Abstract
We have previously reported that disruption of a maize root-expressed 9-lipoxygenase (9-LOX) gene, ZmLOX3, results in dramatic increase in resistance to diverse leaf and stalk pathogens. Despite evident economic significance of these findings, the mechanism behind this increased resistance remained elusive. In this study, we found that increased resistance of the lox3-4 mutants is due to constitutive activation of induced systemic resistance (ISR) signaling. We showed that ZmLOX3 lacked expression in leaves in response to anthracnose leaf blight pathogen Colletotrichum graminicola, but was expressed constitutively in the roots, thus, prompting our hypothesis: the roots of lox3-4 mutants are the source of increased resistance in leaves. Supporting this hypothesis, treatment of wild-type plants (WT) with xylem sap of lox3-4 mutant induced resistance to C. graminicola to the levels comparable to those observed in lox3-4 mutant. Moreover, treating mutants with the sap collected from WT plants partially restored the susceptibility to C. graminicola. lox3-4 mutants showed primed defense responses upon infection, which included earlier and greater induction of defense-related PAL and GST genes compared to WT. In addition to the greater expression of the octadecanoid pathway genes, lox3-4 mutant responded earlier and with a greater accumulation of H2O2 in response to C. graminicola infection or treatment with alamethicin. These findings suggest that lox3-4 mutants display constitutive ISR-like signaling. In support of this idea, root colonization by Trichoderma virens strain GV29-8 induced the same level of disease resistance in WT as the treatment with the mutant sap, but had no additional resistance effect in lox3-4 mutant. While treatment with T. virens GV29 strongly and rapidly suppressed ZmLOX3 expression in hydroponically grown WT roots, T. virens Δsml mutant, which is deficient in ISR induction, was unable to suppress expression of ZmLOX3, thus, providing genetic evidence that SM1 function in ISR, at least in part, by suppressing host ZmLOX3 gene. This study and the genetic tools generated herein will allow the identification of the signals regulating the induction of resistance to aboveground attackers by beneficial soil microorganisms in the future.
Keywords: beneficial microorganisms, oxylipin, priming, hydrogen peroxide, root-to-shoot signaling, Trichoderma, Colletotrichum graminicola, long distance signaling
Introduction
Plants have evolved primary and secondary defense strategies to recognize and protect themselves against their enemies, rendering them resistant to the majority of their pests (Balmer et al., 2013b; Fu and Dong, 2013). The primary defensive layer is often built upon constitutively present defensive structures or molecules, such as, thorns, needles, trichomes, cell wall barriers, and preformed antimicrobial compounds (Pieterse et al., 2009; Balmer et al., 2013a). However, many microbes and herbivores are able to breach this primary layer of defense. To overcome the attenuation, host plants utilize a secondary defensive strategy that is often associated with sophisticated mechanisms of induced resistance (Van der Ent et al., 2009a). Such inducible defense is often characterized by increased resistance against a broad-spectrum of potential pests both locally and systemically in the distal parts from the site of primary invasion (Spoel and Dong, 2012; Balmer et al., 2013a; Walters et al., 2013). This mechanism, known as systemic acquired resistance (SAR), is employed by plants to restrict pathogen expansion in systemic tissue through necrosis at the local site upon primary infection, and typically characterized by the increased level of salicylic acid (SA) and activation of SA-responsive genes, such as those encoding pathogenesis-related (PR) proteins (Durrant and Dong, 2004; Vlot et al., 2008).
Another class of inducible defense, known as “induced systemic resistance” (ISR) is activated upon colonization of roots by mutualistic microbes (Choudhary et al., 2007; Pozo and Azcon-Aguilar, 2007; Van Wees et al., 2008; Van der Ent et al., 2009a; Shoresh et al., 2010; Balmer et al., 2013b; Walters et al., 2013). A number of beneficial microorganisms are known to induce ISR in monocots and dicots through ethylene (ET) and jasmonate (JA)-dependent signaling pathways (Van der Ent et al., 2009b). Previous studies have shown that colonization of maize roots by Trichoderma spp. triggers systemic resistance to foliar infection by Colletotrichum graminicola. Various elicitors of ISR have been isolated from Trichoderma spp., and a recent study has shown that a secreted hydrophobin-like elicitor, SM1, plays an essential role in the elicitation of ISR in maize and other crops, as Δsm1 mutant lacks the ability to induce ISR (Djonovic et al., 2007).
