Abstract
The proteasome has been implicated in gene transcription through a variety of mechanisms. How the proteasome regulates genome-wide transcription in relation to nutrient signalling pathways is largely unknown. Using chemical inhibitors to compromise the functions of the proteasome and/or TORC1, we reveal that the proteasome and TORC1 synergistically promote the expression of de novo purine and amino acid biosynthetic genes, and restrict the transcription of those associated with proteolysis, starvation and stress responses. Genetic analysis demonstrates that TORC1 negatively regulates both the Yak1 and Rim15 kinases to modulate starvation-specific gene expression mediated by the Msn2/4 and Gis1 transcription factors. Compromising proteasome function induces starvation-specific gene transcription in exponential-phase cells and abrogates the strict control of such expression by Yak1 and Rim15 in rapamycin-treated cells, confirming that the proteasome functions to ensure stringent control of the starvation response by the TOR pathway. Synergy between the two pathways is also exhibited on cell growth control. Rpn4-dependent upregulation of proteasomal genes and a catalytically competent 20S proteasome are essential for yeast cells to respond to reduced TORC1 activity. These data suggest that the proteasome and the TOR signalling pathway synergistically regulate a significant portion of the genome to coordinate cell growth and starvation response.
Keywords: proteasome, TORC1, starvation response, Yak1, Rim15, Rpn4
2. Introduction
Most non-lysosomal/vacuolar protein degradation is carried out by the proteasome in cytosolic and nuclear compartments of eukaryotic cells. Proteasome-mediated degradation, a highly regulated process, can be ubiquitin-dependent or ubiquitin-independent [1,2]. Recently, the 26S proteasome and its subcomplexes have been implicated in the regulation of gene transcription through a variety of mechanisms, including transcription factor (TF) processing and chromatin association (recently reviewed in [3–5]). Processing by the proteasome can restrict the steady-state levels of a TF to limit transcription activation [6–8], convert a TF into a functional state or change its location via limited proteolysis to activate transcription [9–10].
In a number of cases, including Gcn4, and perhaps Gal4 and Ino2/4 [11], proteasome-mediated degradation of these TFs is necessary to stimulate transcription. Inhibiting the proteasome function increases the abundance of these TFs but decreases their transcription activation capabilities. Although the detailed mechanism is not fully understood, it is proposed that these TFs are marked as ‘spent’ after transcription initiation, trapped with the chromatin and unable to stimulate new rounds of transcription. Proteasome-mediated proteolysis destroys such TFs, resets the promoter and allows ‘fresh’ activators to initiate a new round of transcription [5].
Apart from processing TFs directly, the proteasome or its subcomplexes have been shown to associate with chromatin to restrict permissive transcription [12] or to promote transcription initiation, elongation or termination [13–17]. Association of the proteasome or its subcomplexes with chromatin is widespread in the yeast genome [18,19]. Furthermore, the proteasome could function at multiple levels within the same pathway, such as processing TFs into an active state and associating with TF target genes to promote transcription [18].
Although the proteasome has been demonstrated to regulate transcription of many genes, how the proteasome modulates genome-wide transcription in relation to nutrient signalling pathways is largely unknown. Several studies in yeast have examined the transcriptional effects of chemical inhibition of proteasome function [20,21]. Treatment with proteasome inhibitors leads to transcriptional downregulation of genes involved in mating, amino acid metabolism and protein synthesis. Among the genes activated are those implicated in protein degradation and stress response. Previously, we have demonstrated that the post-diauxic shift (PDS) TF Gis1 is subjected to proteasome-mediated proteolysis to downregulate its transcription activation capability [8]. Transcription of PDS genes is moderately activated by the proteasome inhibitor (MG132), significantly induced by rapamycin treatment and hyperactivated by treatment with both drugs, suggesting that the proteasome and TORC1 cooperate to modulate PDS gene transcription. Here, we extend the study to the transcriptome level, and reveal that the proteasome and the TOR signalling pathway synergistically regulate transcription of a significant portion of the genome. Bioinformatic, genetic and phenotypic analyses shed new insights into how the two pathways cooperate in gene transcription, cell growth and starvation response.
3. Material and methods
3.1. Strains, plasmids and culture conditions
Yeast deletion strains, generated by Saccharomyces Genome Deletion Project [22], or decreased abundance by mRNA perturbation (DAmP) strains bearing hypomorphic alleles of essential genes made by Breslow et al. [23], were obtained from Open Biosystems. Deletion of RPN4 in pdr5Δ::kanMX4 cells was achieved with HIS3MX6 marker as described previously [24]. The C-terminus of RPN4 was similarly tagged with polyhistidine (6-His) at its genomic locus in the pdr5Δ::kanMX4 cells. Isogenic strains bearing deletions of msn2Δmsn4Δ (msn2/4Δ); gis1Δ; msn2/4Δgis1Δ; rim15Δ; yak1Δ or rim15Δyak1Δ were constructed in the pdr5Δ::HIS3MX6 cells. Overexpression of MSN2 was achieved by placing the MSN2 coding sequence under the control of the tetO7 promoter in pCM190, as previously described for GIS1 overexpression [8]. Yeast extract peptone dextrose (YPD) or supplemented minimal medium (SMM) was used throughout the study. A stock solution (1 mg ml−1) of rapamycin (Sigma) was made up in 90 per cent (v/v) ethanol and 10 per cent (v/v) Tween-20. MG132 (50 mM; Sigma) was prepared in absolute ethanol. Working concentrations were 200 ng ml−1 for rapamycin and 50 µM for MG132 unless otherwise specified.
3.2. Microarray analysis
The pdr5Δ::kanMX4 cells (isogenic to BY4742) were grown to early/mid-exponential phase (OD600 ∼ 0.4) in SMM medium. Cultures were split into four flasks, into which the drug vehicle, rapamycin, MG132 or both drugs were added. Samples were taken at 0, 1, 2 and 3 h after treatment. Total RNA was isolated from cultures as described previously [25]. Genome-wide transcription profiling was carried out using the Yeast2 oligonucleotide arrays (Affymetrix Inc.) according to the manufacturer's instructions. Normalization and statistical analyses of the data generated from three biological replicates were performed using Partek Genomics software (http://www.partek.com) as described previously [8,26]. The average correlation coefficients between the triplicate microarray experiments were between 0.983 and 0.994. In compliance with MIAME guidelines, the data from this study have been deposited in the ArrayExpress repository (http://www.ebi.ac.uk/arrayexpress) at the EBI under accession no. E-MTAB-1550.
3.3. Analysis of individual transcripts
Total RNA (20 µg) from each sample was used for analyses of individual transcript levels, following the procedures described by Engler-Blum et al. [27]. ACT1 was used as the loading control for all samples. Labelled probes were made using Rediprime II DNA labelling system (GE Healthcare). Phosphoimages were scanned using a Typhoon 9000 imager and analysed using ImageQuant TL software (GE Healthcare). Care was taken to limit the exposure time (typically 8–12 h) to ensure that hybridization signals were not saturated. Hybridization signals from the target transcripts were normalized against that of ACT1 for each sample. The levels of the transcripts in the wild-type cells at time 0 h of drug treatment were set to the arbitrary unit 1.
3.4. Western analysis
Anti-myc (Sigma) and anti-tubulin (Cancer Research, UK) antibodies were used in Western analysis to detect the levels of Rpn4-myc and tubulin, respectively, following the protocol described previously [8].
3.5. GIS1 and MSN2 overexpression assays
Yeast transformants bearing the empty vector (pCM190) or the overexpression plasmid (tetO7-GIS1 or tetO7-MSN2) were grown in SMM medium under repressive conditions (plus 20 µg ml−1 of doxycycline) to mid-exponential phase. In order to determine the toxic effects of GIS1 or MSN2 overexpression on cell growth, cells were harvested, washed and resuspended in water to the same density, and spotted in serial dilutions (sevenfold) on SMM medium plates containing 20 µg ml−1 of doxycycline (Dox+) or no doxycycline (Dox−). Cells grown on glucose (2%) were incubated at 30°C for 2 days, and those grown on ethanol (2% v/v) and glycerol (1% v/v) for 7 days.
3.6. Determination of growth rates of proteasomal mutants
Continuous monitoring of cell growth in quadruplicate was carried out using a plate reader (BMG Biotech). To determine the effects of a drug on cell growth, the doubling time of cells grown in medium containing the drug was normalized against that of the same cells grown in the presence of the drug vehicle. Working concentrations were 50 ng ml−1 for rapamycin and 12.5 µM for MG132.
4. Results
4.1. TORC1 and proteasome synergistically regulate the transcription of a significant portion of the genome
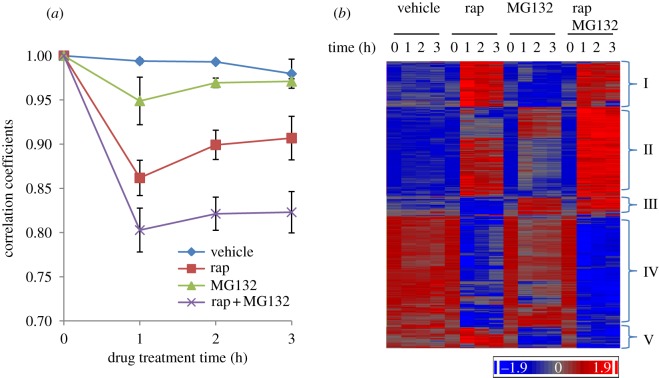
Our previous study [8] indicated that PDS gene transcription mediated by the Gis1 TF is coordinately modulated by the functions of the proteasome and TORC1. To find the extent to which the proteasome and TORC1 cooperate with regulate gene transcription, we treated exponentially growing pdr5Δ cells with the drug vehicle, rapamycin (TORC1 inhibitor), MG132 (the proteasome inhibitor) or both drugs. Samples from biological triplicates were taken at 0, 1, 2 and 3 h post-treatment and microarray experiments carried out using Yeast2 arrays. Transcriptome data analysis was performed using Partek Genomics software, and the results are summarized in electronic supplementary material S1. Comparison of transcriptome data at time 0 with those from subsequent time points revealed that treatment with the drug vehicle did not cause significant changes of whole-genome transcription (figure 1a). Genome-wide transcription was more significantly altered in cells treated with rapamycin than in cells treated with MG132. Addition of both rapamycin and MG132 triggered a more dramatic change in transcriptome than either drug alone (figure 1a).
