Abstract
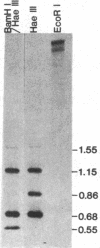
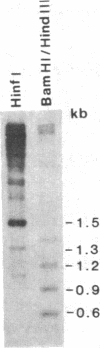
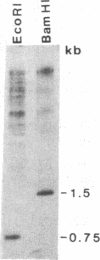
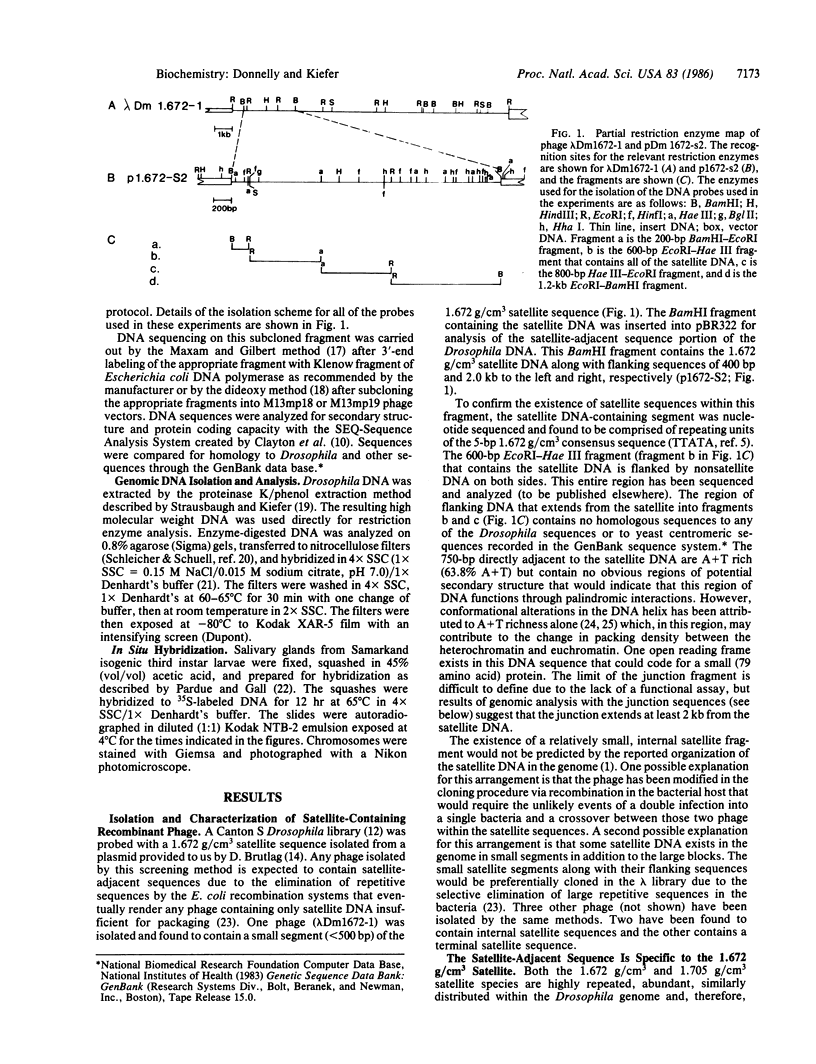
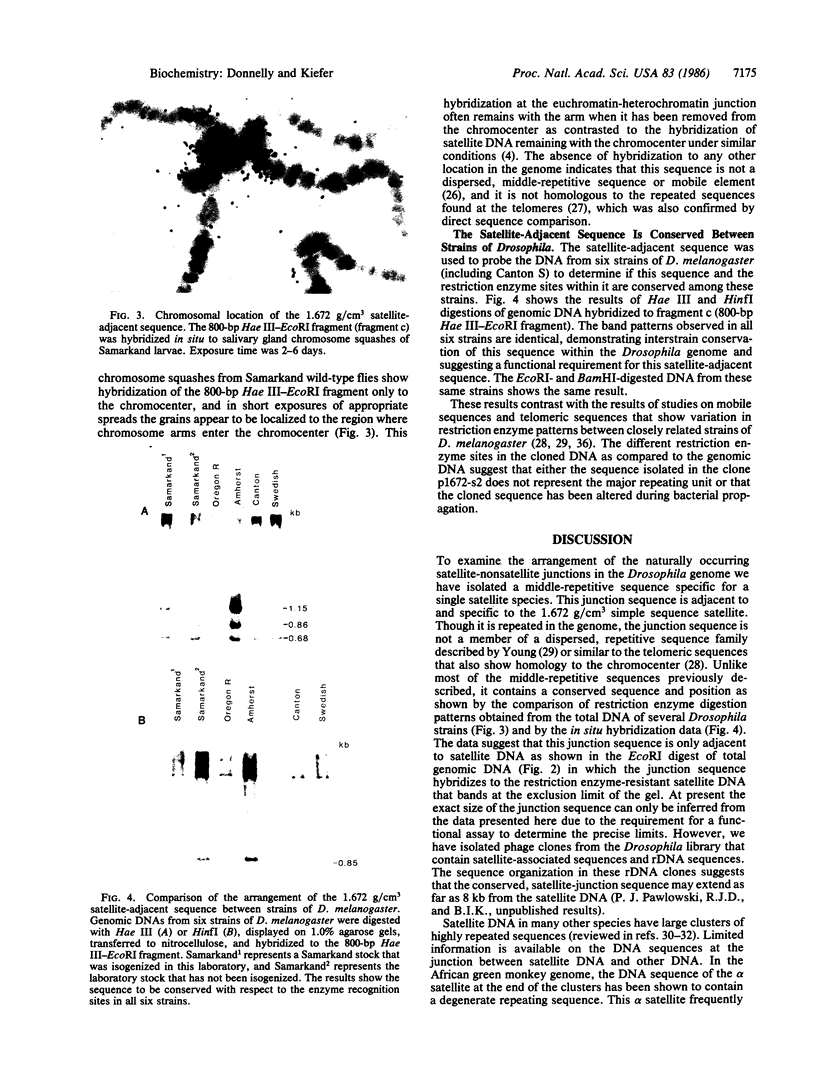
The Drosophila genome contains nearly 2.8 X 10(4) kilobases of satellite DNA. This simple sequence satellite DNA is contained within transcriptionally inactive heterochromatin that is distributed among all chromosomes with a concentration at the centromeres and along the length of the Y chromosome. To investigate the relationship of the satellite DNA with the surrounding sequences, we have isolated a satellite junction sequence that is repetitive and specifically adjacent to the 1.672 g/cm3 satellite DNA. It is conserved between strains of Drosophila melanogaster and localized to the chromocenter of polytene chromosomes. The characteristics of this sequence suggest a functional role involving the specific organization of large regions of chromosomes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Carlson M., Fry K., Hsieh T. S. DNA sequence organization in Drosophila heterochromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1137–1146. doi: 10.1101/sqb.1978.042.01.114. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Fry K., Nelson T., Hung P. Synthesis of hybrid bacterial plasmids containing highly repeated satellite DNA. Cell. 1977 Mar;10(3):509–519. doi: 10.1016/0092-8674(77)90038-1. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Peacock W. J. Sequences of the 1.672 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol. 1979 Dec 15;135(3):565–580. doi: 10.1016/0022-2836(79)90164-5. [DOI] [PubMed] [Google Scholar]
- Carlson M., Brutlag D. Cloning and characterization of a complex satellite DNA from Drosophila melanogaster. Cell. 1977 Jun;11(2):371–381. doi: 10.1016/0092-8674(77)90054-x. [DOI] [PubMed] [Google Scholar]
- Carlson M., Brutlag D. One of the copia genes is adjacent to satellite DNA in Drosophila melanogaster. Cell. 1978 Nov;15(3):733–742. doi: 10.1016/0092-8674(78)90259-3. [DOI] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Finnegan D. J., Rubin G. M., Young M. W., Hogness D. S. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- Fry K., Brutlag D. Detection and resolution of closely related satellite DNA sequences by molecular cloning. J Mol Biol. 1979 Dec 15;135(3):581–593. doi: 10.1016/0022-2836(79)90165-7. [DOI] [PubMed] [Google Scholar]