There are a number of differences between SAR and ISR. JA, ET (Bostock, 2005; Thaler et al., 2012), and volatile organic compounds (VOCs) including green leaf volatiles (GLVs) (Pieterse et al., 2009; Mathys et al., 2012; Weller et al., 2012; Balmer et al., 2013b) are implicated as signals in ISR, while SA and its methyl derivative, along with several other recently identified metabolites might act as the signaling molecules in SAR. It is reported that activation of several PR proteins, e.g., PR1, PR2, and PR5 are not associated with the ISR pathway (Van Wees et al., 1999; Ryu et al., 2004; Ton et al., 2007), and generally ISR is independent of the SA signaling pathway (Pieterse et al., 1996). It has been argued that due to the fitness costs of constitutively expressing ISR or SAR, plants have evolved these inducible mechanisms when encountered with pathogens or insects that break through their first layer of defense (Heil, 2001; Heil and Baldwin, 2002). Thus, a number of negative regulators of these JA- and SA-inducible pathways have likely evolved coordinately (Chini et al., 2007; Hu et al., 2013).upon appropriate stimulation, such us some interaction with pathogens, insects or mutualistic microorganisms, a unique physiological state known as “priming” can be induced. Priming involves subtle regulation of defense related pathways in local and systemic tissues that follow faster and/or greater induction of defense mechanisms upon future encounters (Conrath et al., 2006). Priming protects plants against a broad range of biotic (Conrath et al., 2006) as well as abiotic challenges (Harman et al., 2004; Waller et al., 2005; Baltruschat et al., 2008; Mastouri et al., 2012) and ecological studies have shown that the benefits associated with priming outweighs the cost (van Hulten et al., 2006).
In our previous studies (Gao et al., 2007; Isakeit et al., 2007), we found that disruption of a 9-lipoxygense (9-LOX) of maize, ZmLOX3, resulted in a remarkable increase in resistance to multiple seed, root, leaf, and stem fungal pathogens, including seed contaminating mycotoxin-producing Fusarium verticillioides, southern leaf blight caused by Cochliobolus heterostrophus (Figure 1A) and stalk rots caused by both F. verticillioides and C. graminicola, respectively. Although this resistance is effective against a broad range of pathogens in diverse organs, but regardless of infection, ZmLOX3 transcripts are hardly detectable in the aboveground organs (Gao et al., 2007). Moreover, changes of a set of defense-related genes, including JA/SA/ET-responsive and biosynthetic genes, as well as endogenous levels of JA, SA, and ET were specifically elevated in the roots, but not in the leaves of lox3-4 mutants (Gao et al., 2007). This prompted our major hypothesis tested in this study: the abnormal constitutive activation of all three major defense pathways in the roots of lox3-4 mutant are responsible for the production of yet to be identified signal(s) responsible for constitutively active systemic resistance in all aboveground organs of maize against distantly-related pathogens.
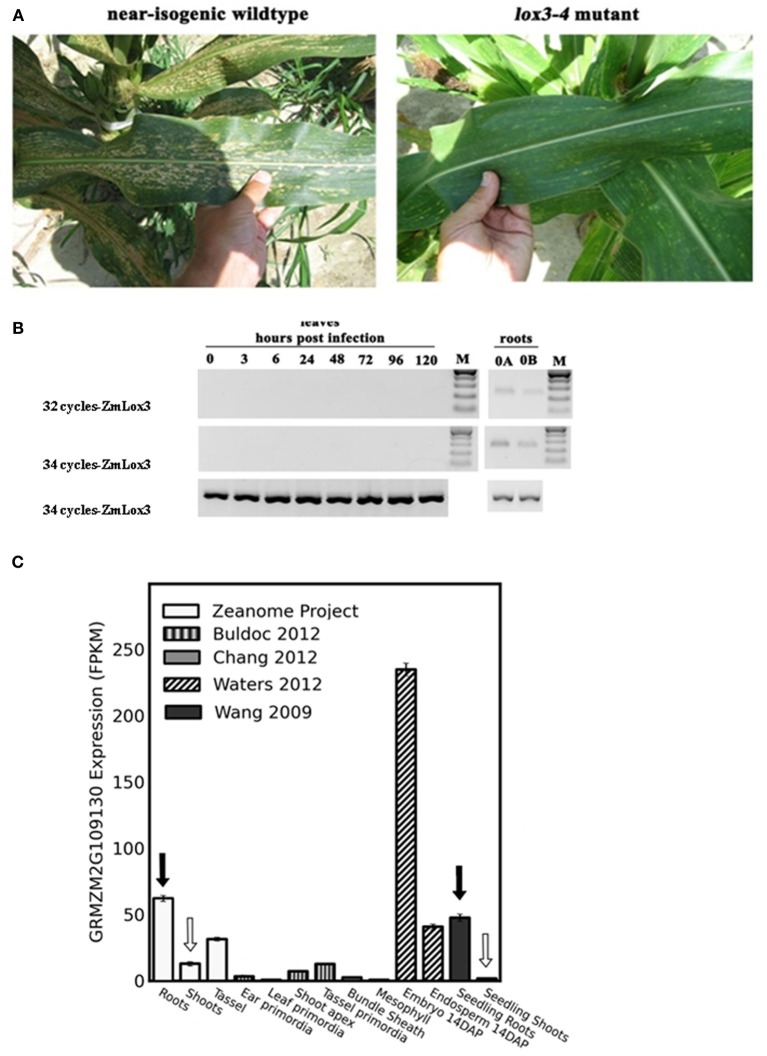
Figure 1.
