle-like clusters, whereas the cytoplasmic signal disappeared, consistent with the existence of a nuclear retention mechanisms preventing export of unspliced pre-mRNAs while the cytoplasmic spliced RNAs were turned over (Fig. 2C). In contrast, MINX_RREin RNAs were still detected in the cytoplasm after SSA treatment. This suggests that Rev mediates nuclear export of unspliced mRNAs associated with stalled spliceosomes, or that these RNAs were exported before the spliceosome assembled on them (Fig. 2C). Consistent with the lack of nuclear retention, the MINX_RREin RNAs did also not accumulate in large speckles when splicing was inhibited in presence of Rev. Furthermore, the fraction of nuclear RNA dots that colocalized with Rev-GFP and CRM1 increased to >90% (Fig. 2D). This is consistent with the fact that the intronic RRE was no longer spliced out and could thus recruit additional molecules of Rev-GFP and CRM1.
FIGURE 2.
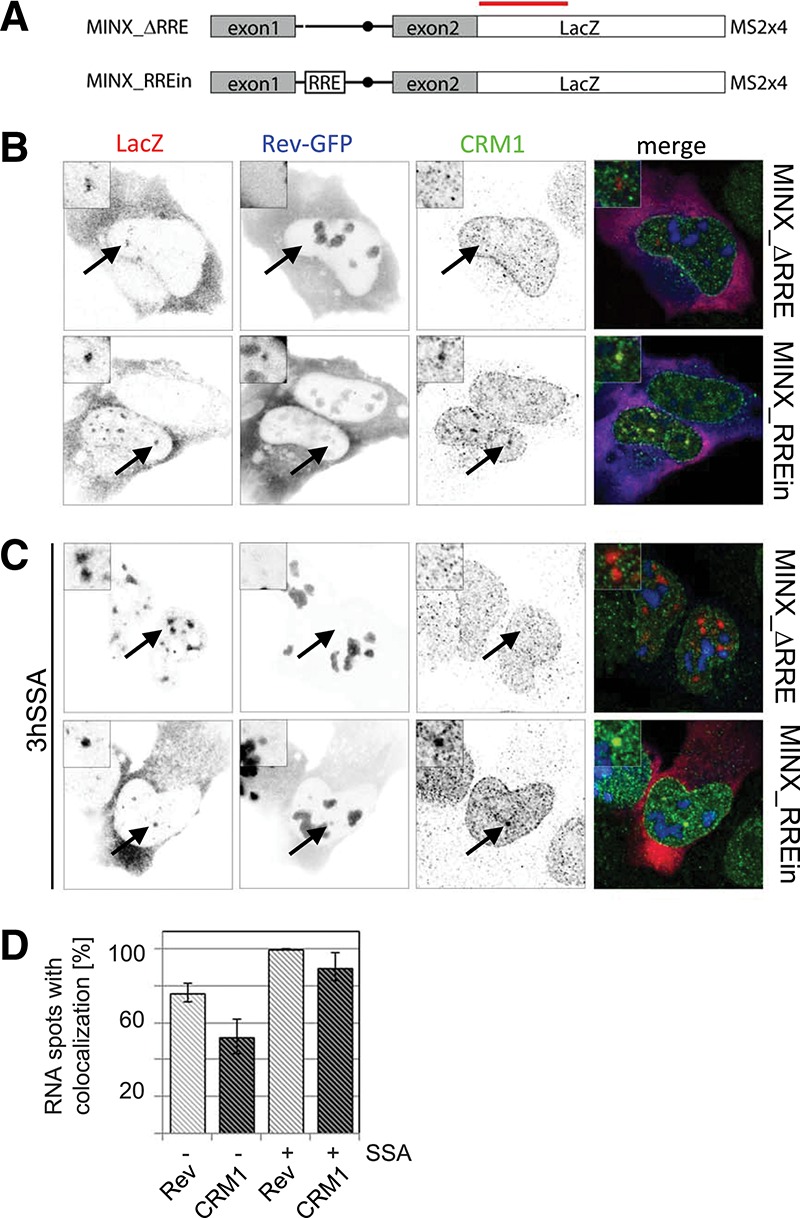
Colocalization of Rev, CRM1, and cotranscriptionally spliced HIV-1 reporter RNAs. (A) Scheme of the chimeric HIV-1/MINX reporters. Transcription was driven by the HIV-1 LTR (data not shown), and the RNAs contained the MINX intron followed by the LacZ sequence. MINX_RREin contains the RRE within the intron. The red bar indicates the position of the LacZ probes used for FISH. (B) Cells were transfected with Tat, Rev-GFP, and the MINX reporters as indicated. The RNA was visualized by FISH, Rev by GFP-fluorescence, and CRM1 by immunofluorescence. In the overlay, the RNA is shown in red, Rev in blue, and CRM1 in green. (C) Same as in B except that the cells were treated with SSA for 3 h prior to fixation and immunolabeling. (D) Bar chart showing the percentage of MINX_RREin RNA dots that colocalize with Rev-GFP (light gray) and CRM1 (dark gray) in the absence and presence of SSA. Thirty to 40 cells were counted in each condition. Arrows indicate the position of the transcription sites.