Figure 1.
(a) Change of correlation coefficients of the transcriptome during drug treatment. These coefficients were calculated between the initial sample and those 1, 2 and 3 h after introduction of the drug. The mean and s.d. were calculated from biological triplicates. (b) Hierarchical clustering of genes whose mean transcript levels were changed more than 1.5-fold by drug treatment.
Out of the 5716 genes detectable with Yeast2 chips, the transcript levels of 3220 open reading frames (ORFs) are changed more than 1.5-fold (p < 0.01) by treatment with rapamycin and/or MG132. Among them, the transcription of 1028 ORFs is regulated more than 1.5-fold by MG132 treatment, contrasting with those of 2565 ORFs similarly altered in rapamycin-treated cells. Comparison of our data with other recent studies revealed that around 70 per cent of the rapamycin-induced genes and 60 per cent of the rapamycin-repressed ORFs were similarly regulated by rapamycin treatment of yeast cells of a different genetic background. By contrast, among the ORFs significantly regulated by MG132 treatment, 42.5 per cent of the upregulated and only 14.5 per cent of the downregulated genes were seen to overlap with those revealed by Dembla-Rajpal et al. [20]. Poorer overlapping between the MG132-repressed gene sets was possibly due to different methodologies used to interrogate the transcriptome in the two studies (Affymetrix genechips versus gene filters by Dembla-Rajpal et al.). An approximately equal number of genes was shown up- or downregulated by MG132 treatment in our study, whereas for Dembla-Rajpal et al. [20] the number of the downregulated ORFs was about one-third of that of the upregulated genes in cells treated with MG132 for 120 min.
Agglomerative hierarchical clustering of the 3220 regulated genes revealed five major clusters (figure 1b). The majority of these genes fall into two clusters: II and IV. Transcription of cluster II genes is activated by rapamycin or MG132 treatment and shows an even greater increase when cells are treated with both drugs. By contrast, the transcript levels of cluster IV genes are significantly decreased in rapamycin-treated cells, and either moderately reduced or barely changed by MG132 treatment. Addition of both drugs led to a more profound decrease in their expression than either drug alone; this was especially true after prolonged drug treatment (figure 1b). Expression of cluster I genes seems to be predominantly activated by rapamycin treatment. Opposite effects of rapamycin and MG132 on transcription are observed in two small clusters (III and V). These data suggest that, despite their independent roles in transcription regulation, TORC1 and the proteasome function synergistically to regulate a significant portion of the yeast transcriptome.
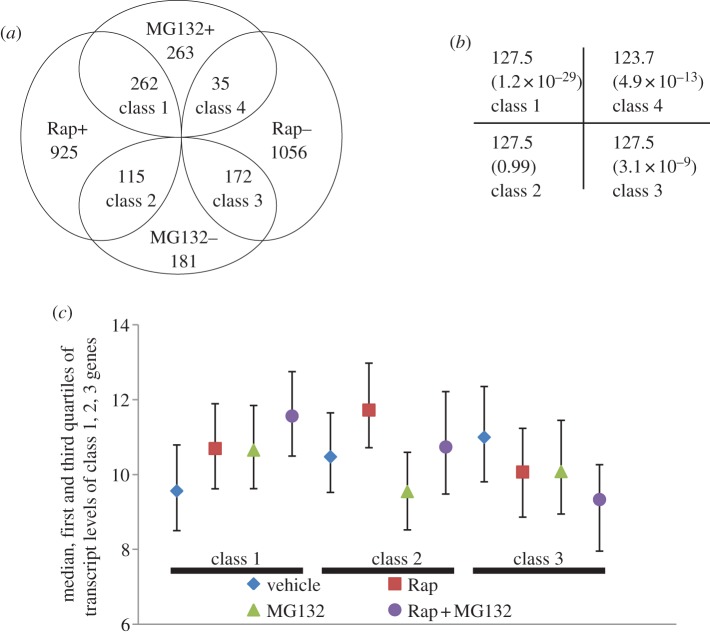
The ORFs that were regulated more than 1.5-fold by both drug treatments were indentified (figure 2a; electronic supplementary material S2). The expected number of each class of overlapping genes and their p-values (shown in brackets) were calculated by chi-squared test (figure 2b). Significantly over-represented were those genes whose transcription was induced (Rap+/MG132+, class 1) or repressed (Rap−/MG132−, class 3) by treatment with either drug. Transcript levels of these genes were more profoundly increased (class 1) or decreased (class 3) in cells treated with both drugs (figure 2c). Rapamycin-induced transcription of a number of genes was downregulated by treatment with the proteasome inhibitor (Rap+/MG132−, class 2; figure 2a,c). The actual number of genes in this category is, however, very close to the expected value (figure 2b). Strikingly, significantly under-represented are those genes whose transcription was repressed by TORC1 inhibition but induced by proteasome inhibition (Rap−/MG132+, class 4; figure 2a,b). Genes whose transcription is downregulated when TORC1 function is inhibited are inferred to be under the positive control of the complex in nutrient-sufficient conditions [29]. By contrast, genes whose transcription is activated in the presence of the proteasome inhibitor can be inferred to be under the negative control of the proteasome. These data suggested that the proteasome and TORC1 pathways do not tend to act antagonistically to regulate gene transcription (class 4) in nutrient-replete cells and that the two pathways function homo-directionally to modulate the transcription of a significant number of genes in the genome (classes 1 and 3).
Figure 2.
(a) The number of genes whose mean transcript level changed more than 1.5-fold in response to rapamycin (Rap) or MG132. ‘+’ and ‘−' denotes up- and downregulation due to drug treatment, respectively. (b) The expected number and p-values displayed for each class of overlapping genes in (a) was revealed by chi-squared test. (c) The median, first and third quartiles of the relative transcript levels for each class of genes revealed in (a). The data represent mean values for samples taken 1 h after drug or vehicle treatment.
4.2. Motifs targeted by a number of transcription factors are enriched in the promoter regions of genes co-regulated by proteasome and TORC1
Gene ontology analysis of the class 1 genes (figure 2a) revealed that they fall into two major functional categories: proteolysis (p-value 1.3 × 10−9) and response to stress (p-value 1.1 × 10−6). The former category was made up of genes implicated in the ubiquitin–proteasome system (UPS; 27 genes) and autophagy/vacuolar function (10 genes). The latter category consisted of those genes involved in starvation and stress response (53 genes). Analysis of the promoter sequences of these ORFs revealed several enriched motifs (p < 0.0001), which resembled the different consensus sequences targeted by a number of TFs, including Rpn4, Msn2/Msn4 (Msn2/4), Gis1 and Hsf1 (table 1). Each of these TFs was also shown to regulate the expression of at least 10 per cent of the class 1 genes by YEASTRACT [30].
Table 1.
Enriched motifs in the promoters of co-regulated genes by TORC1 and proteasome. Question marks denote ‘yet to be identified’.
| category | motifs | p-value | TF | consensus |
|---|---|---|---|---|
| Rap + MG132 + (class 1) | GTGGCAAA | 6.4 × 10−16 | Rpn4 | GGTGGCAA |
| GGTGGCAA | 1.2 × 10−13 | Rpn4 | ||
| AGGGG | 1.6 × 10−13 | Msn2/Msn4 | AGGGG | |
| CGCCAC | 1.2 × 10−8 | ? | ? | |
| TTCTAGAA | 4.7 × 10−7 | Hsf1 | TTCNNGAA | |
| CCGCCA | 1.5 × 10−6 | ? | ? | |
| AAGGGAT | 3.6 × 10−5 | Gis1 | TWAGGGAT | |
| Rap + MG132-(class 2) | CTTATC | 1.3 × 10−11 | Gln3 | CTTATC |
| Gat1 | CTTATC | |||
| Rap-MG132-(class 3) | CGCGTC | 4.5 × 10−8 | ? | ? |
| TGACTC | 7.0 × 10−7 | Bas1 | TGACTC | |
| Gcn4 | TGASTCA | |||
| ACGCGT | 1.1 × 10−6 | Mbp1 | ACGCGT | |
| CGCGAA | 3.0 × 10−6 | ? | ? |
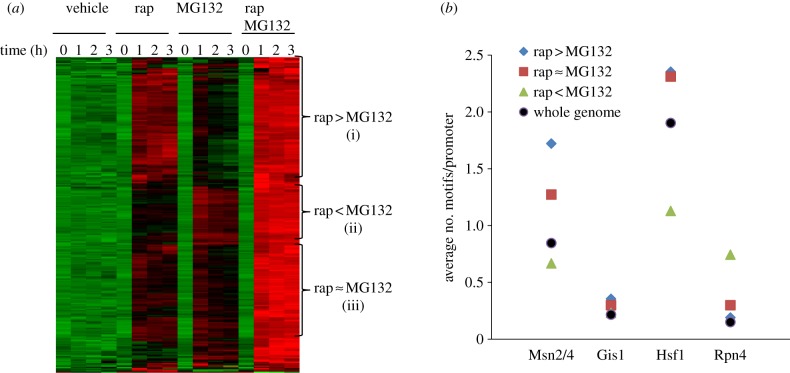
Class 1 genes can be further divided into three subcategories: (i) more significantly activated by rapamycin than by MG132; (ii) more highly activated by MG132 than by rapamycin; and (iii) more or less equally activated by either drug (see figure 3a; electronic supplementary material S3). Category (i) genes were enriched with Msn2/4 and Hsf1 motifs in their promoter sequences (figure 3b). Conversely, the genes in category (ii) were predominantly involved in the UPS (see electronic supplementary material S3), with the Rpn4 motif over-represented and the motifs targeted by Msn2/4 and Hsf1 under-represented in their promoter regions (figure 3b). Rpn4 is both a target and the activator of the 26S proteasome [31]. Msn2 is degraded by the proteasome in the nucleus under stress conditions [7], and Gis1 is subjected to proteasome-mediated limited proteolysis to downregulate its transcription activation capacity [8]. Rapamycin-induced gene expression mediated by Msn2 and Gis1 is strictly regulated by the TORC1-negatively controlled Rim15 kinase [8,32,33]. These results indicated that the functions of TORC1 and the proteasome converge on a number of TFs to keep starvation- and stress-induced gene transcription in control (see also §§4.3–4.5). Their relative roles in regulating transcription may depend on a variety of promoter contexts (figure 3).
