It has been known that mRNA 3′-end formation in yeast proceeds through either an endonucleolytic or an exonucleolytic pathway, but the identity of the endonuclease has not been established. This paper demonstrates that tRNase Z is the endonuclease and shows that the enzyme has roles in both the nucleus and the mitochondrian.
Keywords: tRNA, tRNase Z, 3′-end processing, exonucleases, Saccharomyces cerevisiae
Abstract
Mature tRNA 3′ ends in the yeast Saccharomyces cerevisiae are generated by two pathways: endonucleolytic and exonucleolytic. Although two exonucleases, Rex1 and Rrp6, have been shown to be responsible for the exonucleolytic trimming, the identity of the endonuclease has been inferred from other systems but not confirmed in vivo. Here, we show that the yeast tRNA 3′ endonuclease tRNase Z, Trz1, is catalyzing endonucleolytic tRNA 3′ processing. The majority of analyzed tRNAs utilize both pathways, with a preference for the endonucleolytic one. However, 3′-end processing of precursors with long 3′ trailers depends to a greater extent on Trz1. In addition to its function in the nucleus, Trz1 processes the 3′ ends of mitochondrial tRNAs, contributing to the general RNA metabolism in this organelle.
INTRODUCTION
Mature tRNAs are generated from larger precursors by post-transcriptional processing that involves removing the 5′ and 3′ extensions, addition of CCAOH to the 3′ terminus, and modification of several nucleosides. In most organisms, the tRNA 5′ leader is cleaved off by the ubiquitous endonuclease RNase P, whereas the maturation of 3′ ends is more complex and proceeds by two pathways, endo- and exonucleolytic (for review, see Mörl and Marchfelder 2001; Evans et al. 2006; Späth et al. 2007; Hartmann et al. 2009). In bacteria, a predominant multistep reaction involves a downstream endonucleolytic cleavage by RNase E, followed by exonucleolytic digestion by several partially redundant nucleases, mainly RNases T and PH but also RNases II, D, and PNPase (Li and Deutscher 1996, 2002; Ow and Kushner 2002; Wen et al. 2005). However, CCA-less precursors in Bacillus subtilis are processed endonucleolytically by tRNase Z (Pellegrini et al. 2003; Wen et al. 2005). Conversely, the major pathway for eukaryotic tRNA 3′-end processing is endonucleolytic, while trimming by exonucleases serves as an alternative (Garber and Gage 1979; Hagenbuchle et al. 1979; Castaño et al. 1985; Engelke et al. 1985; Frendewey et al. 1985; Manam and Van Tuyle 1987; Stange and Beier 1987; Furter et al. 1992; Oommen et al. 1992; Papadimitriou and Gross 1996; Han and Kang 1997; Nashimoto 1997; Mayer et al. 2000; Schiffer et al. 2002).
Despite several years of studies, tRNA processing in eukaryotic cells still presents some dilemmas that are slowly being resolved. Some tRNA precursors contain introns, located 3′ of the anticodon, which are spliced-out by cleavage and religation. This process has been assumed for many years to occur in the nucleus, but recent data strongly indicate that in Saccharomyces cerevisiae it takes place in the cytoplasm at the surface of the mitochondria (Yoshihisa et al. 2003, 2007). Another complication is the uncertain order of different processing steps. The processing of the 5′ end most often precedes the removal of 3′ extensions. This has been demonstrated, for example, in S. cerevisiae, where mutations in RNase P block maturation of both 5′ and 3′ ends (Lee et al. 1991; O'Connor and Peebles 1991; Furter et al. 1992; Lygerou et al. 1994). Nevertheless, several exceptions to this order of processing have been reported, including some tRNAs in mouse and wheat germ extracts (Rooney and Harding 1986; Arends and Schön 1997), and in vivo processing of yeast tRNATrp (Kufel and Tollervey 2003). tRNA splicing and end processing are not strictly ordered, but in yeast wild-type cells, the rate of splicing is slower, and the presence of the 5′ leader has been reported to slow down intron removal; therefore, mainly unspliced and end-processed intermediates are detected (O'Connor and Peebles 1991; Kufel and Tollervey 2003; Hiley et al. 2005). This could also be related to the site of pre-tRNA splicing outside of the nucleus. It appears more likely that tRNA ends are processed in the nucleus prior to its export to the cytoplasm for splicing, although an alternative scenario is also possible, when intron-containing and unprocessed precursors leave for the cytoplasm and are re-imported to the nucleus after splicing to undergo end processing, CCA addition, and aminoacylation. Several lines of evidence argue that yeast tRNAs are subject to retrograde movement from the cytoplasm to the nucleus (Shaheen and Hopper 2005; Takano et al. 2005; Whitney et al. 2007; Hopper and Shaheen 2008). It is hypothesized that the main purpose of this process is tRNA quality control and regulation of tRNA availability for translation; however, the complete sequence of events is still unknown.
The endonuclease responsible for tRNA 3′-end processing has long been elusive, and tRNase Z has only recently been shown to carry out this reaction in organisms belonging to all three domains of life (for review, see Vogel et al. 2005; Späth et al. 2007). Yet, its function in vivo has only been demonstrated in B. subtilis, Drosophila melanogaster, human mitochondria, and most recently in Schizosaccharomyces pombe (Pellegrini et al. 2003; Dubrovsky et al. 2004; Brzezniak et al. 2011; Zhang et al. 2013). In addition to 3′-tRNase activity, tRNase Z has been demonstrated to participate in mRNA decay in Escherichia coli (Perwez and Kushner 2006). tRNase Z (ELAC1/2) enzymes belong to the family of metal-dependent β-lactamases (Tavtigian et al. 2001; Schiffer et al. 2002) and exist as two subgroups: small ELAC1 proteins (250–350 amino acids) are found in all three domains, whereas larger ELAC2 proteins (750–900 amino acids) are present exclusively in eukaryotes. Drosophila, Caenorhabditis elegans, S. cerevisiae, and S. pombe tRNase Z protein belong to the ELAC2 class, whereas Arabidopsis thaliana and Homo sapiens have both ELAC1- and ELAC2-type tRNase Z enzymes (for review, see Vogel et al. 2005). It has been predicted that long ELAC2 proteins are routed to organelles. According to biochemical analyses of plant tRNase Z and crystal structures of tRNase Z from B. subtilis and Thermotoga maritima, the short enzyme versions act as a homodimer with two monomers arranged head-to-head to form an active site cleft that accommodates single-stranded RNA (de la Sierra-Gallay et al. 2005; Ishii et al. 2005; Späth et al. 2005).
Yeast tRNase Z, called Trz1, has been demonstrated to have 3′-tRNase activity in vitro using recombinant protein (Takaku et al. 2003; Schilling et al. 2005). Based on fluorescence localization of GFP and YFP fusions, it was shown to localize in the nucleus and mitochondria (Hazbun et al. 2003; Huh et al. 2003). However, its function in vivo has been difficult to verify due to the existence of the alternative exonucleolytic pathway. This exonucleolytic backup activity becomes more evident when the default endonucleolytic 3′ processing is inhibited, e.g., in the absence of the yeast homolog of La phosphoprotein, Lhp1, which, among other functions, binds 3′ poly(U) tracts of newly synthesized tRNA precursors to ensure their proper folding, protect them against exonucleolytic digestion, and stimulate endonucleolytic maturation (Stefano 1984; Van Horn et al. 1997; Yoo and Wolin 1997; Chakshusmathi et al. 2003; Copela et al. 2006). At least some of the exonucleases involved in the alternative pathway have been recently identified. Out of the RNase D family of 3′-5′ exonucleases, Rex1 has been shown to have a major tRNA 3′-end processing activity (van Hoof et al. 2000a; Copela et al. 2008; Ozanick et al. 2009). The remaining Rex proteins, Rex2 and Rex3, play a role in the processing of U4 snRNA and MRP RNA, respectively, and combination of Rex2/3 and Rex1/2/3 have redundant functions in trimming the 3′ ends of 5S rRNA and U5 snRNA, respectively. Although the exosome, a multicomponent complex with 3′-5′ exonuclease activity, is regarded to be involved in the 3′-end formation of sn/snoRNAs but not of tRNAs (Briggs et al. 1998; Allmang et al. 1999; van Hoof et al. 2000b), the nuclear-specific exosome component, Rrp6, has been recently shown to have an effect on the 3′-end maturation of some tRNAs in the absence of Lhp1 (Copela et al. 2008).
To demonstrate the participation of Trz1 in tRNA 3′-end formation and to clarify the relative contribution of this endonuclease and different exonucleases to this process, we examined the status of tRNA precursors upon Trz1 depletion in combinations with deletion of Rrp6 and Rex1-3. We observed that the majority of pre-tRNAs underwent maturation by either the endo- or exonucleolytic pathway, but the selection of the particular processing mode was not clear. Precursors with unusually long 3′ trailers, however, were preferentially processed by the endonucleolytic cleavage, whereas precursors with short 3′ extensions were less dependent on Trz1. These data suggest the propensity directing different tRNA molecules to either pathway.
RESULTS
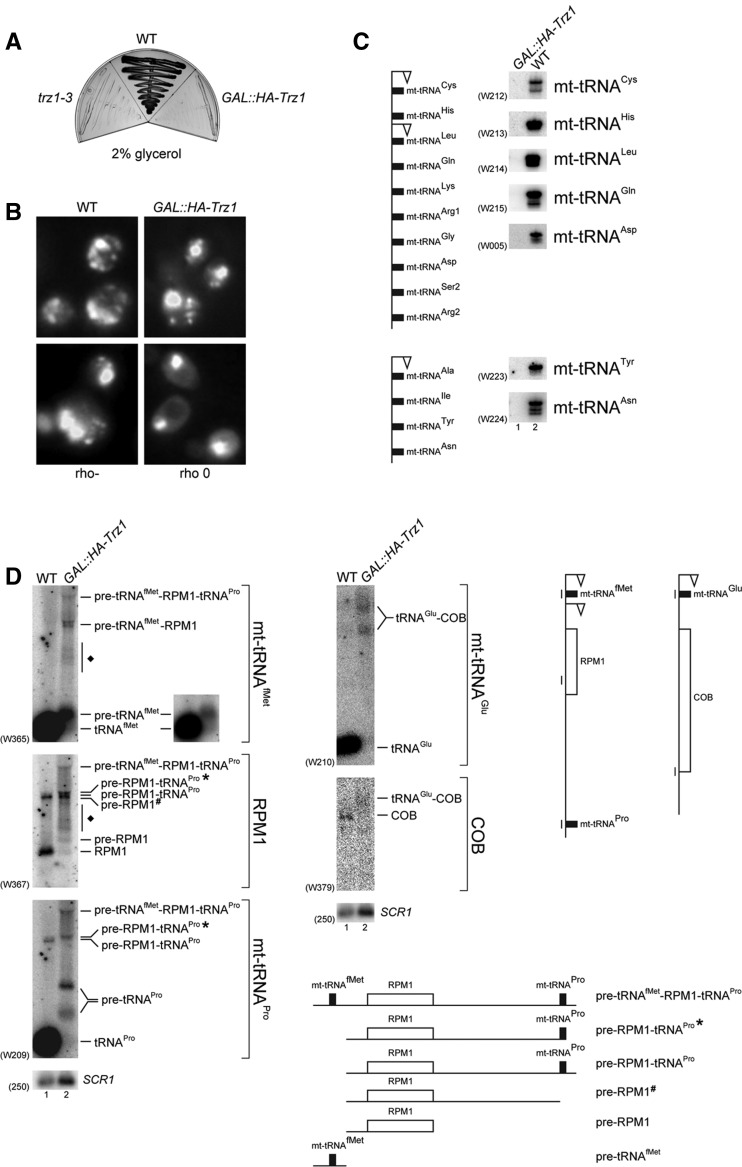
Yeast tRNase Z, Trz1, localizes to both the nucleus and mitochondria and has 3′-tRNA endonuclease activity in vitro
Several ELAC2 proteins are predicted by sorting servers (iPSort and Predotar) to be located in the mitochondria or chloroplasts. Yeast Trz1 does not contain a typical N-terminal mitochondrial import sequence, but some mitochondrial proteins carry internal targeting sequences (Neupert 1997). Based on fluorescence localization of GFP and YFP fusions in two global studies (Hazbun et al. 2003; Huh et al. 2003), Trz1 was reported to localize to the nucleus and mitochondria. To confirm these results, we carried out biochemical fractionation of yeast extract from a Trz1-TAP strain. The cell extract was fractionated by centrifugation in sucrose and Ficoll gradients to obtain a pure nuclear fraction (Doi and Doi 1974; Dove et al. 1998), whereas mitochondria were purified according to the method in Daum et al. (1982); Sperka-Gottlieb et al. (1988). Total, mitochondrial, nuclear, and the remaining cytosolic fractions were analyzed by Western blotting using peroxidase-anti-peroxidase antibodies to detect Trz1-TAP and the following specific antibodies against controls to estimate the purity of each fraction: nucleolar Nop1, mitochondrial Mdh1, and cytoplasmic Hxk2. This approach showed that Trz1 was clearly present in the nuclear and mitochondrial fractions (Fig. 1A, lanes 3 and 4), with some residual amount in the fraction remaining after purification of nuclei (Fig. 1A, lane 2). The latter is possibly a contamination; however, we cannot exclude that a low amount of Trz1 is also present in the cytoplasm. Submitochondrial localization of Trz1-TAP was assessed by fractionation of the mitochondria (Tokatlidis 2000). Mdh1, Tim23, and Tom70 were used as markers for localization in the matrix, inner membrane, and outer membrane, respectively. Western blotting for Trz1-TAP and using specific antibodies against the markers showed that Trz1 was exclusively present in the mitochondrial matrix (Fig. 1B).