Mobility of Rev and CRM1 in the nucleus and at the HIV-1 transcription site
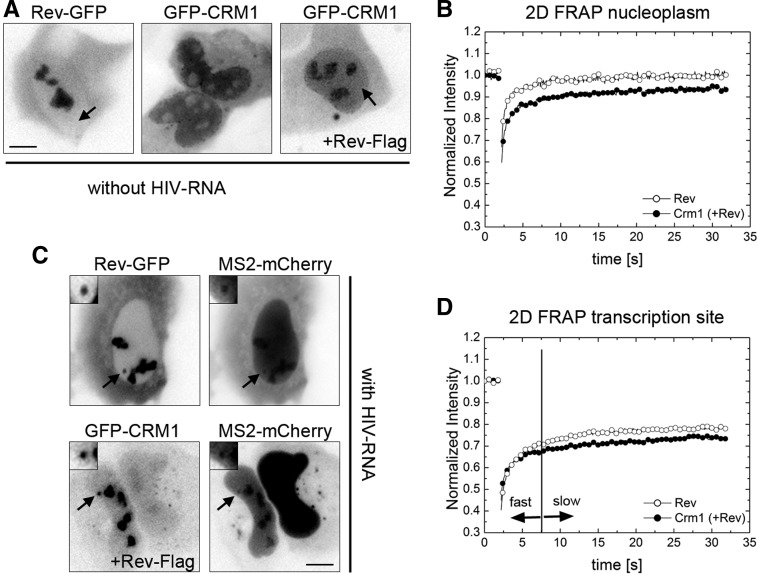
We next investigated the mobility of the proteins by fluorescence recovery after photobleaching (FRAP). First, we investigated the mobility of Rev-GFP and GFP-CRM1 in the nucleoplasm and therefore used cells that did not express any RRE-containing RNAs. In agreement with a previous study (Daelemans et al. 2005), CRM1 redistributed to the nucleoli when Rev-Flag was coexpressed (Fig. 3A, cf. middle and right panels; Supplemental Fig. 2). This allowed us to identify cells transfected with both plasmids by inspection of the GFP-CRM1 localization pattern. We then performed FRAP in arbitrary regions of the nucleoplasm and recorded the recovery for 30 sec after the bleach (fast 2D FRAP approach; see Materials and Methods). The recovery of Rev-GFP was fast, showing full recovery after 10 sec (Fig. 3B), and thus suggested that Rev diffuses rapidly across the nucleoplasm. FRAP of GFP-CRM1 in presence of coexpressed Rev-Flag was also fast; however, the curve recovered only to 85% of its prebleach value, indicating that an immobile fraction of ∼15% remained after the observation period of 30 sec (Fig. 3B, data not shown). This showed that CRM1 also diffuses rapidly, but a small fraction appears to be stably bound to some slowly moving nuclear component.
FIGURE 3.