Figure 3.
(a) Hierarchical clustering of genes significantly upregulated by either rapamycin (rap) or MG132. The means from biological triplicates were used for clustering. Three groups of genes were represented by (i) rap > MG132 for those more strongly activated by rap than by MG132, (ii) rap < MG132 for those less significantly activated by rap than by MG132 and (iii) rap ≈ MG132 for those more or less equally upregulated by either drug. (b) The average number of Msn2/4, Gis1, Hsf1 and Rpn4 motifs in the promoter regions of the three groups of genes. The average number of these motifs in the whole genome is also included for comparison.
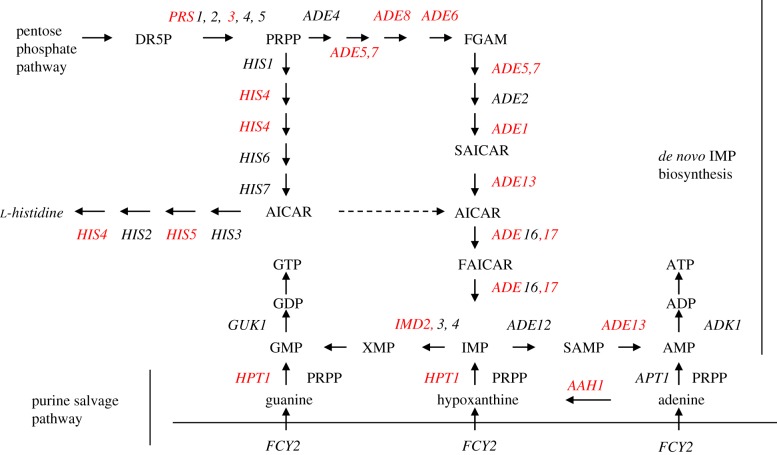
Significantly enriched among the genes downregulated by treatment with rapamycin or MG132 (class 3, figure 2a) are those involved in the biosynthesis of cellular nitrogen compounds (p-value, 2.1 × 10−10), especially de novo IMP biosynthesis and amine metabolic process. Purine nucleotides are generated through the purine biosynthesis pathway, using phosphoribosyl pyrophosphate from the pentose phosphate pathway or 5-aminoimidazole-4-carboxamide-1-β-D-ribofuranoside (AICAR) diverted from the histidine biosynthetic pathway as substrates. The alternate pathway for purine nucleotide formation is the purine salvage pathway, using purine bases or nucleosides from the environment. Treatment with either rapamycin or MG132 led to transcriptional repression of many ADE genes and a number of genes involved in purine salvage (labelled red in figure 4) and in one-carbon metabolism (SHM2, MTD1, GCV1 and GCV2). Concurrently repressed were amino acid biosynthetic genes (ARG1, ARG3, ARG5,6, ARG8, ARO3, ARO4, LEU1, LEU2, LEU9, ILV3, MET3, MET14 and SAM2), ribosomal protein genes (RPL6B, RPS7B, RPL9A, RPS11B, RPL12A, RPS14B and RPS28B) and a number of genes implicated in cell cycle regulation (ACM1, ALK2, CLB1, CLB6, CLN1, GIC1, NRM1, PCL1, PCL2, TOS1, TOS2, TOS4, SHE1, SIM1, SWE1, YHP1 and YOX1). The most over-represented 6-mer in the promoter regions of this class of ORFs was CGCGTC (table 1), but there is no TF that is known to target this motif (http://www.yeastract.com/consensuslist.php). The other significantly enriched sequences, TGACTC and ACTGCT (table 1), were targeted by the Bas1 and Mbp1 TFs, respectively [34–36]. The Bas1 target motif is also similar to the Gcn4-bound element (TGASTCA) [37,38]. Gcn4 is a master regulator of gene expression during amino acid starvation, activating the transcription of more than 500 genes involved in amino acid biosynthesis and purine metabolism [39]. Staschke et al. [28] demonstrated that both Gcn4 and Gln3 are major effectors of the TOR pathway, with each of them inducing and repressing the transcription of a large number of genes during rapamycin treatment. The steady-state level of Gcn4 is subject to translational regulation by TORC1 [40] and proteasome-mediated degradation [41]. Inhibiting the proteasome function, however, increased the level of the Gcn4 protein but reduced the expression of genes activated by Gcn4 in cells grown in minimal medium or starved for amino acids, indicating that degradation of Gcn4 by the proteasome is necessary to stimulate the basal and induced transcription of Gcn4-activated genes [11]. These data suggest that the functions of the proteasome and TORC1 may converge on Gcn4 to regulate the expression of a group of anabolic genes essential to cell growth. This hypothesis needs to be further verified experimentally.
Figure 4.
Genes involved in de novo IMP biosynthesis and purine salvage pathways were downregulated (in red) by treatment with rapamycin or MG132.
Interestingly, among the genes activated by TORC1 inhibition but repressed by MG132 treatment (class 2, figure 2a), ‘amine metabolic process’ (p-value: 9.9 × 10−7) was also the major enriched functional category, including those whose transcription is sensitive to nitrogen catabolite repression (CAR1, CAR2, DUR1,2, DUR3, DAL3, GLT1, MEP1 and MEP2) or elicited upon starvation for amino acids (ALT1, GAP1, DIP5, LYS14, MET28, SAM3, SER3 and SUL1). The other over-represented functional category was ‘response to pheromone’ (p = 0.0005), especially ORFs involved in conjugation with cellular fusion (FUS1, FIG2, AFR1, MFA1, DSE1, SAG1, FUS2, PRM1, AGA1 and RRI2). The most enriched motif in the promoters of this group of genes was CTTATC (table 1), the consensus sequence targeted by the rapamycin-activated TFs, Gln3 and Gat1 [42]. Deletion of GCN4 elevates the basal level of nitrogen catabolite repression (NCR)-sensitive genes in cells grown on nitrogen-rich sources [43], and causes hyperactivation of these genes in rapamycin-treated cells [28,43], indicating that Gcn4 functions to repress the transcription of the NCR-sensitive genes. However, the Gcn4 target motifs were not enriched in the promoter sequences of this class of genes (table 1), suggesting that Gcn4 may repress their transcription indirectly. In this regard, Gcn4, in cells subjected to amino acid starvation, was previously shown to bind Rap1, leading to the inhibition of Esa1 recruitment to the promoters of ribosomal protein genes, and ultimately their transcriptional repression [44].
4.3. TORC1 and proteasome converge on Msn2/4 and Gis1 to regulate the starvation-induced stress response
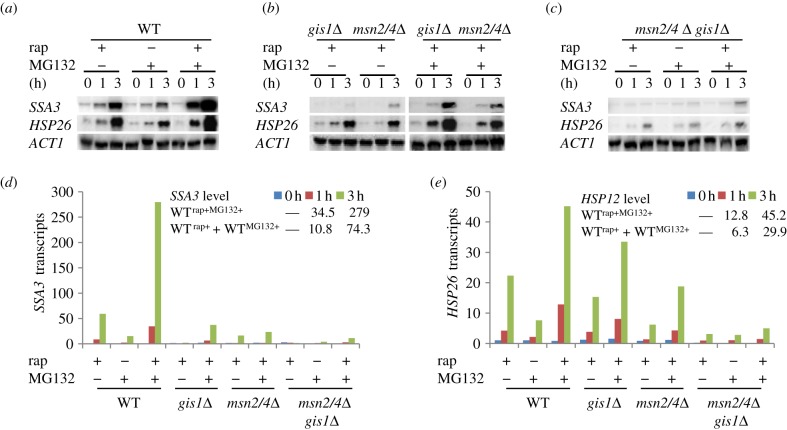
To further confirm that the proteasome and TOR pathways converge on the TFs (table 1) to control starvation-induced gene expression (class 1, figure 2a), we constructed an msn2Δmsn4Δ (msn2/4Δ) double and an msn2/4Δgis1Δ triple mutant in the pdr5Δ deletion background. The transcript levels of two class 1 genes, SSA3 (a PDS gene) and HSP26 (a STRE gene), were assayed in early exponential-phase cells treated with rapamycin and/or MG132 using Northern analysis. The levels of SSA3 and HSP26, after being normalized to that of ACT1 (the loading control), are displayed in figure 5d (for SSA3) and figure 5e (for HSP26). Rapamycin-induced transcription of HSP26 and SSA3 seen in WT cells (lanes 1–3, figure 5a) was significantly reduced in the gis1Δ or msn2/4Δ deletion cells (figure 5b, left) and nearly abolished in the gis1Δmsn2/4Δ triple mutants (lanes 1–3, figure 5c). When compared with that seen in rapamycin-treated cells, the transcription of SSA3 and HSP26 was moderately induced in WT (pdr5Δ) cells treated with the proteasome inhibitor (lanes 4–6, figure 5a). This moderate induction was also dramatically reduced in the gis1Δmsn2/4Δ triple mutants treated with MG132 (lanes 4–6, figure 5c). Hyperactivation of SSA3 and HSP26 was observed when the WT cells were treated with both drugs (lanes 7–9, figure 5a). Such hyperactivation was reduced in either gis1Δ or msn2/4Δ cells (figure 5b, right) and more dramatically decreased in the gis1Δmsn2/4Δ triple mutants similarly treated (lanes 7–9, figure 5c). These data confirmed that the effects of the proteasome and TORC1 on STRE and PDS gene transcription are largely mediated via the Msn2/4 and Gis1 TFs. Furthermore, based on the quantification of SSA3 and HSP26 transcripts (figure 5d and 5e, respectively), we found that the fold-change of either transcript in cells treated with both rapamycin and MG132 is greater than the sum of those in cells treated with either drug (p < 0.01). This is true for both time points taken following drug treatment (1 and 3 h), thus confirming that TORC1 and the proteasome synergistically restrict the expression of starvation-specific transcription.