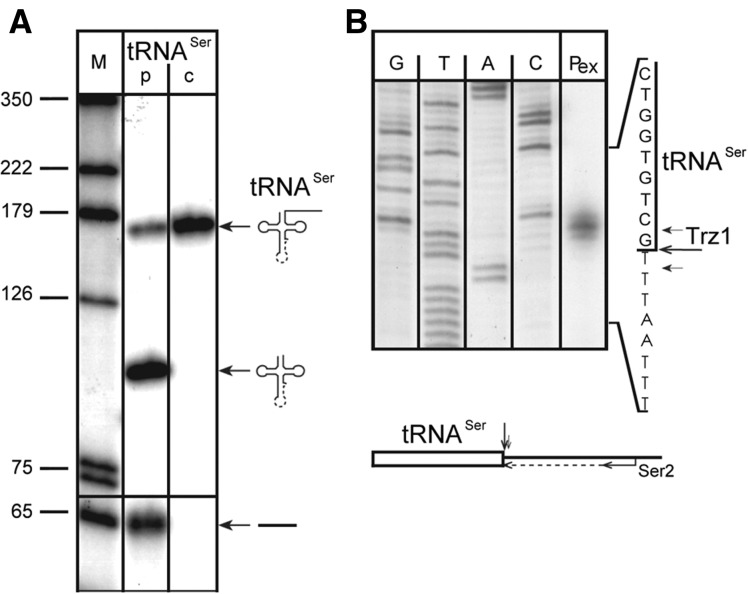
FIGURE 1.
Trz1-TAP is localized in both nucleus and mitochondria. (A) Subcellular fractions of yeast extracts (total cells, cytosol, mitochondria, and nuclei) were analyzed by Western blotting using peroxidase-anti-peroxidase antibodies (PAP). The same blot was probed against nuclear (Nop1), mitochondrial (Mdh1), and cytoplasmic (Hxk2) proteins using specific antibodies. Ponceau S staining is shown in the bottom panel as a loading control. (B) Localization of Trz1-TAP in yeast mitochondrial matrix. Alkali-extracted soluble and membranes fractions as well as total mitochondrial extract were analyzed by Western blotting using PAP antibodies for Trz1 and specific antibodies for markers of different mitochondrial compartments: Mdh1 (matrix, M), Tim23 (inner membrane, IM) and Tom70 (outer membrane, OM). Asterisks mark bands resulting from antibodies’ cross-reactivity.
Previous in vitro data using recombinant Trz1 have demonstrated its 3′-tRNA endonuclease activity against heterologous human or plant precursors with either short or long 3′-end trailer, respectively (Takaku et al. 2003; Schilling et al. 2005). To confirm these results for a matching enzyme-substrate pair, we performed in vitro cleavage of the yeast pre-tRNASer lacking the 5′ leader with recombinant Trz1 as described (Schilling et al. 2005). The reaction generated the intron-containing tRNA and the cleaved-off 3′ trailer (Fig. 2A). Primer extension showed that, as reported for other substrates, yeast tRNA was also cleaved directly 3′ from the discriminator, i.e., the residue directly 5′ of the CCA (Fig. 2B). Short tRNase Z proteins have been demonstrated to act as homodimers (de la Sierra-Gallay et al. 2005; Ishii et al. 2005; Späth et al. 2005). Western blotting using antibodies against the S-tag of yeast recombinant Trz1 cross-linked in the presence of glutaraldehyde revealed it has the capacity to form a homodimer (Supplemental Fig. S1), which suggests that also long ELAC2 proteins may acts as homodimers.
FIGURE 2.
Yeast tRNA precursor is efficiently processed by recombinant Trz1. (A) Processing of an in vitro transcribed yeast nuclear pre-tRNASer by 100 ng of recombinant Trz1. Lane p, processing reaction; lane c, control reaction without the protein. Precursor and products (mature tRNA and a 3′ trailer) are shown schematically on the right; dashed line denotes the intron in tRNASer; DNA size marker (lane M, in nt) is shown on the left. (B) Determination of Trz1 cleavage site in vitro on pre-tRNASer by primer extension. The major cleavage after the discriminator base and two additional, minor cleavages are indicated with arrows. A processing reaction was carried out with nonlabeled pre-tRNA, and the resulting 3′-trailer product was isolated and used as a template in the primer extension reaction. Sequencing reaction (lanes G,A,T,C) performed with the same primer (YC1) is shown on the left. The location of the primer relative to the tRNA is illustrated schematically at the bottom.
Inactivation of Trz1 visibly affects processing of tRNA precursors with long 3′ trailers
The TRZ1 gene encoding putative yeast tRNase Z is essential for cell growth. To analyze the role of Trz1 in tRNA 3′-end processing in vivo, its gene was placed under the control of the regulatable GAL1 promoter and was expressed as an N-terminal fusion with the HA-tag (hemagglutinin). To deplete Trz1, the GAL1::HA-Trz1 strain was transferred from permissive YPGal medium (0 h samples) to repressive YPD medium, and samples were collected at various time-points after the transfer. Depletion was accompanied by a growth defect visible after 10 h, which was more pronounced after 24 h of growth on glucose (Fig. 3A). The level of HA-tagged Trz1 during depletion was detected by the Western blotting using anti-HA antibodies. Trz1 decreased significantly after 12 h, was very low after 24 h, and virtually undetectable after 30 h of depletion (Fig. 3B), which agrees well with the strain growth curve.
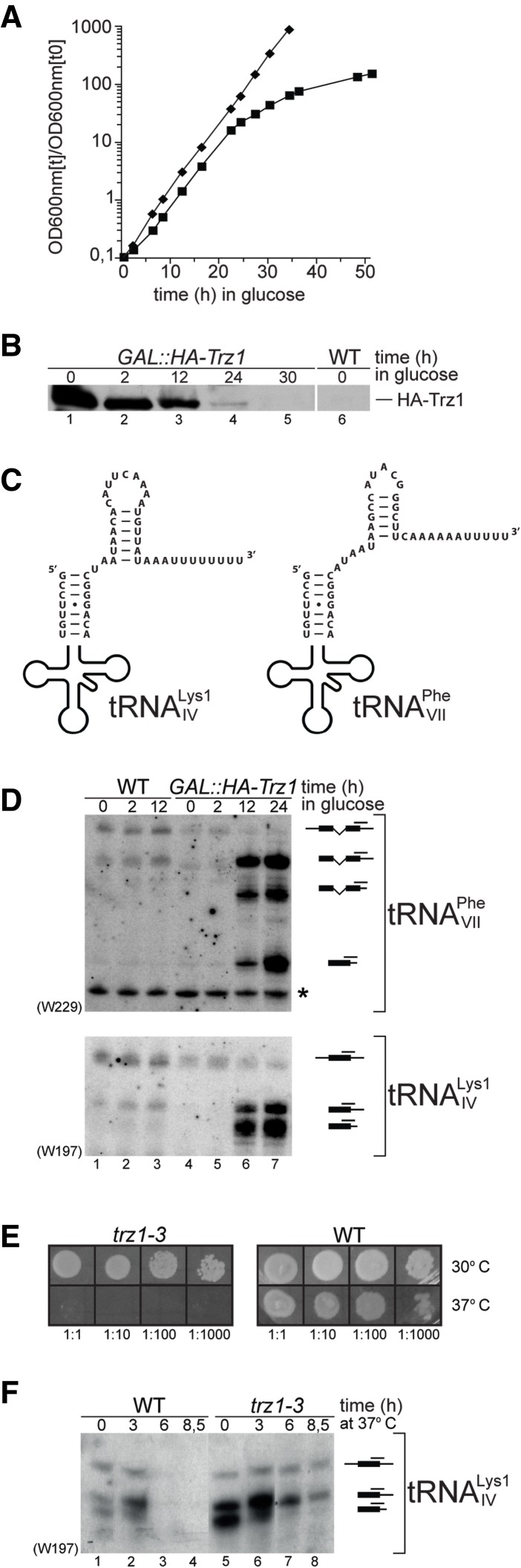
FIGURE 3.
Processing of tRNA precursors with long 3′ trailers is affected in trz1 mutants. (A) Growth curves of wild-type (BMA38, ⧫) and GAL::HA-Trz1 (▪) strains pregrown in YPGal medium (permissive conditions) and shifted to YPD medium (nonpermissive conditions) for the times indicated. Strains were maintained in exponential growth by dilution with fresh medium. Cell densities measured by optical density at 600 nm are shown corrected for dilution. (B) Depletion of Trz1 in the GAL::HA-Trz1 strain grown as described in A; the wild-type strain grown in YPD is shown as a control. Western blotting for proteins extracts prepared from cells harvested at indicated time-points following depletion was probed with anti-HA antibodies. Equal amounts of total protein were loaded in each lane. (C) Potential stem–loop structures in the 3′ extensions of pre-tRNAs with long 3′ trailers. (D,F) Northern analysis of tRNA processing for precursors with long 3′ trailers in the GAL::HA-Trz1 (D) and trz1-3 (F) strains. GAL::HA-Trz1 cells were grown as described in A; trz1-3 cells were pregrown at 25°C (0 h) and transferred to 37°C for the times indicated. RNA was separated on an 8% polyacrylamide gel and hybridized with oligonucleotide probes. Probe names are indicated in parentheses. tRNA species and their graphic representations, with the position of probes used for hybridization, are shown on the right. Asterisk marks cross-hybridization to another RNA. Mature tRNAs are not shown, as specific products of analyzed precursors cannot be detected, and the probes against mature species hybridize to all tRNALys1 or tRNAPhe molecules. (E) Temperature-sensitive phenotype of the trz1-3 mutant. Wild-type and trz1-3 strains were grown on YPS plates at 30°C and 37°C.
The tRNA processing was analyzed in wild-type and GAL1::HA-Trz1 strains by Northern hybridization using probes specific for the intron or mature tRNA sequences, depending on the tRNA tested. For most tRNAs, including tRNALeu3, tRNAIle1, tRNATyr, and tRNALys2, the effects of Trz1 depletion on the level of tRNA precursors were small or moderate (see Fig. 4C, lanes 3–6). This was probably due to the efficient removal of short 3′ extensions by the exonucleolytic pathway when the endonucleolytic cleavage was inhibited.