Fast 2D FRAP of Rev and CRM1 in the nucleoplasm and at the HIV-1 transcription site. (A) U2OS cells were transfected with Rev-GFP alone (left), GFP-CRM1 alone (middle), or with both GFP-CRM1 and Rev-Flag (right). The images show GFP-fluorescence in live cells. The arrows indicate the nucleoplasm. (B) 2D FRAP plots of Rev-GFP (open circles) and GFP-CRM1 (cotransfected with Rev-Flag; closed circles) in the nucleoplasm of U2OS cells. (C) Exo1 cells were transfected with either Tat, Rev-GFP, and MS2-mCherry, or with Tat, Rev-Flag, GFP-CRM1, and MS2-mCherry. The micrographs show the GFP and Cherry fluorescence in live cells. Arrows indicate the position of the transcription sites. (D) 2D FRAP plots Rev-GFP (open circles) and GFP-CRM1 (closed circles) at the transcription site of Exo1 cells. The vertical line indicates the transition point between the fast and slow fractions. (Scale bar) 5 µm.
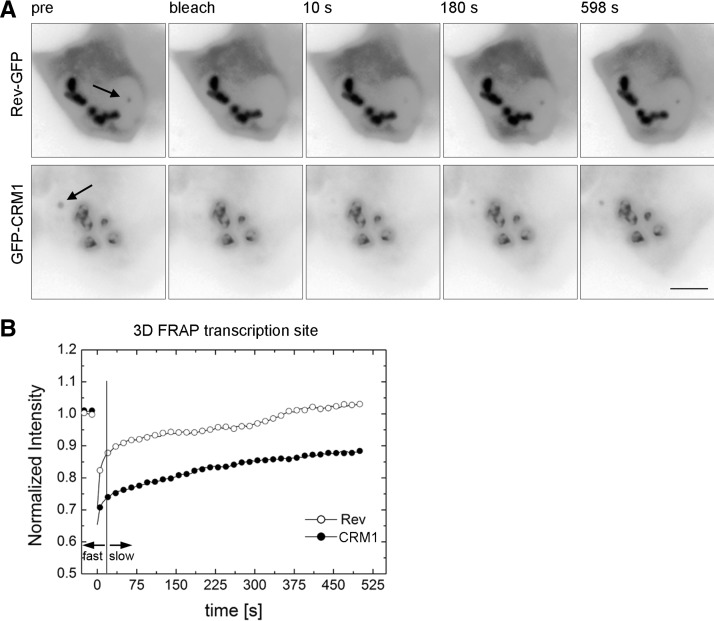
Next, we wanted to investigate the mobility of GFP-CRM1 and Rev-GFP at the HIV-1 transcription sites. To this end, we cotransfected Exo1 cells with Rev-GFP, Tat, and MS2-mCherry, or Rev-Flag, GFP-CRM1, Tat, and MS2-mCherry (Fig. 3C). The FRAP of both Rev-GFP and GFP-CRM1 showed a fast initial recovery during the first seconds after the bleach, which was comparable to the nucleoplasmic FRAPs (Fig. 3D). However, both curves contained a larger immobile fraction when compared to the nucleoplasmic FRAPs (35% and 45% for Rev and CRM1, respectively) (cf. Fig. 3B,D). This fraction suggests that a large percentage of these proteins is stably bound at the transcription site. However, these FRAP experiments were performed for a relatively short observation period of 30 sec, and thus the recovery on longer time scales could not be determined. We therefore used a 3D FRAP approach (see Materials and Methods), which allows to record the recovery for 500 sec (Fig. 4). Again, the FRAP of both proteins showed two distinct components: a fast initial component of a few seconds, followed by a slower one (Fig. 4B). The fast component of Rev-GFP was larger than the one of GFP-CRM1, but did not last longer than 15 sec. Interestingly, Rev-GFP showed a complete recovery over the post-bleach period of 500 sec (Fig. 4B). Likewise, most of GFP-CRM1 also recovered during this period but showed a residual immobile fraction of 30%. In summary, our FRAP data suggests that Rev and CRM1 move rapidly in the nucleus as their fluorescence signal recovers in a few seconds. At the transcription site, an additional slow fraction is observed, which lasts for several minutes and indicates a stable interaction with slowly moving objects, most likely the nascent RNAs.