Figure 5.
(a–c) SSA3 and HSP26 transcripts detected in WT (a), gis1Δ and msn2/4Δ (b), and gis1Δmsn2/4Δ (c) cells treated with MG132 and/or rapamycin. (d,e) Normalized transcript levels of SSA3 (d) and HSP26 (e) to that of ACT1 in WT and mutant cells. The value at time 0 of treatment with rapamycin in WT cells was set to an arbitrary unit of 1.
4.4. Inhibition of the proteasome function abolished the stringent control of Msn2/4- and Gis1-dependent transcription by both Rim15 and Yak1 kinases
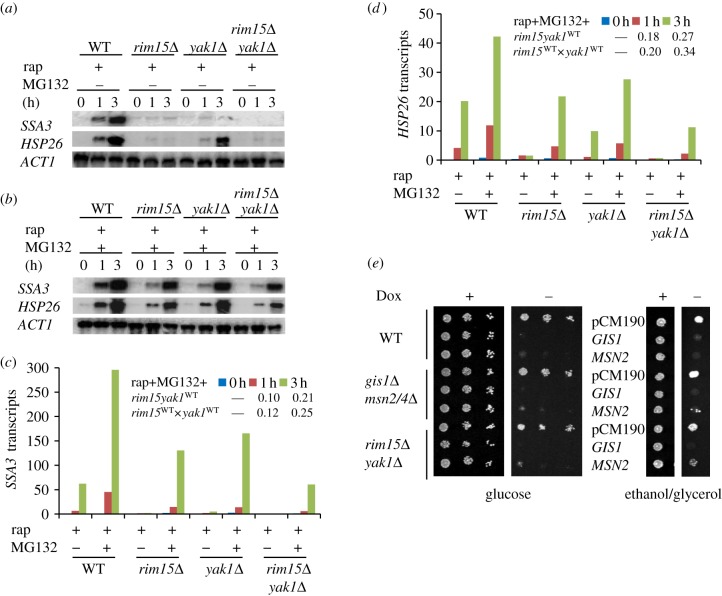
Rapamycin-induced transcription mediated by Msn2/4 and Gis1 was previously shown to be strictly dependent on the Rim15 kinase [8,32,33]. Zhang & Oliver [8] have demonstrated that the strict control of SSA3 transcription by Rim15 in TORC1-inhibited cells was abolished when the function of the proteasome was compromised (see also figure 6a,b). Similar to SSA3, the level of HSP26 transcripts, which were barely detectable in rapamycin-treated rim15Δ cells (figure 6a), was significantly increased by concurrent treatment with both rapamycin and MG132 (figure 6b), confirming that stringent control of STRE and PDS gene transcription by Rim15 requires the function of the proteasome. These data also suggest that other regulators that are negatively controlled by TORC1 may promote STRE and PDS gene expression via pathways that are parallel or compensatory to that of Rim15. Because the localization of the Yak1 kinase to the nucleus is negatively controlled by TORC1 [45] and Yak1 is necessary for Msn2-mediated transcription in response to glucose starvation [46], we tested the hypothesis that Yak1 is one of the other regulators activating STRE/PDS gene transcription in TORC1-inhibited cells. As shown in figure 6a, rapamycin-induced HSP26 and SSA3 transcription seen in wild-type cells was dramatically reduced (for HSP26) or nearly abolished (for SSA3) in the yak1Δ deletion cells. Similar to that observed in the rim15Δ cells, concurrent treatment with both drugs triggered a significant increase in both transcripts in the yak1Δ deletion cells (figure 6b). The degree of transcriptional activation, however, is lower in the yak1Δ or rim15Δ cells than that seen in the WT cells (figure 6c,d).
Figure 6.
(a,b) SSA3 and HSP26 transcripts detected in WT (BY4742pdr5Δ), rim15Δ, yak1Δ and rim15Δyak1Δ cells treated (a) with rapamycin or (b) with both rapamycin and MG132. (c,d) Normalized transcript levels of (c) SSA3 and (d) HSP26 to that of ACT1 and their level in WT cells at time 0 of treatment with rapamycin was set to an arbitrary value of 1. (e) Toxicity of GIS1 or MSN2 overexpression to cell growth on glucose (left) or ethanol/glycerol (right). GIS1 or MSN2 controlled by the tetO7 promoter is switched on in the absence of doxycycline (Dox−) and off in the presence of the drug (+, 20 µg ml−1).
To find the relationship between Rim15 and Yak1, a rim15Δyak1Δ double mutant was constructed in the pdr5Δ deletion background. Transcription activation of HSP26 and SSA3 by rapamycin treatment was completely abolished in the rim15Δyak1Δ double mutants, as was the case in the rim15Δ single mutant (figure 6a). Additional MG132 treatment induced the transcription of both genes in the rim15Δyak1Δ double mutants to a lesser extent than that seen in the rim15Δ or yak1Δ single mutants (figure 6b). At both post-treatment time points (1 and 3 h), the relative level of SSA3 or HSP26 in the rim15Δyak1Δ double mutants, when compared with that in the wild-type cells (rim15yak1WT), was close to the product of its relative levels in the rim15Δ and yak1Δ single mutants (rim15WT × yak1WT; figure 6c,d). This implies that, in TORC1-inhibited cells, Yak1 and Rim15 may promote STRE/PDS gene transcription in parallel pathways. Although much reduced, significant amounts of HSP26 and SSA3 transcripts were detected in the rim15Δyak1Δ cells treated with both drugs (figure 6b), suggesting the presence of yet-to-be-identified regulators acting together with Rim15 and Yak1 to support STRE/PDS gene expression. These observations indicated that upon TORC1 inhibition, multiple regulators, including Yak1 and Rim15, are activated or de-repressed to promote STRE/PDS gene transcription. The function of the proteasome, by limiting the abundance of the Msn2 and Gis1 TFs [7,8], is essential to restrict STRE/PDS gene transcription in exponential-phase cells (figure 5) and to ensure strict control of such transcription by the TORC1-negatively controlled regulators (figure 6).
To further characterize the physiological implications of the proteasome-mediated degradation of Msn2 and Gis1, GIS1 or MSN2 was over-expressed under the control of the tetO promoter. Previous studies have demonstrated that overexpression of MSN2 or GIS1 is toxic to growth [7,8,47]. Overexpression of MSN2 or GIS1 also resulted in growth arrest of gis1Δmsn2/4Δ and rim15Δyak1Δ mutant cells grown on glucose (figure 6d, left). Similar phenotypes were observed when cells were grown on ethanol/glycerol (figure 6d, right), although cells overexpressing GIS1 displayed a more severe growth defect than those overexpressing MSN2, especially in the gis1Δmsn2/4Δ and rim15Δyak1Δ mutants (figure 6d, left and right). These data suggest that proteasome-mediated proteolysis of the starvation-specific TFs (Msn2 and Gis1) ensures not only optimal cell growth but also proper transition from exponential growth to the stationary phase.