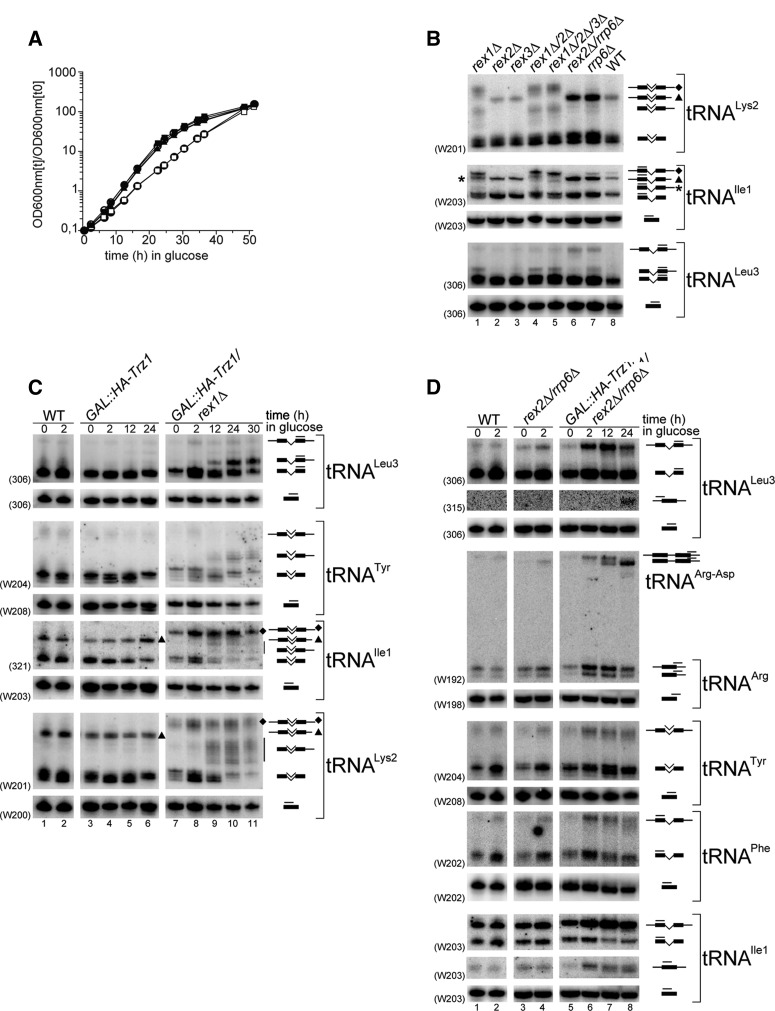
FIGURE 4.
Both Trz1 and Rex1 contribute to tRNA 3′ processing. (A) Growth curves of GAL::HA-Trz1 (▪), GAL::HA-Trz1/rex1Δ (▴), GAL::HA-Trz1/rrp6Δ (□), GAL::HA-Trz1/rex2Δ/rrp6Δ (•), and GAL::HA-Trz1/trf4Δ/rex1Δ (○). Description is as in Figure 3A. Strains with overlapping curves (GAL::HA-Trz1, GAL::HA-Trz1/rex1Δ, and GAL::HA-Trz1/trf4Δ/rex1Δ or GAL::HA-Trz1/rrp6Δ and GAL::HA-Trz1/rex2Δ/rrp6Δ) exhibit similar growth rates. (B–D) Northern analysis of tRNA processing in mutants lacking different combinations of nucleases: strains deleted for Rex1-3 and Rrp6 (B); GAL::HA-Trz1 and GAL::HA-Trz1/rex1Δ strains (C); GAL::HA-Trz1/rex2Δ/rrp6Δ strains (D). Respective wild-type (WT) strains were used as controls. GAL::HA-Trz1 strains were grown as described in Figure 3. Probe names are indicated in parentheses; tRNA species with the position of probes used for hybridization are shown on the right. Asterisks in B denote 3′-ptRNAIle1 in rex1Δ mutants, that has a similar migration as 5′/3′-ptRNAIle1 in other strains; diamonds and triangles in B and C mark extended and standard 5′/3′- ptRNAIle1 or 5′/3′-ptRNALys2 that have different migration in rex1Δ and other strains.
B. subtilis CCA-less tRNA precursors have been proposed to be primarily tRNase Z substrates due to the presence of a stable secondary structure in their long 3′ extensions that prevents exonucleolytic processing (Pellegrini et al. 2003; Wen et al. 2005). Yeast pre-tRNAs typically contain rather short 3′ trailers, comprised of encoded nucleotides (from 0 to 10, most commonly from 3 to 6) plus an oligo(U) tract of heterogeneous length, with the exception of intronless tRNAIV Lys1 and intron-containing tRNAVII Phe (Roman numerals refer to the chromosomal location of genes encoding particular tRNAs) with unusually long 3′ trailers, 25 and 26 nt (excluding the oligo(U) tract), respectively. Interestingly, these two precursors have the capacity to form 3′-terminal structures (Fig. 3C). Predicting that 3′ trailers of these two tRNA molecules will not be trimmed efficiently by exonucleases, we tested the level of their precursors using probes specific for 3′-extended species (W197 and W229). In both cases, 3′-extended pre-tRNAs strongly accumulated following Trz1 depletion for 12 or more hours (Fig. 3D). The largest of these species represent 5′-matured 3′-unprocessed precursors (3′-ptRNA), as estimated by their migration beneath the primary transcript and the fact that they were not detected by probes specific for 5′ leader sequences of these precursors (data not shown). The ladder of heterogeneous bands below the major 3′-extended RNA most likely corresponds to exonucleolytic processing intermediates, except for a precursor larger than the mature tRNAVII Phe, which stands for a spliced-out but 3′-unprocessed species (3′-tRNA). In contrast, the absence of Rex1 or Rrp6 alone had little or no effect on the 3′ processing of tRNAIV Lys1 and tRNAVII Phe, as there was no visible accumulation of the 3′-ptRNAs in rex1Δ or rrp6Δ strains (see Fig. 5A,B).
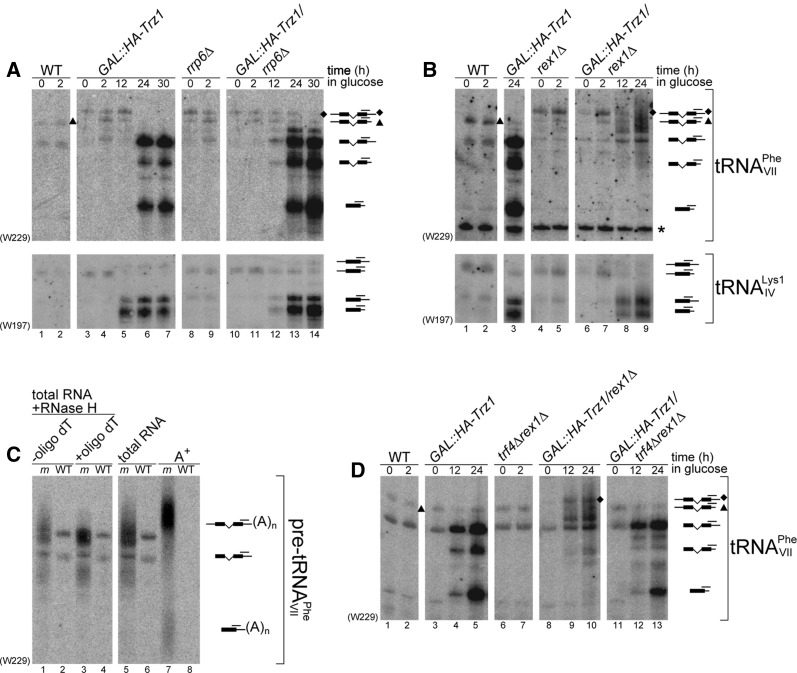
FIGURE 5.
tRNA precursors with long 3′ trailers are markedly affected and polyadenylated by Trf4 in cells lacking both 3′ processing activities. (A,B) Northern analysis of tRNAVII Phe and tRNAIV Lys1 in wild-type, GAL::HA-Trz1, rrp6Δ and GAL::HA-Trz1/rrp6Δ (A) or wild-type, GAL::HA-Trz1, rex1Δ and GAL::HA-Trz1/rex1Δ strains (B). (C) Pre-tRNAVII Phe are polyadenylated in GAL::HA-Trz1/rex1Δ cells. Northern hybridization for tRNAVII Phe of total RNA (lanes 1–4) from GAL::HA-Trz1/rex1Δ mutant (m samples) treated with RNase H in the absence (−) and presence (+) of oligo(dT); or of the total or poly(A)+ RNA fraction (A+) (lanes 5–8). Respective wild-type (WT) strains were used as controls. (D) Polyadenylation of tRNAVII Phe in the GAL::HA-Trz1/rex1Δ strain is carried out by Trf4. Northern analysis of tRNAVII Phe in wild-type, GAL::HA-Trz1, trf4Δ/rex1Δ, GAL::HA-Trz1/rex1Δ and GAL::HA-Trz1/trf4Δ/rex1Δ strains. GAL::HA-Trz1 strains were grown as described in Figure 3. Probe names are indicated in parentheses. tRNA species with the position of probes used for hybridization are shown on the right. Diamonds and triangles in A, B, and D denote 5′/3′-ptRNAVII Phe that have different migration in wild-type, rex1Δ and rrp6Δ strains; asterisk marks cross-hybridization to another RNA.
To confirm these results, a frameshift trz1-3 mutant was generated by inserting the second ATG codon in the TRZ1 gene upstream of the original translation start. Transcriptome microarray analysis of the mutant revealed that, as expected, the level of TRZ1 mRNA was significantly reduced (11-fold) in the mutant compared to the wild type (B Späth and A Marchfelder, unpubl.). This strain showed a marked temperature-sensitive phenotype at 37°C, particularly when cells were grown in the YPS medium (Fig. 3E). The basis of the temperature-sensitive lethality of trz1-3 cells is unknown. Strong accumulation of 3′-unprocessed tRNAIV Lys1 precursors in trz1-3 mutant, similar to that seen when Trz1 was depleted, was observed already at the permissive temperature (Fig. 3F, lane 5). Following the transfer to 37°C, the level of 3′-ptRNAs decreased significantly in the wild-type strain, possibly due to their different processing kinetics at higher temperature, but were still present in the mutant (Fig. 3F, lanes 6–8), consistent with delayed 3′ processing.
These data show that out of several tested pre-tRNAs, only two with longer 3′ trailers show a distinctive 3′-end processing defect in the absence of Trz1. This may be due to an inefficient exonucleolytic pathway, inhibited by secondary structures comprised within their longer 3′ extensions.
Contribution of endo- and exonucleolytic pathways to maturation of tRNA 3′ ends
It has been reported that Rex1 is involved in the exonucleolytic maturation of pre-tRNAArg3 encoded by dicistronic tRNAAsp-Arg3 (van Hoof et al. 2000a) and, more recently, together with Rrp6, in the 3′-end processing of several other tRNAs (Copela et al. 2008; Ozanick et al. 2009). Furthermore, REX2 is a high-copy suppressor of a temperature-sensitive trz1 allele, REX1, REX2, or REX3 are high-copy suppressors of an RRP6 deletion, while deletion of REX1 or REX3 genes in rrp6Δ cells, at least in some genetic backgrounds, results in a synthetic lethal phenotype (van Hoof et al. 2000a; Chen et al. 2005; Abruzzi et al. 2007). These facts strongly suggest that these nucleases function in redundant or alternative pathways.