FIGURE 4.
3D FRAP of Rev-GFP and GFP-CRM1 at the transcription site. (A) Exo1 cells were transfected with either Tat and Rev-GFP (upper), or with Tat, GFP-CRM1, and Rev-Flag (lower). FRAP was performed with the 3D FRAP method. Micrographs show selected time points: prebleach and 10 sec, 180 sec, and 598 sec post-bleach. Arrows indicate the position of the transcription sites. (B) Plot of the recovery curves of Rev-GFP and GFP-CRM1 at the transcription site using the 3D FRAP method. The vertical line indicates the transition point between the fast and slow fractions. (Open circles) Rev-GFP; (closed circles) GFP-CRM1; (scale bar) 5 µm.
Functional interpretation of FRAP curves at the HIV-1 transcription sites
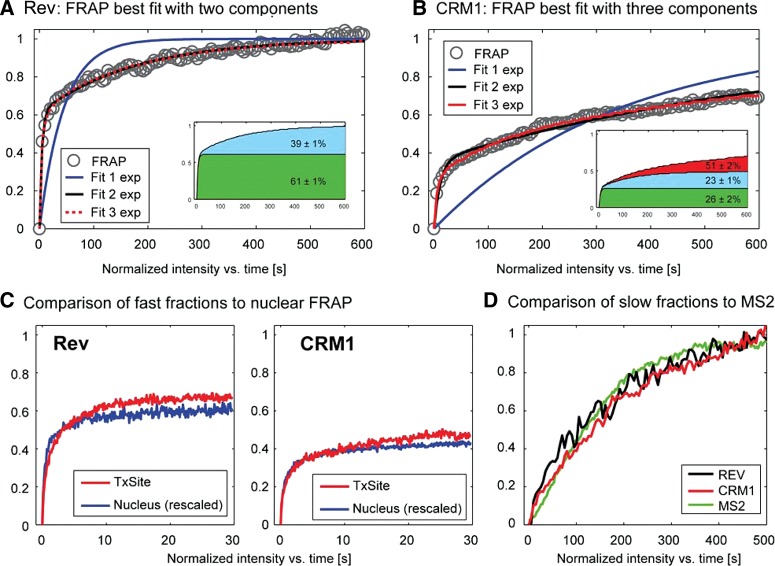
Because of the lack of validated models for the case of a localized array of newly synthesized binding sites (Mueller et al. 2010), we did not model the FRAP data to infer binding rates. Instead, we turned to a simpler analysis and fitted the FRAP with a sum of exponentials. Although the extracted rate constants bear no direct biological meaning in terms of binding rates, this analysis provided us with an estimate of the expected number of functional components in the FRAP curves. We found that neither curve could be described by a single exponential (Fig. 5). Fitting with two exponentials yielded a good description for the FRAP curve of Rev (Fig. 5A), whereas the CRM1 FRAP was best described with three exponentials (Fig. 5B). Accordingly, we assigned a fast and slow component for Rev at the HIV-1 transcription sites; but for CRM1, a fast, a slow and a very slow component were predicted (Fig. 5A,B, insets; Supplemental Table 1). The very slow component was similar to the one observed in the nucleoplasm, suggesting that these were most likely of identical origin and not related to the Rev-dependent recruitment of CRM1 on the HIV-1 RNAs.
FIGURE 5.
Analysis of Rev-GFP and GFP-CRM1 3D FRAPs at the transcription site. (A,B) The FRAP data of Rev-GFP and GFP-CRM1 were fitted with a monoexponential, a double, or a three exponential function. Insets show the sizes of the slow, medium, and fast fraction. (C) Comparison of the nucleoplasmic FRAP of Rev and CRM1 with the fast component of Rev-GFP and GFP-CRM1 recoveries at the transcription site. The FRAP data were rescaled for better comparison. (D) Plot of the recoveries of the slow component of Rev-GFP (black), CRM1-GFP (red), and MS2-GFP (green) at the HIV-1 transcription sites. The curves were rescaled from 0 to 1 for better comparison.