4.5. Transcriptional upregulation of genes coding for proteasomal subunits is mediated by Rpn4 in TORC1-inhibited cells
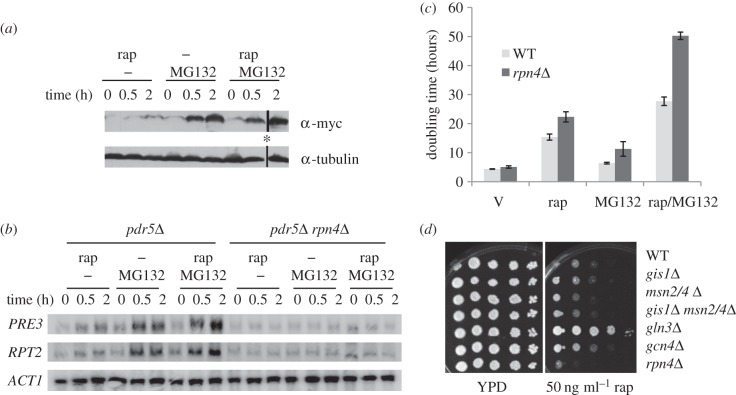
The enrichment of the ‘proteolysis’ functional category (figure 2a) and the Rpn4 motif (table 1) in the class 1 genes suggested that rapamycin- or MG132-induced transcription of genes encoding proteasomal subunits is mediated by Rpn4. To confirm this, the endogenous RPN4 reading frame was tagged with myc at its C-terminus and the steady-state level of Rpn4 assayed in cells treated with either or both drugs. As shown in figure 7a, the level of Rpn4 was only marginally increased in rapamycin-treated cells and significantly elevated in MG132-treated cells. Treatment with both drugs triggered a slightly more dramatic increase in Rpn4 than MG132 alone, especially at 2 h post-treatment. Correspondingly, the transcription of two proteasomal genes, PRE3 and RPT2, was moderately upregulated in rapamycin-treated cells, significantly activated in MG132-treated cells and more dramatically activated by treatment with both drugs (again more evident at 2 h post-treatment; figure 7b). Transcriptional activation of PRE3 and RPT2 was abolished in the rpn4Δ deletion cells in all treatment conditions (figure 7b), confirming that the transcriptional activation of proteasomal genes in TORC1-inhibited cells is mediated through Rpn4, which is similar to the situation seen in MG132-treated cells. To discover the physiological significance of this regulation, wild-type and rpn4Δ cells were grown in SMM medium containing sublethal concentrations of rapamycin (50 ng ml−1) and/or MG132 (12.5 µM). As demonstrated in figure 7c, the doubling time of the wild-type cells is significantly extended by rapamycin treatment, moderately increased by MG132 treatment and greatly extended in the presence of both drugs. The rpn4Δ deletant, when treated with the drug vehicle, exhibited marginally slower growth (approx. 15%) than WT cells similarly treated. This slow-growth phenotype was more pronounced in the presence of either or both drugs (figure 7c). These results indicate that Rpn4-mediated regulation of proteasomal genes and the function of the proteasome are both necessary for cells to adapt to conditions where TORC1 activity is reduced. Similarly, in comparison with WT cells, the rpn4Δ deletant displayed enhanced rapamycin sensitivity when grown in rich medium, as opposed to reduced rapamycin sensitivity shown by the gln3Δ or gcn4Δ mutants (figure 7d). Deletion of MSN2/4 and/or GIS1 did not significantly impact on cell growth in the presence of rapamycin (figure 7d), further highlighting the importance of Rpn4-dependent regulation of proteasome abundance in response to compromised TORC1 function.
Figure 7.
(a) The Rpn4 protein level in WT (pdr5Δ) cells treated with rapamycin (rap) and/or MG132. Endogenous RPN4 was tagged with myc in WT (pdr5Δ) cells. Mid-exponential cells were treated with rap (200 ng ml−1) and/or MG132 (50 µM) for 0, 0.5 and 2 h. *Note that, in the original SDS-PAGE gels, the protein sample representing 2 h of treatment with both drugs was loaded into the well adjacent to the sample denoting 0 h of treatment with rapamycin. (b) PRE3 and RPT2 transcript levels in WT (pdr5Δ) and rpn4Δ deletion cells treated with rap and/or MG132 for 0, 0.5 and 2 h. (c) The doubling time of WT (pdr5Δ) and rpn4Δ deletion cells grown in SMM medium containing drug vehicle (V), rap (50 ng ml−1), MG132 (12.5 µM) or both. (d) Rapamycin hypersensitivity of rpn4Δ deletion cells in YPD medium.
4.6. The core functions of the 20S proteasome is essential for yeast cells to respond to compromised TORC1 activity
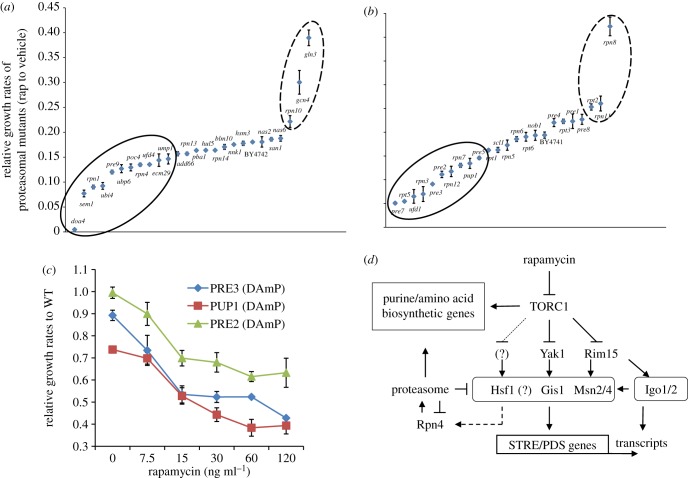
To find out what aspects of proteasome function are required for yeast cells to cope with compromised TORC1 activity, a number of non-essential proteasomal mutants were tested for their sensitivity to low levels of rapamycin. The maximum growth rate of mutant cells in YPD medium containing 50 ng ml−1 of rapamycin was normalized against that of the same cells grown in YPD containing the drug vehicle. The rpn4Δ deletion cells and the gcn4Δ/gln3Δ mutants (figure 7d) were included as negative and positive controls, respectively. When compared with WT cells, deletion of genes coding for subunits of the 19S regulatory particle (SEM1, RPN1) or the 20S core particle (PRE9), or genes involved in proteasome maturation and assembly (ECM29, POC4, UMP1), ubiquitin conjugation (UFD4), or ubiquitin synthesis and recycling (UBI4, DOA4, UBP6) rendered cells more sensitive to rapamycin (figure 8a; p < 0.01, t-test).
Figure 8.
(a,b) Relative sensitivity of proteasomal mutants to mild inhibition of TORC1. (a) Deletion mutants of non-essential genes encoding UPS components and (b) DAmP mutants of essential proteasomal genes were grown in quadruplicates in YPD medium containing 50 ng ml−1 of rapamycin or drug vehicle. Relative sensitivity was calculated by normalizing mutant cells' growth rate in the presence of rapamycin to that in the presence of the drug vehicle. (c) Relative growth rates of the three catalytically compromised DAmP mutants in response to increasing rapamycin concentrations. (d) The working model. STRE/PDS gene expression, mediated by Msn2/4, Gis1 and possibly Hsf1, is controlled by the proteasome and multiple TORC1-negatively regulated proteins, Rim15, Yak1 and others. The arrows and bars denote positive and negative controls, respectively. The question marks and dashed lines refer to unconfirmed or unidentified nodes and regulatory connections.
Pre9 is the only non-essential subunit of the 20S CP. Hypersensitivity of pre9Δ, rpn1Δ, doa4Δ and sem1Δ cells to low levels of rapamycin was reported from previous genome-wide studies [48,49]. By contrast, deletion of RPN10 (coding for a non-ATPase base subunit of the 19S RP) led to rapamycin hyposensitivity (figure 8a). Similar tests were conducted on available mutants of essential proteasomal genes using DAmP strains [23]. In DAmP strains, the mRNA abundance of essential genes is typically reduced by two- to ten-fold [50]. Compared with the wild-type cells, mutants bearing hypomorphic alleles of RPT2, RPN8 or RPN11 (coding for components of the 19S RP) displayed rapamycin hyposensitivity (figure 8b). Conversely, rapamycin hypersensitivity was observed in cells with reduced levels of 19S subunits (Rpn3, Rpt5, Rpn7 and Rpn12), 20S components (Pup1, Pre2, Pre3, Pre5 and Pre7) or Ufd1, a subunit of the Cdc48–Npl4–Ufd1 complex responsible for recruiting polyubiquitinated proteins to the proteasome (figure 8b). Pup1, Pre2 and Pre3 are the three catalytically active subunits of the 26S proteasome, providing the trypsin-like, chymotrypsin-like and caspase-like activities, respectively [51–53]. The three DAmP proteasomal mutants were subjected to rapamycin dosage response assays (figure 8c). When there is no rapamycin in the medium, the PUP1DAmP and PRE3DAmP mutants grew more slowly than the wild-type cells, whereas the PRE2DAmP mutants displayed a similar growth rate as the WT. The relative growth rates of the mutants (to the WT) decreased rapidly with increasing rapamycin concentrations (up to 15 ng ml−1; figure 8c). A further significant change of relative growth rate was only observed for the PUP1DAmP mutant cells when the rapamycin concentration was increased above 15 ng ml−1 (figure 8c). These data confirmed that the growth (rate) of yeast cells is synergistically regulated by the functions of TOR and the proteasome, and that the core functions of the 26S proteasome were essential for yeast cells to adapt to reduced TORC1 activity. Interestingly, the three catalytically compromised DAmP mutants displayed a much faster cell growth than the wild-type cells when treated with 2 µg ml−1 of cycloheximide (electronic supplementary material S4), indicating that cells with reduced proteasomal activities respond differently to TOR inhibition than to translation inhibition. Faster growth of these proteasomal mutants in the presence of cycloheximide has been reported before [54], and the underlying mechanisms remain to be investigated.
5. Discussion
Appropriate regulation of gene transcription is vitally important for cells to both grow when conditions are favourable and to survive when exposed to stressful conditions, including nutrient starvation. The TOR signalling pathway controls cell growth by stimulating anabolic processes and suppressing a variety of stress response programmes [29]. In this study, we have shown that the proteasome regulates a significant portion of the yeast transcriptome synergistically with the TOR signalling pathway. Transcriptional synergy between the proteasome and TORC1 was confirmed by their unidirectional regulation of starvation-specific gene expression and by their cooperative actions in determining the transcription of the proteasome genes. The synergistic effects of the two pathways are also exhibited by their collaboration in cell growth control. Recently, a multi-laboratory systems biology study integrated ‘Omics’ data analysis on two laboratory yeast strains, CEN.PK 113-7D and YSBN2, and indicated that a higher rate of protein turnover and higher proteasomal activity in CEN.PK cells may account for their faster growth [55]. This observation was recently confirmed [56]. Our findings that the function of the proteasome acts synergistically with that of TOR to promote the expression of anabolic genes involved in the de novo biosynthesis of purines and amino acids (table 1) and to restrict the starvation and stress response (figure 2) could provide a further mechanistic explanation for this difference (figure 8d). The combination of TORC1 and proteasome inhibitors was shown to act synergistically to cause cell death in pre-B acute lymphocytic leukaemia [57] and to inhibit cell growth in human oesophageal adenocarcinoma [58], suggesting that the cooperative nature of the two pathways in controlling cell growth may be conserved from yeast to mammals.