Analysis of strains lacking Rex proteins showed that the level of 3′-unprocessed forms of tRNALys2, tRNAIle1, and tRNALeu3 clearly accumulated in rex1Δ, rex1Δ/rex2Δ, and rex1Δ/rex2Δ/rex3Δ but not in single rex2Δ or rex3Δ mutants (Fig. 4B), as previously reported (Copela et al. 2008). In turn, the absence of Rrp6, led to the increase not only of the 3′-ptRNA but also of the full-length 5′/3′-ptRNA (5′- and 3′-unprocessed), indicating a delay also in the 5′-processing pathway (Fig. 4B). Similar phenotypes, albeit to varying degrees, were observed for several other, but not all, tRNAs tested, including tRNATrp and tRNAPhe (data not shown). These data confirm the involvement of Rex1 and, to a lesser extent, Rrp6 in the 3′ exonucleolytic maturation of tRNAs. As reported previously, conspicuously longer full-length 5′/3′-ptRNAs were observed in rex1Δ strains for some tRNA species, including tRNALys2, tRNAIle1, tRNAPhe, and tRNAVII Phe (Figs. 4B, 5B; Copela et al. 2008). The significance of this effect is unclear at present, but it is possible that, unlike primary transcripts detectable in wild-type cells for other tRNAs, these precursors undergo initial processing by Rex1 (Copela et al. 2008).
Since processing of tRNAs with usual 3′ trailers was not visibly affected by the lack of Trz1 alone, we created mutants combining defects in the two pathways, i.e., GAL1::HA-Trz1/rex1Δ, GAL1::HA-Trz1/rrp6Δ, GAL1::HA-Trz1/rex2Δ, and GAL1::HA-Trz1/rex2Δ/rrp6Δ strains (Fig. 4A; Supplemental Fig. S2), and analyzed the pattern of pre-tRNAs by Northern hybridization. When both endo- and exonucleolytic processing was inhibited by depletion of Trz1 in a strain lacking Rex1 3′-unprocessed precursors of several intron-containing tRNAs, including tRNALeu3, tRNALys2, tRNATyr, and tRNAIle1, clearly accumulated, while the fully mature intron-containing form was markedly decreased (Fig. 4C). These precursors were not visible in the GAL1::HA-Trz1 strain and detectable at low level in the rex1Δ mutant (see Fig. 4B). For some tRNAs, particularly tRNALys2 and tRNATyr, the 3′-unprocessed precursors showed a high degree of heterogeneity, indicating a strong impediment of exonucleolytic trimming.
In contrast, depletion of Trz1 in rex2Δ, rex3Δ, or rrp6Δ strains had little or no obvious effect on pre-tRNA with short 3′ extensions (Supplemental Fig. S2). However, the GAL1::HA-Trz1/rex2Δ/rrp6Δ strain compared to single- or double-mutants showed a more pronounced accumulation of the full-length 5′/3′-extended precursors for some tRNAs, namely tRNALeu3, tRNATyr, tRNAPhe, tRNAIle1, and a dicistronic ptRNAArg-Asp (Fig. 4D). The identity of the elevated 5′/3′-ptRNAs was confirmed for ptRNAArg-Asp and tRNALeu3 using probes specific for their 5′ and 3′ regions (Supplemental Fig. S2; data not shown). In addition, the level of 5′- and 3′-unprocessed but spliced tRNAs (shown for tRNAIle1 and tRNALeu3 in Fig. 4D) was also increased in this strain, an effect that was not observed in other mutants tested, including GAL1::HA-Trz1/rex1Δ. These results suggest some level of redundancy between Rrp6 and Rex2 in tRNA maturation. Alternatively, accumulation of unprocessed pre-tRNAs in mutants lacking Rrp6 may be attributed to their degradation by this exonuclease, as tRNA precursors were reported recently to be widespread exosome substrates (Gudipati et al. 2012; Schneider et al. 2012).
Also, the processing of tRNAs with unusually long 3′ trailers, tRNAIV Lys1 and tRNAVII Phe, that showed a marked defect already on depletion of Trz1 alone, was clearly exacerbated by the additional lack of Rrp6, with the stronger accumulation of processing intermediates (Fig. 5A).
Together, these results clearly show that Trz1 is responsible for endonucleolytic tRNA 3′-end processing that is complemented by exonucleolytic trimming mainly by Rex1, with the auxiliary participation of Rrp6 and, to an even lesser extent, Rex2.
Interestingly, in the GAL1::HA-Trz1/rex1Δ strain, 3′-extended precursors became heterogeneous and diffused, particularly in the case of tRNAVII Phe, indicating that these 3′-ptRNAs may carry poly(A) or oligo(A) tails (Fig. 5B). We, therefore, tested the polyadenylation status of pre-tRNAVII Phe in the GAL1::HA-Trz1/rex1Δ and wild-type strains by the RNase H-directed deadenylation assay in the presence of oligo(dT) and by purification of the poly(A)+ RNA fraction. The diffused 3′-extended tRNAVII Phe species present in GAL1::HA-Trz1/rex1Δ cells were shortened and became more prominent after RNase H treatment (Fig. 5C, cf. lanes 1 and 3). Also, abundant longer 3′-extended heterogeneous precursors were enriched in the poly(A)+ fraction from the mutant but not the wild-type (Fig. 5C, cf. lane 7, marked “m,” and 8, marked “WT”), showing that these species carry poly(A) tails.
Several noncoding RNAs and their precursors become polyadenylated during the nuclear surveillance by the alternative poly(A) polymerase Trf4, a component of the TRAMP complex (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005). Trf4 substrates include unstable, hypomodified pre-tRNAi Met (Kadaba et al. 2004) and probably defective precursors generated by Rex1-mediated trimming, as accumulation of unprocessed tRNA species was detected in trf4Δ and rrp6Δ/trf4Δ mutants and polyadenylation of pre-tRNAi Met and pre-tRNACAC Val was observed in the absence of Rex1 (Copela et al. 2008; Ozanick et al. 2009). To assess whether 3′-extended pre-tRNAVII Phe is polyadenylated by Trf4, we constructed the GAL1::HA-Trz1/rex1Δ/trf4Δ strain (see Fig. 4A) and tested the pattern of pre-tRNAVII Phe intermediates (Fig. 5D). In contrast to heterogeneous and diffused species present in the GAL1::HA-Trz1/rex1Δ mutant, the precursor status following depletion of Trz1 in rex1Δ/trf4Δ cells strongly resembled pre-tRNAs in the single GAL1::HA-Trz1 strain, which argues that their polyadenylation in the absence of both Trz1 and Rex1 is carried out by Trf4. Surprisingly, the unusual, longer full-length pre-tRNAVII Phe observed in the absence of Rex1 was replaced by the standard-sized precursor in cells additionally devoid of Trf4. This was not the case for other 5′/3′-ptRNAs extended in the rex1Δ mutant, including tRNAIle1 and tRNALys2 (Supplemental Fig. S2; Copela et al. 2008). This observation suggests that oligoadenylation and trimming by Trf4 and Rex1, respectively, may be involved in the initial steps of 3′-end maturation of some pre-tRNAs, possibly those with atypical 3′ features.
These data show that blocking tRNA 3′-end processing may result in polyadenylation of accumulated precursors mediated by Trf4, probably as an integral step of their quality control.
Trz1 functions in mitochondrial RNA processing
S. cerevisiae contains only one tRNase Z protein, which localizes to both the nucleus and mitochondria. Mitochondrial function of Trz1 is supported by the fact that temperature-sensitive mutations in TRZ1 were shown to result in a petite phenotype (Chen et al. 2005). Also, other trz1 mutants, including GAL1::HA-Trz1 and trz1-3 strains, are respiratory-deficient, as they failed to grow on media containing a nonfermentable carbon source (glycerol) (Fig. 6A; Supplemental Fig. S3). To establish whether their genotype is mit−, rho−, or rho0 (i.e., with point mutations, deletions, or totally lacking mtDNA, respectively), we analyzed DAPI-stained wild-type, GAL1::HA-Trz1, rho−, and rho0 strains under a fluorescence microscope. In addition to the bright nuclear spot, a dappled signal derived from mitochondrial staining was observed in wild-type and rho− cells, as expected, and also in the GAL1::HA-Trz1 mutant but not in rho0 yeast (Fig. 6B). This result was confirmed by detecting specific regions of mtDNA (e.g., tRNAfMet-RPM1-tRNAPro, 15S-tRNATrp, COX2) in GAL1::HA-Trz1 cells by PCR, which were absent in the rho0 strain (Supplemental Fig. S3). Interestingly, all regions tested that contain only tRNA genes were not retained in the mtDNA of this strain. In agreement with this observation, Northern hybridization with several probes for mitochondrial-encoded tRNAs showed that those originating from tRNA-only transcriptional units (e.g., tRNACys-tRNAArg2 and tRNAAla-tRNAAsn) were missing, and larger precursors were also not observed (Fig. 6C). These data clearly show that the GAL1::HA-Trz1 strain has a mitochondrial rho− genotype with large deletions in mtDNA. Partial loss of mtDNA in this case may result from the altered level of Trz1 or from the presence of the N-terminal HA tag that interferes with the mitochondrial localization of Trz1. However, the GAL1::TRZ1 and Tet::TRZ1 strains lacking this tag were also respiratory-deficient, and depletion of Trz1 in these mutants was very ineffective (Supplemental Fig. S3; data not shown). mtDNA rearrangements resulting in a rho− phenotype may arise due to a strong translational defect in the absence of pre-tRNA processing. It is commonly observed that mutations leading to altered expression of mitochondrial genes affect the integrity of the mitochondrial genome (Lipinski et al. 2010). The annotated Trz1 sequence lacks an apparent standard mitochondrial targeting signal, but the fact that expression of TRZ1 variants carrying changes upstream of the Trz1 open reading frame results in a petite phenotype suggests a possibility that localization information is contained in this sequence, maybe through an alternative translation start site.
FIGURE 6.
Synthesis of mitochondrial transcripts depends on Trz1. (A) trz1 mutants are respiratory-deficient. Wild-type (WT), GAL::HA-Trz1, and trz1-3 strains were grown on plates containing glycerol as a sole carbon source. (B) DAPI staining to visualize nuclear and mitochondrial DNA in wild-type (WT), GAL::HA-Trz1, rho−, and rho0 cells by fluorescent microscopy. (C) Northern analysis of mature mt-tRNAs in the mitochondrial RNA fraction of wild-type (WT) and GAL::HA-Trz1 strains grown at permissive conditions (galactose). (D) Northern analysis of tRNAfMet-RPM1-tRNAPro and tRNAGlu-COB1 mitochondrial transcription units using total RNA of wild-type (WT) and GAL::HA-Trz1 strains grown at permissive conditions. Probe names are indicated in parentheses, identities of RNA transcripts are shown on the right, graphic representations of corresponding mitochondrial transcription units as in Tzagoloff and Myers (1986); Foury et al. (1998); Schonauer et al. (2008), with the position of probes used for hybridization, are depicted on the left (C) or on the right (D). RNAs marked with diamonds represent aberrant RPM1 precursors or processing intermediates. Promoters are depicted as flags. SCR1 was used as a loading control. Note that the mature tRNAfMet in the tRNAfMet-RPM1-tRNAPro unit can originate from two precursors, pre-tRNAfMet or tRNAfMet-RPM1-tRNAPro (Stribinskis et al. 1996).
Mitochondrial localization of S. cerevisiae Trz1 implies its function in this organelle, most likely also in tRNA 3′-end processing. In contrast to nuclear genes, mitochondrial genes are organized in polycistronic transcription units that give rise to at least 13 primary transcripts containing mRNAs, tRNAs, and rRNAs (Christianson and Rabinowitz 1983; Tzagoloff and Myers 1986; Foury et al. 1998; Schafer 2005). Individual transcripts are liberated by endonucleolytic processing, including cleavages at the 5′ ends of tRNAs by RNase P and at tRNA 3′ ends, probably by Trz1 (Lipinski et al. 2010). It has been recently demonstrated to be the case for human and S. pombe mitochondrial RNAs, where ELAC2/tRNase Z or Trz2, respectively, carry out mt-tRNA 3′-end processing (Brzezniak et al. 2011; Zhang et al. 2013).