We then compared the fast components of the transcription site with the recoveries obtained in the nucleoplasm (Fig. 5C). Nucleoplasmic FRAP curves were renormalized and overlaid with the fast component of the transcription site (Fig. 4C). For both Rev and CRM1, we found good agreement between these curves, arguing that the fast component of the transcription sites reflects the mobility observed for the nucleoplasmic FRAP, e.g., the freely mobile molecules. We then compared the slow components of Rev and CRM1 to the mobility of the HIV-1 RNAs as measured by FRAP of MS2-GFP (Boireau et al. 2007). Previous work showed that this is largely determined by the de novo synthesis of MS2 stem–loops and represents a direct measure of the HIV-1 transcriptional activity (Boireau et al. 2007). For better comparison, we excluded the fast components by renormalizing the Rev and CRM1 FRAP curves to zero after 20 sec and to 1 at the end of the recovery (Fig. 5D). The curves showed good agreement with each other and with the MS2-GFP FRAP, suggesting that Rev and CRM1 rapidly bind HIV-RNAs during their synthesis and leave the transcription site as a single, RNA-bound complex.
DISCUSSION
HIV-1 viral progression relies on the export of unspliced RNA to the cytoplasm, where it becomes packed into viral particles and eventually leaves the cell. HIV-1 has developed mechanisms to secure export of its unspliced RNA despite the fact that cellular pre-mRNAs, undergoing splicing, are generally retained in the nucleus. It is well established that the HIV-1 Rev protein recognizes the RRE localized in intron-containing viral RNAs and recruits the cellular export factor CRM1 to mediate translocation across the pore. Nucleocytoplasmic export of unspliced HIV-1 RNA has been the subject of many studies that addressed the biochemical or structural characteristics of the HIV-1 RNA/Rev/CRM1 export complex (Yi et al. 2002; Yedavalli et al. 2004; Daugherty et al. 2010; Sherer et al. 2011). These studies provide important insights into the mechanisms of nucleocytoplasmic transport, but the spatiotemporal relationship between Rev, CRM1, and unspliced HIV-1 RNA remains unknown. Likewise, the stability of viral and cellular export complexes is not known, despite the fact that this is an important parameter for HIV-1 as the formation of a stable complex is important to ensure efficient export and to prevent splicing.
Here, we investigated the recruitment of Rev and CRM1 to the transcription sites of HIV-1 reporters that differ in their splicing pattern. We first used the Exo1 cell lines expressing a HIV-1 pre-mRNA that contains the HIV splice donor (SD1) and the A7 splice acceptor sites and that splices post-transcriptionally (Boireau et al. 2007). We showed by FISH and immunofluorescence that Rev and CRM1 are both recruited early to the RNA transcription sites. It was previously reported that Rev function is abolished in the context of strong 3′ splice sites, probably because Rev binding and splicing are then in kinetic competition (Chang and Sharp 1989; Hammarskjöld et al. 1994; Kammler et al. 2006). Post-transcriptional splicing thus allows binding of Rev and CRM1 to the unspliced RNA during transcription and helps to prevent a kinetic competition between splicing and export complex formation. Nevertheless, we found that Rev and CRM1 are recruited to more than half the transcription site of a reporter that splices cotranscriptionally, despite the fact that this RNA is normally spliced very efficiently. When splicing was inhibited by SSA, almost all RNA spots contained Rev and CRM1. This increase is consistent with a model in which binding of Rev and CRM1 would be in a kinetic competition with splicing. In agreement with this interpretation, Rev and tri-snRNP binding to unspliced HIV-1 RNAs have been proposed to be mutually exclusive (Kjems and Sharp 1993), and SSA stalls spliceosome assembly at an A-like stage, before productive incorporation of the U5/U4:U6 tri-snRNP (Schmidt et al. 2009; Roybal and Jurica 2010). In addition, SSA-stalled pre-mRNAs resemble the complexes assembled on HIV-1 pre-mRNAs in nuclear extract (Dyhr-Mikkelsen and Kjems 1995). Thus, we conclude that rapid binding of Rev during transcription and cotranscriptional assembly of an export-competent complex acts to prevent premature splicing, as it has been observed in vitro (Kjems et al. 1991), and enhances export of unspliced RNAs.