Several lines of evidence suggest that Hsf1 may cooperate with Msn2/4 and Gis1 to regulate gene expression in the starvation-induced stress response (figure 8d). First, moderate levels of HSP26 and SSA3 transcripts were detected in the msn2/4Δgis1Δ cells treated with rapamycin and MG132 (figure 5c). Besides STRE and PDS motifs, the heat shock element is also enriched in the promoters of class 1 genes (table 1 and figure 3b), and is present in the promoters of both HSP26 and SSA3. Second, the Hsf1 and Msn2/4 TFs were shown to cooperate in regulating the expression of HSP26 in starvation and stress conditions [59], and the three classes of stress response TFs (Msn2/4, Gis1 and Hsf1) were commonly activated in the long-lived sch9Δ, ras2Δ and tor1Δ mutants [60]. Previous studies by our group and others indicated that Msn2 and Gis1 are targets of the proteasome [7,8,61]. Recently, Hu & Mivechi [62] revealed that the mammalian homologue of Hsf1, the main regulator of the heat shock response in mammals, is degraded by the proteasome, indicating that proteasome-mediated degradation of the stress response TFs may be a common mechanism adopted by cells to reduce the severity of the stress response. By contrast, TORC1 promotes nuclear export of Msn2 [63] through the Tap42-PP2A signalling branch [64,65]. The Rim15 kinase, shown to coordinate Msn2/4- and Gis1-mediated transcription with post-transcriptional mRNA protection [32,66,67], is retained in the cytoplasm by TORC1 activity via Sch9 and 14-3-3 proteins [68,69]. Similarly, cytoplasmic retention of Yak1 is mediated by the yeast 14-3-3 protein, Bmh1 [45]. Our transcript analysis indicated that Yak1 and Rim15 act in parallel pathways to promote transcription mediated by Msn2/4 and Gis1 (figure 6). These data support the hypothesis that while the proteasome restricts starvation and stress response by controlling the levels of the stress response TFs, the TOR signalling pathway, via different downstream signalling branches, negatively modulates a number of regulators (Rim15, Yak1 and others) by retaining them in the cytoplasm. Upon TORC1 inhibition, these regulators translocate into the nucleus and cooperate to activate the starvation-induced transcription programme mediated by the Msn2/4, Gis1 and possibly Hsf1 proteins (figure 8d). The fidelity of the starvation-induced transcription programme requires the proteasome function to prevent activation of starvation-specific genes in exponentially growing cells (figures 1b and 5a) and to avoid excessive activation of stress response genes in TORC1-inhibited cells (figures 1b and 6b), in order to ensure optimum cell growth (figures 6c and 7c) and possibly a speedy return to exponential growth when starved cells are refed with nutrients [70].
Transcriptional activation of proteasomal genes is mediated by Rpn4 in TORC1-inhibited cells (figure 7a,b). Rpn4 is a short-lived protein and is degraded by the proteasome [31], thus providing a negative feedback loop to determine the proteasome abundance in the cell [71,72]. Loss of RPN4 causes slower cell growth, hypersensitivity to rapamycin (figure 7c,d) and decreased cell viability when exposed to alkylating agents, arsenic, UV, DTT or cadmium [73–75], suggesting that upregulation of proteasome abundance through Rpn4 is a general mechanism adopted by yeast cells to adapt to starvation and stress. Interestingly, transcription of RPN4 itself is regulated by a range of stress response TFs, including Hsf1, the multi-drug-resistance-related factors Pdr1 and Pdr3, and Yap1, a TF essential for response to oxidation, toxic metals and MMS [75–77]. Transcription of HSF1, YAP1 and RPN4 is significantly increased by rapamycin treatment (see electronic supplementary material S1), suggesting that Rpn4-mediated transcriptional increase in proteasomal genes may, at least partially, result from activation of the stress response network in TORC1-inhibited cells (figure 8d).
We have demonstrated that the proteasome and TOR pathways converge on Msn2/4, Gis1 and Rpn4 to regulate starvation-induced gene expression. How the functions of the two pathways cooperate to coordinately regulate the transcription of the ADE and amino acid biosynthetic genes (class 3, figure 2a) or the NCR-sensitive genes in a complementary manner (class 2, figure 2a) is less well understood. Over-represented motifs identified in the promoter regions of these genes suggest that the two pathways may converge on Gcn4/Bas1/Mbp1 and Gln3/Gat1, respectively, to modulate their expression (table 1). MG132 treatment leads to transcriptional downregulation of both set of genes, indicating that the proteasome function is necessary for the basal level of their expression. Proteasome-mediated degradation of Gcn4 is required for expression of Gcn4-activated genes [11]. Whether the function of the proteasome is needed for the expression of Gcn4-repressed genes [28] is not known. Equally, it cannot be ruled out that the proteasome may associate with chromatin to influence their transcription, as demonstrated for Spt23- and Mga2-regulated genes involved in lipid metabolism [18].
Connections between the UPS and the TOR signalling pathway were reported previously. Chotechuang et al. [78] observed that downregulation of the UPS system induced by high-protein diet requires the inhibition of AMPK and the activation of the mTOR pathways. Furthermore, the inhibition of the proteasome function represses mTOR signalling and protein translation in colon cancer cells [79]. Recently, DEPTOR, the endogenous inhibitor of mTORC1 and mTORC2, was shown to be degraded by the proteasome [80,81], mediated by the SCFβTrCP E3 ubiquitin ligase [82–84], indicating that the proteasome is directly involved in modulating the function of mTOR. Although there is no Saccharomyces cerevisiae homologue for the mammalian DEPTOR protein, these and our own studies have provided a platform for future investigations of the complex interactions between the proteasome and TOR pathways in gene expression, cell growth and the stress response. As rapamycin does not inhibit all the functions of TOR [85], the combination of the proteasome inhibitors with TOR active-site inhibitors [86], ATP-competitive inhibitors [87] or nitrogen starvation should be included to further interrogate the complex relationship between the two pathways.
6. Acknowledgements
We thank Dr Pinar Pir and Dr Leo Zeef for their help with the initial transcriptome data analysis. Z.Q. thanks the Cambridge Overseas Trust and Lucy Cavendish College for financial support. This work was also supported by a BBSRC grant (no. BB/C505140/2) and a UNICELLSYS grant awarded to S.G.O. by the EC.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
References
- 1.Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 10.1146/annurev.genet.30.1.405 (doi:10.1146/annurev.genet.30.1.405) [DOI] [PubMed] [Google Scholar]
- 2.Jariel-Encontre I, Bossis G, Piechaczyk M. 2008. Ubiquitin-independent degradation of proteins by the proteasome. Biochim. Biophys. Acta 1786, 153–177 10.1016/j.bbcan.2008.05.004 (doi:10.1016/j.bbcan.2008.05.004) [DOI] [PubMed] [Google Scholar]
- 3.Kwak J, Workman JL, Lee D. 2011. The proteasome and its regulatory roles in gene expression. Biochim. Biophys. Acta 1809, 88–96 10.1016/j.bbagrm.2010.08.001 (doi:10.1016/j.bbagrm.2010.08.001) [DOI] [PubMed] [Google Scholar]
- 4.Bhat KP, Greer SF. 2011. Proteolytic and non-proteolytic roles of ubiquitin and the ubiquitin proteasome system in transcriptional regulation. Biochim. Biophys. Acta 1809, 150–155 10.1016/j.bbagrm.2010.11.006 (doi:10.1016/j.bbagrm.2010.11.006) [DOI] [PubMed] [Google Scholar]
- 5.Geng F, Wenzel S, Tansey WP. 2012. Ubiquitin and proteasomes in transcription. Annu. Rev. Biochem. 81, 177–201 10.1146/annurev-biochem-052110-120012 (doi:10.1146/annurev-biochem-052110-120012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muratani M, Tansey WP. 2003. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 4, 192–201 10.1038/nrm1049 (doi:10.1038/nrm1049) [DOI] [PubMed] [Google Scholar]
- 7.Durchschlag E, Reiter W, Ammerer G, Schuller C. 2004. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J. Biol. Chem. 279, 55 425–55 432 10.1074/jbc.M407264200 (doi:10.1074/jbc.M407264200) [DOI] [PubMed] [Google Scholar]
- 8.Zhang N, Oliver SG. 2010. The transcription activity of Gis1 is negatively modulated by proteasome-mediated limited proteolysis. J. Biol. Chem. 285, 6465–6476 10.1074/jbc.M109.073288 (doi:10.1074/jbc.M109.073288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. 1994. The ubiquitin-proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell 78, 773–785 10.1016/S0092-8674(94)90482-0 (doi:10.1016/S0092-8674(94)90482-0) [DOI] [PubMed] [Google Scholar]
- 10.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. 2000. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577–586 10.1016/S0092-8674(00)00080-5 (doi:10.1016/S0092-8674(00)00080-5) [DOI] [PubMed] [Google Scholar]
- 11.Lipford JR, Smith GT, Chi Y, Deshaies RJ. 2005. A putative stimulatory role for activator turnover in gene expression. Nature 438, 113–116 10.1038/nature04098 (doi:10.1038/nature04098) [DOI] [PubMed] [Google Scholar]