To determine whether mt-tRNA 3′-end processing is affected by the lack of Trz1, we tested the level of mitochondrial transcripts in the GAL1::HA-Trz1 strain; the most informative data were obtained for units enclosing tRNAfMet-RPM1-tRNAPro and tRNAGlu-COB1 genes (Fig. 6D). The effects on mtRNA processing were comparable prior to and following Trz1 depletion (Supplemental Fig. S3), supporting the notion that Trz1 may not be efficiently imported to the mitochondria in this strain and that the rho− phenotype results from RNA maturation defects.
Analysis of the processing of the tRNAfMet-RPM1-tRNAPro transcript in the GAL1::HA-Trz1 mutant revealed a complex pattern of precursors, intermediates, or degradation products. Their identities, particularly of wild-type or unprocessed/aberrant RPM1 or tRNAPro precursors, as well as those enclosing RPM1 and both or either tRNAs (e.g., pre-tRNAfMet-RPM1-tRNAPro, pre-tRNAfMet-RPM1, pre-RPM1-tRNAPro and its derivatives), were assigned based on their relative sizes, hybridization pattern, and previous reports (Stribinskis et al. 1996, 2001; Schonauer et al. 2008). GAL1::HA-Trz1 cells showed a clear accumulation of larger, unprocessed tRNAPro precursors or intermediates, but little or no mature tRNAPro was detected (Fig. 6D). In addition, a marked decrease in mature RPM1, the RNA subunit of mitochondrial RNase P, accompanied by the accumulation of higher-molecular weight RNA species, was observed, in agreement with previous suggestions that endonucleolytic cleavages by tRNase Z contribute to the release of the mature RPM1 (Stribinskis et al. 2001; Schonauer et al. 2008). Also, the appearance of the 3′-extended pre-RPM1-tRNAPro* and the absence of pre-RPM1-tRNAPro (detected by both RPM1 and tRNAPro probes) in the mutant is consistent with a processing deficiency by Trz1.
It is apparent that, in the GAL1::HA-Trz1 mutant, not only Trz1 activity is compromised but also that of RNase P, suggesting that 5′ processing of pre-tRNAs may be affected. However, the existence of the RPM1# precursor lacking tRNAPro (detected only with a probe specific for RPM1) argues that some 5′ tRNA processing by RNase P still takes place, even with a significantly reduced amount of mature RPM1. Therefore, accumulation of unprocessed pre-tRNAPro is most likely a direct effect of Trz1 dysfunction, especially considering that similar RNA species are not generated in strains with defective RNase P (Stribinskis et al. 1996, 2001; Schonauer et al. 2008).
In contrast to tRNAPro, both precursor and mature tRNAfMet were visible but significantly reduced in GAL1::HA-Trz1 cells. The residual amount of mature tRNAfMet in the absence of Trz1 could be explained by the existence of two promoters in the tRNAfMet-RPM1-tRNAPro unit, located upstream of tRNAfMet and RPM1 genes (Stribinskis et al. 1996). In addition, a major tRNAfMet precursor in the mitochondrial RNase P mutant represents 5′-extended and 3′-end mature species. Assuming that yeast mitochondrial tRNase Z also favors 5′-processed substrates (Kunzmann et al. 1998; Brzezniak et al. 2011), it is likely that tRNAfMet is transcribed from the upstream promoter also as an independent species and may, to some extent, undergo Trz1-independent exonucleolytic trimming.
In the case of the second transcriptional unit tested, larger precursors, but little or no mature tRNAGlu or COB1 mRNA, were detected in the GAL1::HA-Trz1 strain (Fig. 6D). Larger precursors containing pre-tRNAs and the adjacent mRNAs were also observed upon inactivation of S. pombe mitochondrial tRNAse Z, Trz2 (Zhang et al. 2013).
To investigate further which exonucleases cooperate with Trz1 in the maturation of mitochondrial transcripts, we generated GAL1::HA-Trz1 strains lacking Nuc1, Rex2, or Pet127. Nuc1 is a major mitochondrial nuclease with RNase and DNase activities (Vincent et al. 1988), Pet127 is probably the main nonspecific 5′-3′ exonuclease in mitochondria and is involved in 5′ processing of several mtRNAs, including RPM1 (Wiesenberger and Fox 1997; Ellis et al. 2005; Fekete et al. 2008; Schonauer et al. 2008), whereas 3′-5′ exonuclease Rex2, in addition to its functions in the nucleus, was shown to have mitochondrial localization (Hanekamp and Thorsness 1999; Kumar et al. 2002; Huh et al. 2003). In addition, physical or genetic interactions were demonstrated between Trz1 and Nuc1 or Rex1, respectively (Chen et al. 2005; Benschop et al. 2010). The main 3′-5′ exonucleolytic activity in yeast mitochondria, responsible for mtRNA processing and turnover, is provided by the degradosome (mtEXO), composed of RNase Dss1 and helicase Suv3 (Dziembowski et al. 2003; Lipinski et al. 2010). Importantly, three mitochondrial RNA processing activities, RNase P, RNase Z, and mtEXO, were recently reported to form a stable RMT (RNA processing, Metabolism, Translation) supercomplex (Daoud et al. 2012). However, yeast suv3Δ or dss1Δ strains exhibit a particularly high level of mitochondrial genome instability due to inhibition of translation (Dziembowski et al. 1998), and it was not practical to combine these deletions with a dysfunction of Trz1, which also results in a similar mt phenotype. In addition, mtEXO was reported not to be involved in tRNA processing (Dziembowski et al. 2003), and its presence in the RMT supercomplex probably reflects its participation in maturation of rRNA and/or mRNA molecules, as rRNA subunits, some ribosomal proteins, and rRNA or mRNA processing factors are also part of this complex (Daoud et al. 2012).
The effects of NUC1, REX2, or PET127 deletion in the GAL1::HA-Trz1 strain were analyzed for the tRNAfMet-RPM1-tRNAPro unit prior to Trz1 depletion since dysfunction of Trz1 in mitochondrial RNA processing was clearly observed in these conditions (Fig. 7). The combined deficiencies of Trz1 and Rex2 resulted in a stronger accumulation of precursors and processing intermediates and in the appearance of additional species (detected with a probe specific for RPM1), whereas deletion of NUC1 changed the pattern and relative abundance of medium-sized intermediates present in the GAL1::HA-Trz1 cells (detected with a probe against tRNAPro). A higher level of tRNAPro and tRNAfMet precursors in the double GAL1::HA-Trz/rex2Δ strain indicates that some exonuleolytic trimming by Rex2 may be involved in mt-tRNA processing. In turn, while the absence of Pet127 alone affected 5′-end maturation of RPM1, yielding, as reported, a 5′-extended precursor (Ellis et al. 2005; Schonauer et al. 2008), the double GAL1::HA-Trz1/pet127Δ mutant showed a decrease in the level of mature tRNAfMet and most of the precursors and intermediates observed upon inactivation of Trz1. It is not clear why the residual tRNAfMet was further reduced in GAL1::HA-Trz1 cells lacking 5′-3′ exonuclease Pet127, but this observation may support the proposed link between 5′ and 3′ processing of mitochondrial transcripts (Wegierski et al. 1998; Stribinskis et al. 2001).
FIGURE 7.
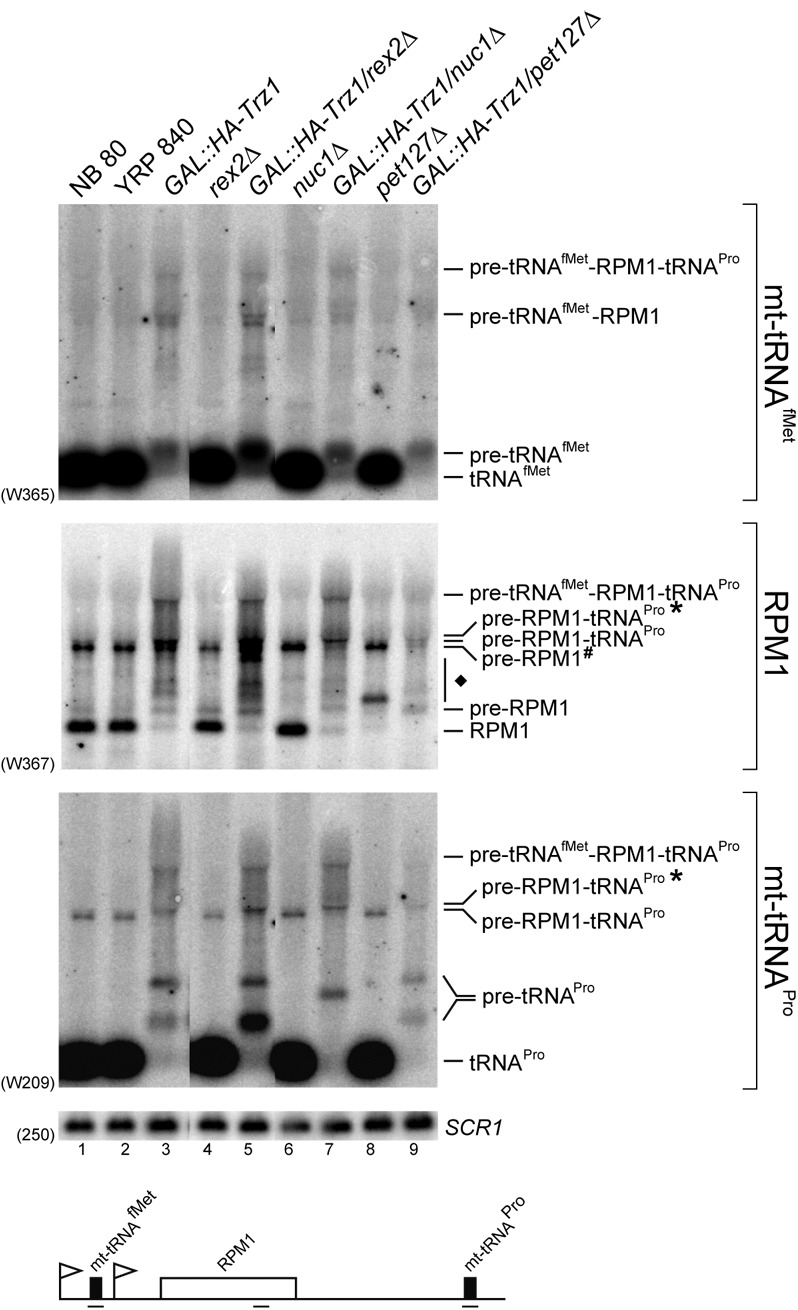
Mitochondrial nucleases contribute to the overall processing of mtRNAs. Northern analysis of tRNAfMet-RPM1-tRNAPro transcription units using total RNA of wild-type, rex2Δ, nuc1Δ, pet127Δ, GAL::HA-Trz1 single-mutants and GAL::HA-Trz1/rex2Δ, GAL::HA-Trz1/nuc1Δ, and GAL::HA-Trz1/pet127Δ double-mutants. Corresponding wild-type strains, NB80 for pet127Δ mutants and YRP840 for the remaining mutants, were used as controls. Probe names are indicated in parentheses, identities of RNA transcripts are shown on the right. Graphic representation of the tRNAfMet-RPM1-tRNAPro transcription unit and major detected precursors and intermediates is shown below. SCR1 was used as a loading control.
These results suggest that processing of mt-tRNAs probably does not entail a robust involvement of exonucleases, with a possible minor contribution of Rex2. Exonucleases are likely to play a more regular role in maturation of other mitochondrial transcripts, following endonucleolytic cleavages by RNase P and Trz1. In addition, these activities may participate, possibly in a redundant manner, in elimination of a broad variety of processing intermediates and by-products.
Taken together, these data confirm that Trz1 also acts as a 3′ tRNA endonuclease in mitochondria, probably contributing to the release of individual mitochondrial transcripts (Schafer 2005). Since Trz1 dysfunction also impairs RNase P activity, the combined defect may result in a profound processing phenotype of most mitochondrial RNAs. Defects in maturation of mt-tRNAs observed in the absence of Trz1 most likely also result in serious impediment of mitochondrial translation, which is a frequent basis for the respiratory-deficient phenotype (Myers et al. 1985; Lipinski et al. 2010). The more universal function of Trz1 in the metabolism of most, if not all, mitochondrial RNAs explains even better the rho− genotype of strains with altered expression of this protein.