- Hilliker A. J., Appels R., Schalet A. The genetic analysis of D. melanogaster heterochromatin. Cell. 1980 Oct;21(3):607–619. doi: 10.1016/0092-8674(80)90424-9. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. L. A protein that preferentially binds Drosophila satellite DNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):726–730. doi: 10.1073/pnas.76.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinger L., Varshavsky A. Protein D1 preferentially binds A + T-rich DNA in vitro and is a component of Drosophila melanogaster nucleosomes containing A + T-rich satellite DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7152–7156. doi: 10.1073/pnas.79.23.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Maresca A., Singer M. F. Deca-satellite: a highly polymorphic satellite that joins alpha-satellite in the African green monkey genome. J Mol Biol. 1983 Mar 15;164(4):493–511. doi: 10.1016/0022-2836(83)90047-5. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. A bent helix in kinetoplast DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):279–283. doi: 10.1101/sqb.1983.047.01.033. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Dawid I. B. Chromosomal locations of two DNA segments that flank ribosomal insertion-like sequences in Drosophila: flanking sequences are mobile elements. Chromosoma. 1981;83(1):29–43. doi: 10.1007/BF00286014. [DOI] [PubMed] [Google Scholar]
- Pardue M. L., Gall J. G. Nucleic acid hybridization to the DNA of cytological preparations. Methods Cell Biol. 1975;10:1–16. doi: 10.1016/s0091-679x(08)60727-x. [DOI] [PubMed] [Google Scholar]
- Peacock W. J., Lohe A. R., Gerlach W. L., Dunsmuir P., Dennis E. S., Appels R. Fine structure and evolution of DNA in heterochromatin. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1121–1135. doi: 10.1101/sqb.1978.042.01.113. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. F. Highly repeated sequences in mammalian genomes. Int Rev Cytol. 1982;76:67–112. doi: 10.1016/s0074-7696(08)61789-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Spradling A. C., Rubin G. M. Drosophila genome organization: conserved and dynamic aspects. Annu Rev Genet. 1981;15:219–264. doi: 10.1146/annurev.ge.15.120181.001251. [DOI] [PubMed] [Google Scholar]