The colocalization experiments showed that Rev and CRM1 both localize to the transcription sites of HIV-1 RNA but they are not informative regarding the affinity and dynamic binding of the proteins to the RNA. Furthermore, little or no information is currently available regarding the stability of export complexes in the cell nucleus. This is important in the context of viral export, as continuous dissociation and reassociation of Rev to the viral RNA would allow spliceosome assembly and splicing of the RNA. We used FRAP to measure the mobility of Rev and CRM1 in the nucleus and at the viral sites of transcription. Fitting the data showed that two exponentials correctly described the recovery of Rev, whereas that of CRM1 required three exponentials. Comparison with the mobility of the proteins in the nucleoplasm indicated that the fast component corresponded to the free protein, whereas the slow component represented the RNA-bound form. Moreover, the slow component of Rev and CRM1 nicely overlapped with each other and with the recovery of the RNA itself. It was previously shown that recovery of the RNA corresponds to the time of its synthesis, processing, and release (Boireau et al. 2007). This therefore suggests that Rev and CRM1 bind RNA during its synthesis and then leave the transcription site as a complex with the RNA.
Taken together, our data are consistent with a model in which Rev and CRM1 bind cotranscriptionally to the viral RNA in competition with splicing to form a stable complex that can rapidly be released into the nucleoplasm to allow for efficient export.
MATERIALS AND METHODS
Cells and plasmids
Plasmids encoding Tat, Rev-GFP, and GFP-CRM1 were previously described (Boireau et al. 2007; Molle et al. 2007). To generate MINX_RREin construct, the RRE was excised from pBRRE-350 (Askjaer et al. 1998) and inserted into the intron of HIV-1/MINX MS2 × 4 (Schmidt et al. 2011) using the BstEII restriction site. Flag-Rev was constructed by Gateway cloning. To generate stable cells expressing MINX_RREin, U2OS cells were cotransfected with the MINX_RREin plasmid together with a plasmid encoding for a hygromycin resistence, treated with hygromycin for several weeks, and stable clones were selected. U2OS cells stably expressing the HIV-1 reporter pExo-MS2 × 24 (Exo1 cells) and the MINX_ΔRRE (termed MINX_MS2ex2) were described previously (Boireau et al. 2007; Schmidt et al. 2011). All cells were cultured in DMEM containing 10% FBS, 1% penicillin/streptomycin, and 132 µg/mL hygromycin.
Fluorescence in situ hybridization (FISH)
Cells were grown on coverslips and transfected with JetPrime or Lipofectamine 2000 reagent. The following day, cells were fixed in 4% paraformaldehyde/2% sucrose/PBS for 20 min at room temperature, washed twice in PBS, and permeabilized in 70% ethanol overnight at 4°C. To inhibit splicing cells were treated with SSA (100 ng/mL) for 3 h before fixation.
For FISH, cells were first rehydrated in 2× SSC and washed in 2× SSC/formamide (Sigma), then incubated overnight at 37°C in hybridization mix (1× SSC, 10% dextran sulphate, 0.02% RNA grade BSA, 2 mM Vanadyl-R-C, 40 µg tRNA, and MS2-Cy3 probe or a mix of 14 LacZ probes). MS2 was hybridized in 20% formamide, and LacZ was hybridized in 40% formamide. The oligonucleotide probes were briefly denatured in formamide at 95°C and then added to the hybridization mix. After overnight incubation, cells were washed twice for 30 min at 37°C in formamide/2× SCC and afterward in 1× SCC for 30 min. Finally, the coverslips were mounted in VectaShield. Oligonucleotides were labeled with Cy3 monoreactive dyes (GE Healthcare); the sequences were described previously (Boireau et al. 2007; Schmidt et al. 2011).