DISCUSSION
Synthesis of tRNA molecules in eukaryotic cells might appear as an unnecessarily sophisticated process. Only the processing of their 5′ end is a straightforward one-enzyme reaction carried out by a ubiquitous ribonucleoprotein complex, which is composed of one RNA and at least nine proteins in the yeast S. cerevisiae (for review, see Evans et al. 2006). Remarkably, intron removal and 3′-end maturation are far more complex. Not only do splicing and end-processing have no fixed order, but they also occur in different cellular compartments (the cytoplasm and nucleus, respectively). In addition, 3′-end maturation can proceed in two seemingly independent pathways: endonucleolytic cleavage, which was proposed to constitute a major pathway, and exonucleolytic trimming acting as an alternative. Such redundancy is most likely to have evolved to ensure the most productive synthesis of mature tRNAs.
Recently, Rex1 has been reported to be the main exonuclease involved in trimming tRNA 3′ ends and to function in removing the CCA from end-processed ptRNAs (Copela et al. 2008). We confirmed that Rex1, with a minor contribution from Rrp6 and Rex2, is required for tRNA 3′-end maturation in the exonucleolytic pathway. More importantly, our results showed that yeast tRNase Z, Trz1, carries out endonucleolytic cleavage, removing tRNA 3′ trailers in vitro as well as in vivo. Maturation of standard precursors probably proceeds via both pathways and exonucleolytic digestion also occurs to some degree in wild-type cells and not only when endonucleolytic cleavage is inhibited. Thus, no default 3′-end processing scenario seems to exist. The exceptions are two precursors with unusually long 3′ trailers that are predominantly matured by Trz1, where deletion of exonucleases has little or no consequence. It is possible that a putative stem–loop structure forming within long 3′ trailers of these species, that may inhibit exonuclease activity, biases the choice of pathway toward the endonucleolytic cleavage, while precursors with particularly short trailers are preferentially trimmed by exonucleases. Interestingly, processing of mitochondrial pre-tRNAs, which possess longer 3′ trailers, was affected more severely than processing of those in the nucleus (Dubrovsky et al. 2004). Another option is that ptRNAs with different 3′ ends display diverse affinities for binding Lhp1, which favors endonucleolytic cleavage and protects against digestion by exonucleases. However, the relative contribution of both pathways is not always straightforward to estimate, since their simultaneous inhibition has additive, but varying, effects on the processing of different tRNAs.
Blocking either or both enzymes generating mature 3′ ends, Rex1 and Trz1, in most cases leads to accumulation of 3′-unprocessed precursors. In contrast, in cells lacking Rrp6 and, more visibly, Trz1 and both Rrp6 and Rex2, the increased level of some 5′/3′-ptRNAs, symptomatic of the defect in 5′-end processing, was also detected. Although inhibition of 3′ cleavage when RNase P processing is reduced has been observed repeatedly (Lee et al. 1991; Lygerou et al. 1994; Kufel and Tollervey 2003), the reverse has not been reported. Moreover, tRNase Z has been shown to efficiently cleave only 5′–processed pre-tRNAs, and human tRNase Z activity in vitro is inhibited by the presence of longer 5′ extensions (Kunzmann et al. 1998; Nashimoto et al. 1999; Schierling et al. 2002; Dubrovsky et al. 2004). In turn, our data suggest that the roles of Rex1 and Rex2 proteins and Rrp6 in generating mature tRNA 3′ ends do not fully overlap, but that Rrp6 together with Rex2 may also affect the kinetics or efficiency of the 5′ leader cleavage by RNase P. It is difficult to explain the basis of this effect, but the existence of a link between 3′ and 5′ tRNA processing is quite realistic, being manifested when some aspects of end maturation and/or tRNA quality control are defective. Alternatively, considering that the exosome was recently reported to degrade a large fraction of pre-tRNAs (Copela et al. 2008; Gudipati et al. 2012; Schneider et al. 2012), the lack of Rrp6 together with Rex2 may affect efficient removal of unprocessed precursors in the case where their proper 3′-end maturation is blocked.
Furthermore, our results confirm that pre-tRNAVII Phe, and most likely, other tRNA precursors as well, become polyadenylated by Trf4, probably as a component of the TRAMP complex, when their 3′-end processing is inhibited. This is consistent with a notion of tRNA quality control in which inhibition of tRNA processing results in their degradation by the action of the TRAMP/exosome complexes, as is the case for unstable tRNA molecules with defective modifications or unprocessed tRNA species in the absence of Rex1 (Kadaba et al. 2004; Copela et al. 2008; Ozanick et al. 2009). This effect is more pronounced when both pathways, endonucleolytic by Trz1 and exonucleolytic by Rex1, are blocked (this work).
To date, all studies on tRNA 3′-end maturation in mitochondria have identified endonucleolytic but no exonucleolytic activities (see references in Mörl and Marchfelder 2001). We observed that Trz1 also processes precursors to mt-tRNAs in yeast and, via these cleavages, generates several individual mitochondrial transcripts, including that encoding the RNA component of mtRNase P. Together, these two endonucleases might be responsible for the release of the majority of mtRNAs from polycistronic precursors.
The processing phenotypes observed in yeast lacking Trz1 and additional nucleases, Rex2, Nuc1, or Pet127, that may contribute to the synthesis of mitochondrial transcripts, revealed a complex pattern of RNA processing in yeast mitochondria. They are consistent with the tRNA punctuation model, where endonucleolytic release of mature mt-tRNAs from their precursors by RNases P and Z initiate 5′ and 3′ processing of other mtRNAs. The analysis of mt-tRNAfMet indicates the possibility that at least some mitochondrial tRNAs may undergo limited exonucleolytic 3′-end processing; however, in agreement with previous reports, our study confirms that the major activity in mitochondria is provided by endonucleases (Chen and Martin 1988; Papadimitriou and Gross 1996).
In S. cerevisiae, which contains only one long tRNase Z protein (ELAC2), it has to be targeted to both the nucleus and mitochondria. Several other proteins, which function in tRNA processing in two locations (nucleus/cytoplasm or mitochondria), are also encoded by a single nuclear gene (Martin and Hopper 1994). In turn, organisms like Homo sapiens, with both long and short tRNase Z versions, could, in principle, use the shorter enzyme to catalyze tRNA 3′ processing in the nucleus and the longer one to generate organellar tRNA 3′ ends. However, in a recent study, the short human tRNase Z (ELAC1) was shown to be cytosolic, whereas the long ELAC2 was present in both the nucleus and mitochondria and participated in mt-tRNA 3′-end processing (Brzezniak et al. 2011). Defects in this process, particularly when combined with mt-RNaseP deficiency, also resulted in accumulation of larger unprocessed mitochondrial transcripts.
Yeast Trz1 is required for cell viability, and therefore its essential function is most likely not to be in tRNA processing, as an exonucleolytic backup system already exists. We predict that Trz1 might also be involved in the processing of other noncoding RNA classes, rRNAs being the most likely substrates, or in the decay and regulation of specific mRNAs.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains used in this work are listed in Supplemental Table S2. The transformation procedure was as described (Gietz et al. 1992). rho0 and rho− strains were generated by ethidium bromide treatment of BMA38. The GAL1::HA-TRZ1 strain was constructed using a one-step PCR procedure (Longtine et al. 1998) by transforming cells with the PCR product amplified using the pFA6a-His3Mx6-pGAL1 plasmid and primers W303a and W303b. Deletion of REX1 in the trf4Δ strain was prepared by the same PCR strategy using the pAG32 plasmid (Goldstein and McCusker 1999) and primers Rex1HigF and Rex1HigR. The remaining mutants carrying the GAL1::HA-TRZ1 allele were made using genomic DNA from the GAL1::HA-TRZ1 strain as a template and primers W133 and W134. The ATG-frameshift mutant trz1-3 was generated using the pSE358-Trz1 plasmid obtained by subcloning the TRZ1 ORF together with upstream 1.1 kb and downstream 108 bp from plasmid p426-Trz1 (Chen et al. 2005) into pSE358 (Elledge and Davis 1988). The Quick Change Multi Site-Directed Mutagenesis Kit (Stratagene) with “ATG-frameshift” primer was used to insert a T, creating an additional ATG translation start codon 10 nt upstream of the original one and resulting in a frameshift for the protein translated from the new ATG. The sequence of the resulting clone was confirmed by sequencing, the plasmid was introduced into the YL03-47 strain with the disrupted chromosomal copy of TRZ1, and the p426-Trz1 with the wild-type TRZ1 copy was shuffled out by plating cells on 5-FOA medium. The temperature-sensitive phenotype of the resulting trz1-3 mutant was tested by growth at 37°C. Cells were grown at 25°C either in YPD or YPGal medium (1% yeast extract, 2% Bacto-peptone, 2% glucose, or 2% galactose, respectively). The strain containing a conditional temperature-sensitive trz1-3 allele was grown at 24°C in YPS medium (1% yeast extract, 2% Bacto-peptone, 2% sucrose) up to mid-exponential phase and transferred to 37°C. Depletion of Trz1 in the GAL::HA-TRZ1 strains was achieved by transfer of yeast cultures from YPGal to YPD medium. Growth curves of strains containing GAL::HA-TRZ1 or trz1-3 alleles were performed by transferring the cells from the permissive (YPGal or 24°C) to the restrictive (YPD or 37°C) conditions, and the growth was followed with maintaining the cells in exponential growth by dilution with prewarmed medium. At chosen time-points, cell aliquots were collected to isolate RNA samples.
RNA methods
Total RNA from yeast cells was isolated using a hot phenol procedure (Schmitt et al. 1990). The fraction of mitochondrial RNA was isolated from purified mitochondria according to Sperka-Gottlieb et al. (1988). Northern hybridization was essentially as described (Tollervey and Mattaj 1987). Eight micrograms of total RNA was separated on 8% denaturing polyacrylamide-urea gels, transferred onto nylon membranes, and hybridized. Oligonucleotide probes used are listed in Supplemental Table S1. Polyadenylated RNAs were isolated using the Poly(A) Purist Mag kit (Ambion). Five-tenths microgram of poly(A)+ RNA and 2 μg of total RNA were separated on 6% polyacrylamide gels. Deadenylation of RNA was performed as described (LaCava et al. 2005) using 20 μg of RNA that had been hybridized to 10 pM of oligo(dT)20.
Cell fractionation and Western blotting
Mitochondria were isolated as described (Daum et al. 1982; Sperka-Gottlieb et al. 1988). Cells were harvested by centrifugation, washed once with distilled water, suspended to a concentration of 0.5 g (wet weight)/mL in 0.1 M Tris-SO4, pH 9.4, 10 mM DTT, and incubated for 10 min at 30°C. The pellet was washed once with 1.2 M sorbitol and suspended in 1.2 M sorbitol, 20 mM KH2PO4, pH 7.4 to give 0.15 g (wet weight)/mL. Zymolyase 20T (final concentration 5 mg/g of cells, wet weight) was added, and the cells were spheroplasted at 30°C. Spheroplasts were harvested by centrifugation for 3 min at 3500 rpm and washed twice with 1.2 M sorbitol at room temperature, suspended in 0.6 M mannitol, 10 mM Tris-C1, pH 7.4, 0.1% bovine serum albumin, 1 mM PMSF (homogenization buffer) to a concentration of 0.15 g of spheroplasts (wet weight)/mL, chilled on ice, and homogenized by 10–15 strokes in a Potter homogenizer. From this point on, all operations were carried out at 4°C, and all buffers were ice-cold. The homogenate was diluted with one volume of the homogenization buffer and centrifuged for 5 min at 3500 rpm. The supernatant was collected, and the pellet was rehomogenized as before and recentrifuged at 3500 rpm. The supernatants were combined, and crude mitochondria were sedimented by centrifugation at 9000 rpm for 10 min. The pellet was resuspended in the homogenization buffer, and the suspension was centrifuged for 5 min at 4000 rpm to remove residual cell debris. The supernatant was saved and centrifuged at 9000 rpm for 10 min. The mitochondrial pellet was subjected to further analyses.