- Steffensen D. M., Appels R., Peacock W. J. The distribution of two highly repeated DNA sequences within Drosophila melanogaster chromosomes. Chromosoma. 1981;82(4):525–541. doi: 10.1007/BF00295011. [DOI] [PubMed] [Google Scholar]
- Strausbaugh L. D., Kiefer B. I. Genetic modulation of RNA metabolism in Drosophila. III. Requirement for an rDNA-deficient X chromosome in YbbSuVar-3-mediated increases in RNA synthesis. Genetics. 1979 Oct;93(2):411–422. doi: 10.1093/genetics/93.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel E., Dunsmuir P., Rubin G. M. Polymorphisms in the chromosomal locations of elements of the 412, copia and 297 dispersed repeated gene families in Drosophila. Cell. 1979 Jun;17(2):429–439. doi: 10.1016/0092-8674(79)90169-7. [DOI] [PubMed] [Google Scholar]
- Tartof K. D., Hobbs C., Jones M. A structural basis for variegating position effects. Cell. 1984 Jul;37(3):869–878. doi: 10.1016/0092-8674(84)90422-7. [DOI] [PubMed] [Google Scholar]
- Truett M. A., Jones R. S., Potter S. S. Unusual structure of the FB family of transposable elements in Drosophila. Cell. 1981 Jun;24(3):753–763. doi: 10.1016/0092-8674(81)90101-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Miklos G. L. Genetic dissection of heterochromatin in Drosophila: the role of basal X heterochromatin in meiotic sex chromosome behaviour. Chromosoma. 1977 Apr 19;60(3):283–296. doi: 10.1007/BF00329776. [DOI] [PubMed] [Google Scholar]
- Young B. S., Pession A., Traverse K. L., French C., Pardue M. L. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983 Aug;34(1):85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
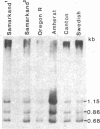
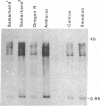
- Young M. W. Middle repetitive DNA: a fluid component of the Drosophila genome. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6274–6278. doi: 10.1073/pnas.76.12.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]