Immunofluorescence
Cells were fixed and permeabilized in 0.1% Triton-X100 (Sigma)/PBS or in 70% ethanol overnight and blocked in 2% BSA/0.01% Triton/PBS for 5 min at room temperature. Antibodies were diluted in 2% BSA/0.01% Triton/PBS, and cells were incubated with the antibody solution for 60 min at room temperature. Cells were washed three times in 0.01% Triton/PBS for 10 min and then incubated for 30 min with α-rabbit Alexa 647 antibody. After incubation, cells were washed three times in 0.01% Triton/PBS and mounted using VectaShield. Antibody against CRM1 (Santa Cruz) was diluted 1:50; and antibody against Flag (M2; Sigma) was diluted 1:500.
Microscopy
Fluorescent images of fixed cells were captured on a LSM 510 META confocal microscope (63×, NA 1.4, Zeiss). Each color was acquired sequentially. DAPI was excited at 405 nm, GFP at 488 nm, and Cy3 and Alexa 647 at 541 nm and 633 nm, respectively. Detection windows were 420–470 nm (DAPI), 500–550 nm (GFP), 570–640 nm (Cy3), and LP 640 nm (Alexa 647).
For counting the number of Rev and CRM1 colocalized RNA spots, we acquired confocal images in all three channels (Rev-GFP, RNA-Cy3, and CRM1-Alexa647) and inspected colocalization at the transcription site by a line scan. Colocalization was judged positive when the intensity distribution of Rev-GFP and CRM1, respectively, increased concomitantly with the intensity distribution of the RNA spots.
Image processing
All images were carefully gray-level corrected and prepared with ImageJ and Adobe Photoshop. The MS2-mCherry signal in Figure 3C (upper) was further γ-corrected to better visualize the transcription site.
FRAP experiments
FRAP was performed on a Nikon TE200 microscope with a 100×, NA 1.45 objective as described previously (Boireau et al. 2007). For the fast 2D-FRAP approach, 20 prebleach frames were acquired at an exposure time of 100 msec, then a region of interest (ROI) 1.6 μm in diameter was bleached for 250 msec with high laser intensity pulse, and 300 post-bleach frames were collected for another 30 sec. To obtain slow recoveries at the transcription site, we used a 3D FRAP protocol similar to the one described in Boireau et al. (2007) with a bleach spot size of 1.6 µm. Z-stacks were acquired every 5 sec, and the transcription site was bleached with a high intensity laser beam. Recovery was recorded for 500 sec with a lag time of 5 sec between stacks.
The immobilized fractions (IF) were calculated according to the following:
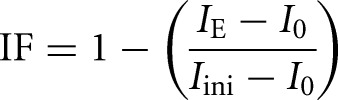 |
where IE denotes the end value of the recovered fluorescence intensity; I0 is the first post-bleach fluorescence intensity; and Iini is the initial (prebleach) fluorescence intensity.
FRAP curves were fit with a sum of exponentials according to the following equation:
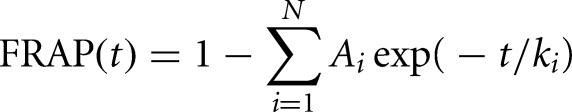 |
(1) |
where N denotes the number of exponentials; ki are their corresponding rate constants; and the Ai are their relative fractions.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
The help of Maria Carmo-Fonseca, José Rino, and the members of the Bertrand and Kjems teams are gratefully acknowledged. We thank the Montpellier RIO Imaging (MRI) facility for the microscopy support and advice. This work was supported by the Danish Research Council for Independent Research, SIDACTION and ANRS. I.N. was supported by Aarhus University and an EMBO fellowship. FRM supported F.M. and Sidaction supported U.S.