Mitochondria were fractionated as described (Tokatlidis 2000). Yeast mitochondria (corresponding to the yield of 100 μg of proteins) were resuspended in 200 μL of 20 mM Hepes, pH 7.4, vortexed, and incubated on ice for 30 min. Mitoplasts were collected by centrifugation for 5 min at 14,000g at 4°C, resuspended in 100 µL of 100 mM Na2CO3, 1 mM PMSF, and incubated for 30 min on ice for alkali extraction. The supernatant (matrix) and the pellet (membranes) were separated by centrifugation at 70,000 rpm (TL100 rotor) for 30 min. The matrix proteins were TCA-precipitated, and fractions were resolved by SDS-PAGE, transferred to nitrocellulose membranes, and analyzed by Western blotting.
Nuclei were prepared by lysis of spheroplasts in the presence of 18% Ficoll and enrichment by differential centrifugation, following the procedure based on Doi and Doi (1974); Dove et al. (1998). Yeast were spheroblasted as described above, resuspended in 20 mM KH2PO4, pH 6.45, 1.2 M sorbitol, 18% Ficoll 400, 0.5 M MgCl2, 1 mM PMSF, and homogenized in a Potter homogenizer. All operations were carried out at 4°C using ice-cold buffers. Cells were diluted to 20 mM KH2PO4, pH 6.45, 1.2 M sorbitol, 9% Ficoll 400, 0.5 M MgCl2, unbroken cells were removed by centrifugation at 4000 rpm for 5 min, and supernatant was centrifuged for 25 min at 10,000 rpm. The pellet containing nuclei was resuspended in 20 mM KH2PO4, pH 6.45, 0.3 M sucrose, 16.6% Ficoll 400, 0.5 M MgCl2, and loaded onto a discontinuous sucrose gradient (2, 1.8, 1.5, 1.3, and 1.2 M) in 20 mM KH2PO4, pH 6.45, 9% Ficoll 400, 0.5 M MgCl2, and centrifuged at 25,000 rpm (SW40 rotor) for 1 h. Interphase between 1.5 M and 1.8 M sucrose was collected, dialyzed to 20% Ficoll, 20 mM KH2PO4, pH 6.5, 1 mM MgCl2, and loaded onto a continuous Ficoll 400 gradient (50%-40%-30%) in 20 mM KH2PO4, pH 6.5, 1 mM MgCl2. Samples were centrifuged at 20,000 rpm (SW28 rotor) for 3 h. Nuclei pellets were suspended in the protein sample buffer and analyzed by SDS-PAGE and Western blotting.
Western blotting was performed using anti-HA antibody (Roche) to detect HA-Trz1; peroxidase-anti-peroxidase antibody (Sigma) to detect Trz1-TAP; specific monoclonal A66 antibody against Nop1 (Aris and Blobel 1988) and specific anti-Mdh1, anti-Hxk2 anti-Tom70, and anti-Tim23 antibodies. Anti-mouse or anti-rabbit horseradish peroxidase-conjugated antisera (Calbiochem) were used as secondary antibodies.
Cell staining and microscopy
Approximately 0.1 OD600 of cells was stained with 4′,6-diamidino-2-phenylindole (DAPI, final concentration 0.1 μg/mL) following a 2 min incubation in 70% ethanol at room temperature. Images were obtained with the epifluorescence Nikon Eclipse E800 microscope (100× objective) using the Hamamatsu digital CCD camera.
In vitro processing
Recombinant Trz1 was obtained as described using constructs pET32a-Trz1 (Schilling et al. 2005) and pET29a-Trz1, where TRZ1 ORF was subcloned into the pET29a vector. All tags (S, His, or thioredoxin) were removed using recombinant enterokinase (Novagen) in the case of pET32a-Trz1, whereas protein derived from pET29a-Trz1 includes the S-Tag. The 32P-labeled yeast tRNASer was transcribed in vitro with T7 polymerase using a PCR template generated using primers SceSer3 (carrying a T7 promoter) and SceSer2 and plasmid pM11-tRNACGA Ser (Olsen et al. 1981). The tRNASer contains a 19-nt intron. Processing reactions were performed as described (Kunzmann et al. 1998; Schilling et al. 2005) using 100 ng of the recombinant Trz1. The assay was performed in Trz1-buffer (50 mM Tris, pH 7.1, 5 mM KCl, 5 mM MgCl2, 2 mM DTT, pH 7.1). The location of the cleavage site on tRNASer was determined as described (Kunzmann et al. 1998) by primer extension with a SceSer2 primer.
Glutaraldehyde cross-linking
The protein derived from pET29a-Trz1 (1–2 µg) in a volume of 10 µL was incubated with 0.05% glutaraldehyde in a volume of 10 µL for 1 h at room temperature. The reaction was stopped with 1/10 volume 1 M lysine and subsequently analyzed on a 5% reducing SDS-PAGE. SDS-PAGE was either silver-stained or pET29a-Trz1 protein was detected by Western blotting with the S-protein HRP-conjugate (Novagen).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank David Tollervey (University of Edinburgh, UK) for antibodies against Nop1; Magdalena Boguta (IBB PAS, Warsaw) for antibodies against Mdh1 and Hxk2; Gottfried Schatz (University of Basel) for antibodies against Tim23 and Tom70; Roy Parker (University of Arizona) for rrp6Δ, rex1Δ, rex2Δ, rex3Δ, rex2Δ/rex3, rex1Δ/rex2Δ/rex3Δ, and rex2Δ/rrp6Δ mutants; Olga Puchta (IBB PAS, Warsaw) for the pet127Δ strain; Hildburg Beier (University of Würzburg) for the pM11-tRNASer(CGA) plasmid; and Yang Chen (Myriad Genetics, USA) for the p426-Trz1 plasmid. We also thank Karolina Labedzka (University of Warsaw), Maciej Szymanski (University of Adam Mickiewicz, Poznan), Sylvia Rösch, Sybille Apfel, and Elli Bruckbauer (Universität Ulm) for technical help and Bertrand Séraphin (IGBMC, Illkirch) for helpful discussions. This work was supported by the Wellcome Trust (to J.K.), Polish-Swiss Research Programme [PSPB-183/2010] (to J.K.), the Ministry of Science and Higher Education intramural BW through the Faculty of Biology, University of Warsaw (to E.S.), and the Fritz-Thyssen-Stiftung (to A.M.). Experiments were carried out with the use of CePT infrastructure financed by the European Union–the European Regional Development Fund (Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00).
REFERENCES
- Abruzzi K, Denome S, Olsen JR, Assenholt J, Haaning LL, Jensen TH, Rosbash M 2007. A novel plasmid-based microarray screen identifies suppressors of rrp6Δ in Saccharomyces cerevisiae. Mol Cell Biol 27: 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends S, Schön A 1997. Partial purification and characterization of nuclear ribonuclease P from wheat. Eur J Biochem 244: 635–645 [DOI] [PubMed] [Google Scholar]
- Aris JP, Blobel G 1988. Identification and characterization of a yeast nucleolar protein that is similar to a rat liver nucleolar protein. J Cell Biol 107: 17–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benschop JJ, Brabers N, van Leenen D, Bakker LV, van Deutekom HW, van Berkum NL, Apweiler E, Lijnzaad P, Holstege FC, Kemmeren P 2010. A consensus of core protein complex compositions for Saccharomyces cerevisiae. Mol Cell 38: 916–928 [DOI] [PubMed] [Google Scholar]
- Briggs MW, Burkard KT, Butler JS 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J Biol Chem 273: 13255–13263 [DOI] [PubMed] [Google Scholar]
- Brzezniak LK, Bijata M, Szczesny R, Stepien PP 2011. Human ELAC2/tRNAse Z is responsible for mitochondrial tRNA 3′ processing and acts subsequently to the RNase P endonucleolytic cleavage. RNA Biol 8: 616–626 [DOI] [PubMed] [Google Scholar]
- Castaño JG, Tobian JA, Zasloff M 1985. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase). J Biol Chem 260: 9002–9008 [PubMed] [Google Scholar]
- Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL 2003. A La protein requirement for efficient pre-tRNA folding. EMBO J 22: 6562–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Martin NC 1988. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3′-processing of tRNA precursors. J Biol Chem 263: 13677–13682 [PubMed] [Google Scholar]
- Chen Y, Beck A, Davenport C, Shattuck D, Tavtigian SV 2005. Characterization of TRZ1, a yeast homolog of the human candidate prostate cancer susceptibility gene ELAC2 encoding tRNase Z. BMC Mol Biol 6: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson T, Rabinowitz M 1983. Identification of multiple transcriptional initiation sites on the yeast mitochondrial genome by in vitro capping with guanylyltransferase. J Biol Chem 258: 14025–14033 [PubMed] [Google Scholar]
- Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL 2006. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 12: 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daoud R, Forget L, Lang BF 2012. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res 40: 1728–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum G, Böhni PC, Schatz G 1982. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Biol Chem 257: 13028–13033 [PubMed] [Google Scholar]
- de la Sierra-Gallay IL, Pellegrini O, Condon C 2005. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature 433: 657–661 [DOI] [PubMed] [Google Scholar]
- Doi K, Doi A 1974. Isolation of nuclei from a tetraploid strain of Saccharomyces cerevisiae. J Biochem 75: 1017–1026 [DOI] [PubMed] [Google Scholar]
- Dove JE, Brockenbrough JS, Aris JP 1998. Isolation of nuclei and nucleoli from the yeast Saccharomyces cerevisiae. Methods Cell Biol 53: 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Levinger L, Schiffer S, Marchfelder A 2004. Drosophila RNase Z processes mitochondrial and nuclear pre-tRNA 3′ ends in vivo. Nucleic Acids Res 32: 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A, Malewicz M, Minczuk M, Golik P, Dmochowska A, Stepien PP 1998. The yeast nuclear gene DSS1, which codes for a putative RNase II, is necessary for the function of the mitochondrial degradosome in processing and turnover of RNA. Mol Gen Genet 260: 108–114 [DOI] [PubMed] [Google Scholar]
- Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP 2003. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem 278: 1603–1611 [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Davis RW 1988. A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene 70: 303–312 [DOI] [PubMed] [Google Scholar]
- Ellis TP, Schonauer MS, Dieckmann CL 2005. CBT1 interacts genetically with CBP1 and the mitochondrially encoded cytochrome b gene and is required to stabilize the mature cytochrome b mRNA of Saccharomyces cerevisiae. Genetics 171: 949–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke DR, Gegenheimer P, Abelson J 1985. Nucleolytic processing of a tRNAArg-tRNAAsp dimeric precursor by a homologous component from Saccharomyces cerevisiae. J Biol Chem 260: 1271–1279 [PubMed] [Google Scholar]
- Evans D, Marquez SM, Pace NR 2006. RNase P: Interface of the RNA and protein worlds. Trends Biochem Sci 31: 333–341 [DOI] [PubMed] [Google Scholar]
- Fekete Z, Ellis TP, Schonauer MS, Dieckmann CL 2008. Pet127 governs a 5′→3′-exonuclease important in maturation of apocytochrome b mRNA in Saccharomyces cerevisiae. J Biol Chem 283: 3767–3772 [DOI] [PubMed] [Google Scholar]
- Foury F, Roganti T, Lecrenier N, Purnelle B 1998. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440: 325–331 [DOI] [PubMed] [Google Scholar]
- Frendewey D, Dingermann T, Cooley L, Söll D 1985. Processing of precursor tRNAs in Drosophila. Processing of the 3′ end involves an endonucleolytic cleavage and occurs after 5′ end maturation. J Biol Chem 260: 449–454 [PubMed] [Google Scholar]
- Furter R, Snaith M, Gillespie DE, Hall BD 1992. Endonucleolytic cleavage of a long 3′-trailer sequence in a nuclear yeast suppressor tRNA. Biochemistry 31: 10817–10824 [DOI] [PubMed] [Google Scholar]
- Garber RL, Gage LP 1979. Transcription of a cloned Bombyx mori tRNA2 Ala gene: Nucleotide sequence of the tRNA precursor and its processing in vitro. Cell 18: 817–828 [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH 1992. Improved method for high efficient transformation of intact yeast cells. Nucleic Acids Res 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AL, McCusker JH 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553 [DOI] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48: 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenbuchle O, Larson D, Hall GI, Sprague KU 1979. The primary transcription product of a silkworm alanine tRNA gene: Identification of in vitro sites of initiation, termination and processing. Cell 18: 1217–1229 [DOI] [PubMed] [Google Scholar]