- 12.Szutorisz H, Georgiou A, Tora L, Dillon N. 2006. The proteasome restricts permissive transcription at tissue-specific gene loci in embryonic stem cells. Cell 127, 1375–1388 10.1016/j.cell.2006.10.045 (doi:10.1016/j.cell.2006.10.045) [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. 2002. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296, 548–550 10.1126/science.1069490 (doi:10.1126/science.1069490) [DOI] [PubMed] [Google Scholar]
- 14.Morris MC, Kaiser P, Rudyak S, Baskerville C, Watson MH, Reed SI. 2003. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423, 1009–1013 10.1038/nature01720 (doi:10.1038/nature01720) [DOI] [PubMed] [Google Scholar]
- 15.Malik S, Shukla A, Sen P, Bhaumik SR. 2009. The 19 s proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 284, 35 714–35 724 10.1074/jbc.M109.035709 (doi:10.1074/jbc.M109.035709) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran K, Mahr JA, Spector DH. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J. Virol. 84, 3079–3093 10.1128/JVI.02236-09 (doi:10.1128/JVI.02236-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. 2004. Physical and functional association of RNA polymerase II and the proteasome. Proc. Natl Acad. Sci. USA 101, 5904–5909 10.1073/pnas.0305411101 (doi:10.1073/pnas.0305411101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. 2006. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell 21, 861–871 10.1016/j.molcel.2006.02.020 (doi:10.1016/j.molcel.2006.02.020) [DOI] [PubMed] [Google Scholar]
- 19.Sikder D, Johnston SA, Kodadek T. 2006. Widespread, but non-identical, association of proteasomal 19 and 20 S proteins with yeast chromatin. J. Biol. Chem. 281, 27 346–27 355 10.1074/jbc.M604706200 (doi:10.1074/jbc.M604706200) [DOI] [PubMed] [Google Scholar]
- 20.Dembla-Rajpal N, Seipelt R, Wang Q, Rymond BC. 2004. Proteasome inhibition alters the transcription of multiple yeast genes. Biochim. Biophys. Acta 1680, 34–45 10.1016/j.bbaexp.2004.08.008 (doi:10.1016/j.bbaexp.2004.08.008) [DOI] [PubMed] [Google Scholar]
- 21.Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK. 2002. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl Acad. Sci. USA 99, 1461–1466 10.1073/pnas.032516399 (doi:10.1073/pnas.032516399) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler EA, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 10.1126/science.285.5429.901 (doi:10.1126/science.285.5429.901) [DOI] [PubMed] [Google Scholar]
- 23.Breslow DK, et al. 2008. A comprehensive strategy enabling high-resolution functional analysis of the yeast genome. Nat. Methods 5, 711–718 10.1038/nmeth.1234 (doi:10.1038/nmeth.1234) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Longtine MS, Mckenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 (doi:10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 25.Brown AJ, et al. 2001. Transcript analysis of 1003 novel yeast genes using high-throughput northern hybridizations. EMBO J. 20, 3177–3186 10.1093/emboj/20.12.3177 (doi:10.1093/emboj/20.12.3177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang N, Wu J, Oliver SG. 2009. Gis1 is required for transcriptional reprogramming of carbon metabolism and the stress response during transition into stationary phase in yeast. Microbiology 155, 1690–1698 10.1099/mic.0.026377-0 (doi:10.1099/mic.0.026377-0) [DOI] [PubMed] [Google Scholar]
- 27.Engler-Blum G, Meier M, Frank J, Muller GA. 1993. Reduction of background problems in nonradioactive northern and Southern blot analyses enables higher sensitivity than 32P-based hybridizations. Anal. Biochem. 210, 235–244 10.1006/abio.1993.1189 (doi:10.1006/abio.1993.1189) [DOI] [PubMed] [Google Scholar]
- 28.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN,, Pan T, Edenberg HJ, Wek RC. 2010. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J. Biol. Chem. 285, 16 893–16 911 10.1074/jbc.M110.121947 (doi:10.1074/jbc.M110.121947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Virgilio C, Loewith R. 2006. The TOR signalling network from yeast to man. Int. J. Biochem. Cell Biol. 38, 1476–1481 10.1016/j.biocel.2006.02.013 (doi:10.1016/j.biocel.2006.02.013) [DOI] [PubMed] [Google Scholar]
- 30.Abdulrehman D, et al. 2011. YEASTRACT: providing a programmatic access to curated transcriptional regulatory associations in Saccharomyces cerevisiae through a web services interface. Nucleic Acids Res. 39, D136–D140 10.1093/nar/gkq964 (doi:10.1093/nar/gkq964) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie Y, Varshavsky A. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA 98, 3056–3061 10.1073/pnas.071022298 (doi:10.1073/pnas.071022298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Talarek N, et al. 2010. Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5′-3′ mRNA decay pathway. Mol. Cell 38, 345–355 10.1016/j.molcel.2010.02.039 (doi:10.1016/j.molcel.2010.02.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. 2003. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell 12, 1607–1613 10.1016/S1097-2765(03)00485-4 (doi:10.1016/S1097-2765(03)00485-4) [DOI] [PubMed] [Google Scholar]
- 34.Tice-Baldwin K, Fink GR, Arndt KT. 1989. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 246, 931–935 10.1126/science.2683089 (doi:10.1126/science.2683089) [DOI] [PubMed] [Google Scholar]
- 35.Daignan-Fornier B, Fink GR. 1992. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl Acad. Sci. USA 89, 6746–6750 10.1073/pnas.89.15.6746 (doi:10.1073/pnas.89.15.6746) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. 1993. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science 261, 1551–1557 10.1126/science.8372350 (doi:10.1126/science.8372350) [DOI] [PubMed] [Google Scholar]
- 37.Oliphant AR, Brandl CJ, Struhl K. 1989. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol. Cell Biol. 9, 2944–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavrothalassitis G, Beal G, Papas TS. 1990. Defining target sequences of DNA-binding proteins by random selection and PCR: determination of the GCN4 binding sequence repertoire. DNA Cell Biol. 9, 783–788 10.1089/dna.1990.9.783 (doi:10.1089/dna.1990.9.783) [DOI] [PubMed] [Google Scholar]
- 39.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. 2001. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell Biol. 21, 4347–4368 10.1128/MCB.21.13.4347-4368.2001 (doi:10.1128/MCB.21.13.4347-4368.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherkasova VA, Hinnebusch AG. 2003. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 17, 859–872 10.1101/gad.1069003 (doi:10.1101/gad.1069003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irniger S, Braus GH. 2003. Controlling transcription by destruction: the regulation of yeast Gcn4p stability. Curr. Genet. 44, 8–18 10.1007/s00294-003-0422-3 (doi:10.1007/s00294-003-0422-3) [DOI] [PubMed] [Google Scholar]
- 42.Cooper TG. 2002. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol. Rev. 26, 223–238 10.1111/j.1574-6976.2002.tb00612.x (doi:10.1111/j.1574-6976.2002.tb00612.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sosa E, Aranda C, Riego L, Valenzuela L, DeLuna A, Cantú JM, González A. 2003. Gcn4 negatively regulates expression of genes subjected to nitrogen catabolite repression. Biochem. Biophys. Res. Commun. 310, 1175–1180 10.1016/j.bbrc.2003.09.144 (doi:10.1016/j.bbrc.2003.09.144) [DOI] [PubMed] [Google Scholar]
- 44.Joo YJ, Kim JH, Kang UB, Yu MH, Kim J. 2011. Gcn4p-mediated transcriptional repression of ribosomal protein genes under amino-acid starvation. EMBO J. 30, 859–872 10.1038/emboj.2010.332 (doi:10.1038/emboj.2010.332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin DE, Soulard A, Hall MN. 2004. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL1. Cell 119, 969–979 10.1016/j.cell.2004.11.047 (doi:10.1016/j.cell.2004.11.047) [DOI] [PubMed] [Google Scholar]
- 46.Lee P, Cho BR, Joo HS, Hahn JS. 2008. Yeast Yak1 kinase, a bridge between PKA and stress-responsive transcription factors, Hsf1 and Msn2/Msn4. Mol. Microbiol. 70, 882–895 10.1111/j.1365-2958.2008.06450.x (doi:10.1111/j.1365-2958.2008.06450.x) [DOI] [PubMed] [Google Scholar]
- 47.Pedruzzi I, Burckert N, Egger P, De Virgilio C. 2000. Saccharomyces cerevisiae Ras/cAMP pathway controls post-diauxic shift element-dependent transcription through the zinc finger protein Gis1. EMBO J. 19, 2569–2579 10.1093/emboj/19.11.2569 (doi:10.1093/emboj/19.11.2569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan TF, Carvalho J, Riles L, Zheng XF. 2000. A chemical genomics approach toward understanding the global functions of the target of rapamycin protein (TOR). Proc. Natl Acad. Sci. USA 97, 13 227–13 232 10.1073/pnas.240444197 (doi:10.1073/pnas.240444197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie MW, Jin F, Hwang H, Hwang S, Anand V, Duncan MC, Huang J. 2005. Insights into TOR function and rapamycin response: chemical genomic profiling by using a high-density cell array method. Proc. Natl Acad. Sci. USA 102, 7215–7220 10.1073/pnas.0500297102 (doi:10.1073/pnas.0500297102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuldiner M, et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123, 507–519 10.1016/j.cell.2005.08.031 (doi:10.1016/j.cell.2005.08.031) [DOI] [PubMed] [Google Scholar]
- 51.Chen P, Hochstrasser M. 1996. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell 86, 961–972 10.1016/S0092-8674(00)80171-3 (doi:10.1016/S0092-8674(00)80171-3) [DOI] [PubMed] [Google Scholar]