REFERENCES
- Askjaer P, Jensen TH, Nilsson J, Englmeier L, Kjems J 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J Biol Chem 273: 33414–33422 [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM 1998. Localization of ASH1 mRNA particles in living yeast. Mol Cell 2: 437–445 [DOI] [PubMed] [Google Scholar]
- Boireau S, Maiuri P, Basyuk E, de la Mata M, Knezevich A, Pradet-Balade B, Bäcker V, Kornblihtt A, Marcello A, Bertrand E 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J Cell Biol 179: 291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Sharp PA 1989. Regulation by HIV Rev depends upon recognition of splice sites. Cell 59: 789–795 [DOI] [PubMed] [Google Scholar]
- Cullen BR 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249: 203–210 [DOI] [PubMed] [Google Scholar]
- Daelemans D, Costes SV, Lockett S, Pavlakis GN 2005. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol 25: 728–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, Frankel AD 2010. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat Struct Mol Biol 17: 1337–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhr-Mikkelsen H, Kjems J 1995. Inefficient spliceosome assembly and abnormal branch site selection in splicing of an HIV-1 transcript in vitro. J Biol Chem 270: 24060–24066 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311 [DOI] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Hammarskjöld ML, Li H, Rekosh D, Prasad S 1994. Human immunodeficiency virus env expression becomes Rev-independent if the env region is not defined as an intron. J Virol 68: 951–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3: 576–583 [DOI] [PubMed] [Google Scholar]
- Kammler S, Otte M, Hauber I, Kjems J, Hauber J, Schaal H 2006. The strength of the HIV-1 3′ splice sites affects Rev function. Retrovirology 3: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J, Sträßer K, Podtelejnikov A, Mann M, Jung JU, Hurt E 1999. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J 18: 2593–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J, Sharp PA 1993. The basic domain of Rev from human immunodeficiency virus type 1 specifically blocks the entry of U4/U6·U5 small nuclear ribonucleoprotein in spliceosome assembly. J Virol 67: 4769–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J, Frankel AD, Sharp PA 1991. Specific regulation of mRNA splicing in vitro by a peptide from HIV-1 Rev. Cell 67: 169–178 [DOI] [PubMed] [Google Scholar]
- Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338: 254–257 [DOI] [PubMed] [Google Scholar]
- Meyer BE, Malim MH 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev 8: 1538–1547 [DOI] [PubMed] [Google Scholar]
- Molle D, Maiuri P, Boireau S, Bertrand E, Knezevich A, Marcello A, Basyuk E 2007. A real-time view of the TAR:Tat:P-TEFb complex at HIV-1 transcription sites. Retrovirology 4: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F, Mazza D, Stasevich TJ, McNally JG 2010. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: What do we really know? Curr Opin Cell Biol 22: 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M 1997. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol 7: 767–775 [DOI] [PubMed] [Google Scholar]
- Roybal GA, Jurica MS 2010. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res 38: 6664–6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Im KB, Benzing C, Janjetovic S, Rippe K, Lichter P, Wachsmuth M 2009. Assembly and mobility of exon–exon junction complexes in living cells. RNA 15: 862–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt U, Basyuk E, Robert MC, Yoshida M, Villemin JP, Auboeuf D, Aitken S, Bertrand E 2011. Real-time imaging of cotranscriptional splicing reveals a kinetic model that reduces noise: Implications for alternative splicing regulation. J Cell Biol 193: 819–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherer NM, Swanson CM, Hué S, Roberts RG, Bergeron JR, Malim MH 2011. Evolution of a species-specific determinant within human CRM1 that regulates the post-transcriptional phases of HIV-1 replication. PLoS Pathog 7: e1002395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzfus CM, Madsen JM 2006. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res 4: 43–55 [DOI] [PubMed] [Google Scholar]
- Valencia P, Dias AP, Reed R 2008. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci 105: 3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392 [DOI] [PubMed] [Google Scholar]
- Yi R, Bogerd HP, Cullen BR 2002. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J Virol 76: 2036–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M, Zapp ML, Berget SM 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol Cell Biol 8: 814–821 [DOI] [PMC free article] [PubMed] [Google Scholar]