- Han SJ, Kang HS 1997. Purification and characterization of the precursor tRNA 3′-end processing nuclease from Aspergillus nidulans. Biochem Biophys Res Commun 233: 354–358 [DOI] [PubMed] [Google Scholar]
- Hanekamp T, Thorsness PE 1999. YNT20, a bypass suppressor of yme1 yme2, encodes a putative 3′-5′ exonuclease localized in mitochondria of Saccharomyces cerevisiae. Curr Genet 34: 438–448 [DOI] [PubMed] [Google Scholar]
- Hartmann RK, Gössringer M, Späth B, Fischer S, Marchfelder A 2009. The making of tRNAs and more—RNase P and tRNase Z. Prog Mol Biol Transl Sci 85: 319–368 [DOI] [PubMed] [Google Scholar]
- Hazbun TR, Malmstrom L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol Cell 12: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Hiley SL, Babak T, Hughes TR 2005. Global analysis of yeast RNA processing identifies new targets of RNase III and uncovers a link between tRNA 5′ end processing and tRNA splicing. Nucleic Acids Res 33: 3048–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Shaheen HH 2008. A decade of surprises for tRNA nuclear-cytoplasmic dynamics. Trends Cell Biol 18: 98–104 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ishii R, Minagawa A, Takaku H, Takagi M, Nashimoto M, Yokoyama S 2005. Crystal structure of the tRNA 3′ processing endoribonuclease tRNase Z from Thermotoga maritima. J Biol Chem 280: 14138–14144 [DOI] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Tollervey D 2003. 3′-processing of yeast tRNATrp precedes 5′-processing. RNA 9: 202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Agarwal S, Heyman JA, Matson S, Heidtman M, Piccirillo S, Umansky L, Drawid A, Jansen R, Liu Y, et al. 2002. Subcellular localization of the yeast proteome. Genes Dev 16: 707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunzmann A, Brennicke A, Marchfelder A 1998. 5′ end maturation and RNA editing have to precede tRNA 3′ processing in plant mitochondria. Proc Natl Acad Sci 95: 108–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724 [DOI] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR 1991. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae RNase P. Mol Cell Biol 11: 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 1996. Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell 86: 503–512 [DOI] [PubMed] [Google Scholar]
- Li Z, Deutscher MP 2002. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA 8: 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski KA, Kaniak-Golik A, Golik P 2010. Maintenance and expression of the S. cerevisiae mitochondrial genome—from genetics to evolution and systems biology. Biochim Biophys Acta 1797: 1086–1098 [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Séraphin B, Tollervey D 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev 8: 1423–1433 [DOI] [PubMed] [Google Scholar]
- Manam S, Van Tuyle GC 1987. Separation and characterization of 5′- and 3′-tRNA processing nucleases from rat liver mitochondria. J Biol Chem 262: 10272–10279 [PubMed] [Google Scholar]
- Martin NC, Hopper AK 1994. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie 76: 1161–1167 [DOI] [PubMed] [Google Scholar]
- Mayer M, Schiffer S, Marchfelder A 2000. tRNA 3′ processing in plants: Nuclear and mitochondrial activities differ. Biochemistry 39: 2096–2105 [DOI] [PubMed] [Google Scholar]
- Mörl M, Marchfelder A 2001. The final cut. The importance of tRNA 3′-processing. EMBO Rep 2: 17–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers AM, Pape LK, Tzagoloff A 1985. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J 4: 2087–2092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M 1997. Distribution of both lengths and 5′ terminal nucleotides of mammalian pre-tRNA 3′ trailers reflects properties of 3′ processing endoribonuclease. Nucleic Acid Res 25: 1148–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto M, Wesemann DR, Geary S, Tamura M, Kaspar RL 1999. Long 5′ leaders inhibit removal of a 3′ trailer from a precursor tRNA by mammalian tRNA 3′ processing endoribonuclease. Nucleic Acids Res 27: 2770–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neupert W 1997. Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- O'Connor JP, Peebles CL 1991. In vivo pre-tRNA processing in Saccharomyces cerevisiae. Mol Cell Biol 11: 425–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen I, Dean MF, Harris G, Muir H 1981. Direct transfer of a lysosomal enzyme from lymphoid cells to deficient fibroblasts. Nature 291: 244–247 [DOI] [PubMed] [Google Scholar]
- Oommen A, Li XQ, Gegenheimer P 1992. Cleavage specificity of chloroplast and nuclear tRNA 3′-processing nucleases. Mol Cell Biol 12: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow MC, Kushner SR 2002. Initiation of tRNA maturation by RNase E is essential for cell viability in E. coli. Genes Dev 16: 1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozanick SG, Wang X, Costanzo M, Brost RL, Boone C, Anderson JT 2009. Rex1p deficiency leads to accumulation of precursor initiator tRNAMet and polyadenylation of substrate RNAs in Saccharomyces cerevisiae. Nucleic Acids Res 37: 298–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadimitriou A, Gross HJ 1996. Pre-tRNA 3′-processing in Saccharomyces cerevisiae. Purification and characterization of exo- and endoribonucleases. Eur J Biochem 242: 747–759 [DOI] [PubMed] [Google Scholar]
- Pellegrini O, Nezzar J, Marchfelder A, Putzer H, Condon C 2003. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis. EMBO J 22: 4534–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perwez T, Kushner SR 2006. RNase Z in Escherichia coli plays a significant role in mRNA decay. Mol Microbiol 60: 723–737 [DOI] [PubMed] [Google Scholar]
- Rooney RJ, Harding JD 1986. Processing of mammalian tRNA transcripts in vitro: Different pre-tRNAs are processed along alternative pathways that contain a common rate-limiting step. Nucleic Acids Res 14: 4849–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer B 2005. RNA maturation in mitochondria of S. cerevisiae and S. pombe. Gene 354: 80–85 [DOI] [PubMed] [Google Scholar]
- Schierling K, Rosch S, Rupprecht R, Schiffer S, Marchfelder A 2002. tRNA 3′ end maturation in archaea has eukaryotic features: The RNase Z from Haloferax volcanii. J Mol Biol 316: 895–902 [DOI] [PubMed] [Google Scholar]
- Schiffer S, Rösch S, Marchfelder A 2002. Assigning a function to a conserved group of proteins: The tRNA 3′-processing enzymes. EMBO J 21: 2769–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling O, Spath B, Kostelecky B, Marchfelder A, Meyer-Klaucke W, Vogel A 2005. Exosite modules guide substrate recognition in the ZiPD/ElaC protein family. J Biol Chem 280: 17857–17862 [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D 2012. Transcriptome-wide analysis of exosome targets. Mol Cell 48: 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonauer MS, Kastaniotis AJ, Hiltunen JK, Dieckmann CL 2008. Intersection of RNA processing and the type II fatty acid synthesis pathway in yeast mitochondria. Mol Cell Biol 28: 6646–6657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Späth B, Kirchner S, Vogel A, Schubert S, Meinlschmidt P, Aymanns S, Nezzar J, Marchfelder A 2005. Analysis of the functional modules of the tRNA 3′ endonuclease (tRNase Z). J Biol Chem 280: 35440–35447 [DOI] [PubMed] [Google Scholar]
- Späth B, Canino G, Marchfelder A 2007. tRNase Z: The end is not in sight. Cell Mol Life Sci 64: 2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka-Gottlieb CD, Hermetter A, Paltauf F, Daum G 1988. Lipid topology and physical properties of the outer mitochondrial membrane of the yeast, Saccharomyces cerevisiae. Biochim Biophys Acta 946: 227–234 [DOI] [PubMed] [Google Scholar]
- Stange N, Beier H 1987. A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J 6: 2811–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154 [DOI] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Sulo P, Dang YL, Martin NC 1996. Yeast mitochondrial RNase P RNA synthesis is altered in an RNase P protein subunit mutant: Insights into the biogenesis of a mitochondrial RNA-processing enzyme. Mol Cell Biol 16: 3429–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Sulo P, Ellis SR, Martin NC 2001. Rpm2p: Separate domains promote tRNA and Rpm1r maturation in Saccharomyces cerevisiae mitochondria. Nucleic Acids Res 29: 3631–3637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T 2005. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 309: 140–142 [DOI] [PubMed] [Google Scholar]
- Tavtigian SV, Simard J, Teng DH, Abtin V, Baumgard M, Beck A, Camp NJ, Carillo AR, Chen Y, Dayananth P, et al. 2001. A candidate prostate cancer susceptibility gene at chromosome 17p. Nat Genet 27: 172–180 [DOI] [PubMed] [Google Scholar]
- Tokatlidis K 2000. Directing proteins to mitochondria by fusion to mitochondrial targeting signals. Methods Enzymol 327: 305–317 [DOI] [PubMed] [Google Scholar]
- Tollervey D, Mattaj IW 1987. Fungal small nuclear ribonucleoproteins share properties with plant and vertebrate U-snRNPs. EMBO J 6: 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM 1986. Genetics of mitochondrial biogenesis. Annu Rev Biochem 55: 249–285 [DOI] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000a. Three conserved members of the RNase D family have unique and overlapping functions in the processing of 5S, 5.8S, U4, U5, RNase MRP and RNase P RNAs in yeast. EMBO J 19: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Lennertz P, Parker R 2000b. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol Cell Biol 20: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn DJ, Yoo CJ, Xue D, Shi H, Wolin SL 1997. The La protein in Schizosaccharomyces pombe: A conserved yet dispensable phosphoprotein that functions in tRNA maturation. RNA 3: 1434–1443 [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent RD, Hofmann TJ, Zassenhaus HP 1988. Sequence and expression of NUC1, the gene encoding the mitochondrial nuclease in Saccharomyces cerevisiae. Nucleic Acids Res 16: 3297–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel A, Schilling O, Spath B, Marchfelder A 2005. The tRNase Z family of proteins: Physiological functions, substrate specificity and structural properties. Biol Chem 386: 1253–1264 [DOI] [PubMed] [Google Scholar]
- Wegierski T, Dmochowska A, Jablonowska A, Dziembowski A, Bartnik E, Stepien PP 1998. Yeast nuclear PET127 gene can suppress deletions of the SUV3 or DSS1 genes: An indication of a functional interaction between 3′ and 5′ ends of mitochondrial mRNAs. Acta Biochim Pol 45: 935–940 [PubMed] [Google Scholar]
- Wen T, Oussenko IA, Pellegrini O, Bechhofer DH, Condon C 2005. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis. Nucleic Acid Res 33: 3636–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK 2007. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 18: 2678–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenberger G, Fox TD 1997. Pet127p, a membrane associated protein involved in stability and processing of Saccharomyces cerevisiae mitochondrial RNAs. Mol Cell Biol 17: 2816–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737 [DOI] [PubMed] [Google Scholar]
- Yoo CJ, Wolin SL 1997. The yeast La protein is required for the 3′ endonucleolytic cleavage that matures tRNA precursors. Cell 89: 393–402 [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T 2003. Possibility of cytoplasmic pre-tRNA splicing: The yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T 2007. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 12: 285–297 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhao Q, Huang Y 2013. Partitioning of the nuclear and mitochondrial tRNA 3′-end processing activities between two different proteins in Schizosaccharomyces pombe. J Biol Chem 288: 27415–27422 [DOI] [PMC free article] [PubMed] [Google Scholar]