- 52.Arendt CS, Hochstrasser M. 1997. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl Acad. Sci. USA 94, 7156–7161 10.1073/pnas.94.14.7156 (doi:10.1073/pnas.94.14.7156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heinemeyer W, Fischer M, Krimmer T, Stachon U, Wolf DH. 1997. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J. Biol. Chem. 272, 25 200–25 209 10.1074/jbc.272.40.25200 (doi:10.1074/jbc.272.40.25200) [DOI] [PubMed] [Google Scholar]
- 54.Gerlinger UM, Guckel R, Hoffmann M, Wolf DH, Hilt W. 1997. Yeast cycloheximide-resistant crl mutants are proteasome mutants defective in protein degradation. Mol. Biol. Cell 8, 2487–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Canelas AB, et al. 2010. Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat. Commun. 1, 145. 10.1038/ncomms1150 (doi:10.1038/ncomms1150) [DOI] [PubMed] [Google Scholar]
- 56.Hong KK, Hou J, Shoaie S, Nielsen J, Bordel S. 2012. Dynamic 13C-labeling experiments prove important differences in protein turnover rate between two Saccharomyces cerevisiae strains. FEMS Yeast Res. 12, 741–747 10.1111/j.1567-1364.2012.00823.x (doi:10.1111/j.1567-1364.2012.00823.x) [DOI] [PubMed] [Google Scholar]
- 57.Saunders P, Cisterne A, Weiss J, Bradstock KF, Bendall LJ. 2011. The mammalian target of rapamycin inhibitor RAD001 (everolimus) synergizes with chemotherapeutic agents, ionizing radiation and proteasome inhibitors in pre-B acute lymphocytic leukemia. Haematologica 96, 69–77 10.3324/haematol.2010.026997 (doi:10.3324/haematol.2010.026997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Issaenko OA, Bitterman PB, Polunovsky VA, Dahlberg PS. 2012. Cap-dependent mRNA translation and the ubiquitin-proteasome system cooperate to promote ERBB2-dependent esophageal cancer phenotype. Cancer Gene Ther. 19, 609–618 10.1038/cgt.2012.39 (doi:10.1038/cgt.2012.39) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amoros M, Estruch F. 2001. Hsf1p and Msn2/4p cooperate in the expression of Saccharomyces cerevisiae genes HSP26 and HSP104 in a gene- and stress type-dependent manner. Mol. Microbiol. 39, 1523–1532 10.1046/j.1365-2958.2001.02339.x (doi:10.1046/j.1365-2958.2001.02339.x) [DOI] [PubMed] [Google Scholar]
- 60.Cheng C, Fabrizio P, Ge H, Longo VD, Li LM. 2007. Inference of transcription modification in long-live yeast strains from their expression profiles. BMC Genomics 8, 219. 10.1186/1471-2164-8-219 (doi:10.1186/1471-2164-8-219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quan Z, Oliver SG, Zhang N. 2011. JmjN interacts with JmjC to ensure selective proteolysis of Gis1 by the proteasome. Microbiology 157, 2694–2701 10.1099/mic.0.048199-0 (doi:10.1099/mic.0.048199-0) [DOI] [PubMed] [Google Scholar]
- 62.Hu Y, Mivechi NF. 2011. Promotion of heat shock factor Hsf1 degradation via adaptor protein filamin A-interacting protein 1-like (FILIP-1L). J. Biol. Chem. 286, 31 397–31 408 10.1074/jbc.M111.255851 (doi:10.1074/jbc.M111.255851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beck T, Hall MN. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402, 689–692 10.1038/45287 (doi:10.1038/45287) [DOI] [PubMed] [Google Scholar]
- 64.Duvel K, Santhanam A, Garrett S, Schneper L, Broach JR. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11, 1467–1478 10.1016/S1097-2765(03)00228-4 (doi:10.1016/S1097-2765(03)00228-4) [DOI] [PubMed] [Google Scholar]
- 65.Santhanam A, Hartley A, Duvel K, Broach JR, Garrett S. 2004. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryot. Cell 3, 1261–1271 10.1128/EC.3.5.1261-1271.2004 (doi:10.1128/EC.3.5.1261-1271.2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luo X, Talarek N, De Virgilio C. 2011. Initiation of the yeast G0 program requires Igo1 and Igo2, which antagonize activation of decapping of specific nutrient-regulated mRNAs. RNA Biol. 8, 14–17 10.4161/rna.8.1.13483 (doi:10.4161/rna.8.1.13483) [DOI] [PubMed] [Google Scholar]
- 67.Bontron S, Jaquenoud M, Vaga S, Talarek N, Bodenmiller B, Aebersold R, De Virgilio C. 2013. Yeast endosulfines control entry into quiescence and chronological life span by inhibiting protein phosphatase 2A. Cell Rep. 3, 16–22 10.1016/j.celrep.2012.11.025 (doi:10.1016/j.celrep.2012.11.025) [DOI] [PubMed] [Google Scholar]
- 68.Wanke V, Cameroni E, Uotila A, Piccolis M, Urban J, Loewith R, De Virgilio C. 2008. Caffeine extends yeast lifespan by targeting TORC1. Mol. Microbiol. 69, 277–285 10.1111/j.1365-2958.2008.06292.x (doi:10.1111/j.1365-2958.2008.06292.x) [DOI] [PubMed] [Google Scholar]
- 69.Wanke V, Pedruzzi I, Cameroni E, Dubouloz F, De Virgilio C. 2005. Regulation of G0 entry by the Pho80–Pho85 cyclin-CDK complex. EMBO J. 24, 4271–4278 10.1038/sj.emboj.7600889 (doi:10.1038/sj.emboj.7600889) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bonzanni N, Zhang N, Oliver SG, Fisher J. 2011. The role of proteosome-mediated proteolysis in modulating potentially harmful transcription factor activity in Saccharomyces cerevisiae. Bioinformatics 27, i283–i287 10.1093/bioinformatics/btr211 (doi:10.1093/bioinformatics/btr211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ju D, Xie Y. 2004. Proteasomal degradation of RPN4 via two distinct mechanisms, ubiquitin-dependent and -independent. J. Biol. Chem. 279, 23 851–23 854 10.1074/jbc.C400111200 (doi:10.1074/jbc.C400111200) [DOI] [PubMed] [Google Scholar]
- 72.Wang X, Xu H, Ha SW, Ju D, Xie Y. 2010. Proteasomal degradation of Rpn4 in Saccharomyces cerevisiae is critical for cell viability under stressed conditions. Genetics 184, 335–342 10.1534/genetics.109.112227 (doi:10.1534/genetics.109.112227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jelinsky SA, Estep P, Church GM, Samson LD. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol. Cell Biol. 20, 8157–8167 10.1128/MCB.20.21.8157-8167.2000 (doi:10.1128/MCB.20.21.8157-8167.2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Haugen AC, Kelley R, Collins JB, Tucker CJ, Deng C, Afshari CA, Brown JM, Ideker T, Van Houten B. 2004. Integrating phenotypic and expression profiles to map arsenic-response networks. Genome Biol. 5, R95. 10.1186/gb-2004-5-12-r95 (doi:10.1186/gb-2004-5-12-r95) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Xu H, Ju D, Xie Y. 2008. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 180, 1945–1953 10.1534/genetics.108.094524 (doi:10.1534/genetics.108.094524) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hahn JS, Neef DW, Thiele DJ. 2006. A stress regulatory network for co-ordinated activation of proteasome expression mediated by yeast heat shock transcription factor. Mol. Microbiol. 60, 240–251 10.1111/j.1365-2958.2006.05097.x (doi:10.1111/j.1365-2958.2006.05097.x) [DOI] [PubMed] [Google Scholar]
- 77.Yokoyama H, Mizunuma M, Okamoto M, Yamamoto J, Hirata D, Miyakawa T. 2006. Involvement of calcineurin-dependent degradation of Yap1p in Ca2+-induced G2 cell-cycle regulation in Saccharomyces cerevisiae. EMBO Rep. 7, 519–524 10.1038/sj.embor.7400647 (doi:10.1038/sj.embor.7400647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chotechuang N, Azzout-Marniche D, Bos C, Chaumontet C, audichon C, Tomé D. 2011. Down-regulation of the ubiquitin-proteasome proteolysis system by amino acids and insulin involves the adenosine monophosphate-activated protein kinase and mammalian target of rapamycin pathways in rat hepatocytes. Amino Acids 41, 457–468 10.1007/s00726-010-0765-2 (doi:10.1007/s00726-010-0765-2) [DOI] [PubMed] [Google Scholar]
- 79.Wu WK, Volta V, Cho CH, Wu YC, Li HT, Yu L, Li ZJ, Sung JJY. 2009. Repression of protein translation and mTOR signaling by proteasome inhibitor in colon cancer cells. Biochem. Biophys. Res. Commun. 386, 598–601 10.1016/j.bbrc.2009.06.080 (doi:10.1016/j.bbrc.2009.06.080) [DOI] [PubMed] [Google Scholar]
- 80.Ghosh P, Wu M, Zhang H, Sun H. 2008. mTORC1 signaling requires proteasomal function and the involvement of CUL4-DDB1 ubiquitin E3 ligase. Cell Cycle 7, 373–381 10.4161/cc.7.3.5267 (doi:10.4161/cc.7.3.5267) [DOI] [PubMed] [Google Scholar]
- 81.Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137, 873–886 10.1016/j.cell.2009.03.046 (doi:10.1016/j.cell.2009.03.046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. 2011. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell 44, 317–324 10.1016/j.molcel.2011.09.005 (doi:10.1016/j.molcel.2011.09.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao Y, Xiong X, Sun Y. 2011. DEPTOR, an mTOR inhibitor, is a physiological substrate of SCFβTrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell 44, 304–316 10.1016/j.molcel.2011.08.029 (doi:10.1016/j.molcel.2011.08.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gao D, et al. 2011. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell 44, 290–303 10.1016/j.molcel.2011.08.030 (doi:10.1016/j.molcel.2011.08.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J. 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proc. Natl Acad. Sci. USA 105, 17 414–17 419 10.1073/pnas.0809136105 (doi:10.1073/pnas.0809136105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, Shokat KM, Hunter T. 2009. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. 10.1371/journal.pbio.1000038 (doi:10.1371/journal.pbio.1000038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thoreen CC, et al. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 10.1074/jbc.M900301200 (doi:10.1074/jbc.M900301200) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.