Colletotrichum graminicola susceptibility factor, ZmLOX3, accumulates preferentially in belowground tissue of maize. (A) Southern corn leaf blight disease symptoms observed under natural field conditions in 2006, College Station, TX, USA of lox3-4 mutant and near-isogenic wild-type maize leaves of the FR2128 background. (B) Semi-quantitative reverse-transcriptase PCR analysis of ZmLOX3 transcript accumulation in C. graminicola infected maize leaves and in untreated roots at 32 and 34 cycles. GAPc at 30 cycles was used as an internal control to normalize cDNA template concentrations. (C) ZmLox3 gene expression from several RNA-seq sources assembled through qTeller. Highlight is expression in leaves (white arrows) comparable with roots (black arrows).
Anthracnose leaf blight, one of the most devastating diseases of maize caused by C. graminicola, was employed as the model disease for this study. C. graminicola is a hemibiotroph, with a well-defined transition from biotrophic to necrotrophic stage in maize. C. graminicola induces tremendous changes in the transcriptome of the host plant at the site of inoculation as well as in systemic tissue (Vargas et al., 2012; Balmer et al., 2013b). Defense related genes are upregulated in both local and systemic tissues of the host as early as 12 h post inoculation (hpi) (Vargas et al., 2012; Balmer et al., 2013b), and continue to increase to peak at ~60 hpi when the pathogen switches to the necrotrophic stage of its life cycle. These findings indicate that the pathogen does not suppress the common defense mechanisms of the host. Instead, it has been suggested that C. graminicola switches lifestyles to evade the increasing host defense machinery against biotrophic pathogens (Vargas et al., 2012). Defense, therefore, seems life-style dependent on a subset of distinct biochemical processes. Maize plants accumulate H2O2 in cells adjacent to the fungal hyphae, suggesting its involvement in defense against C. graminicola (Vargas et al., 2012) and in promoting cell wall lignification around the site of pathogen penetration (Torres and Dangl, 2005). H2O2 accumulation is suggested to limit growth of hemibiotrophic fungi such as Septoria tritici during both biotrophic and necrotrophic stages (Shetty et al., 2007), while the pathogenicity of the Magnaporthe oryzae is shown to be directly related to the ability to scavenge and detoxify ROS (Huang et al., 2011).
To test our hypothesis and probe the molecular mechanism underlying the systemic resistance observed in lox3-4 mutant, as well as to identify the potential molecules responsible for ISR-like activity, we performed a series of experiments using the xylem sap collected from lox3-4 mutant roots. We found that WT plants treated with the sap of lox3-4 mutant showed enhanced resistance to C. graminicola, which was almost identical to the levels of ISR induced in WT maize when colonized by the beneficial fungus T. virens, and the resistance levels observed in the untreated lox3-4 mutant. However, no such ISR-like activity was observed in either WT or lox3-4 mutant in response to xylem exudates from WT, suggesting the constitutive nature of ISR signaling in the lox3-4 mutant. The results of comparative gene expression revealed that ISR-inducing strain of T. virens down-regulates ZmLOX3 in host roots. In contrast, the mutant strain, in which ISR-inducing SM1 protein was deleted, was not able to suppress expression of ZmLOX3. These results explain for the first time the molecular mechanism of the remarkable resistance of lox3-4 mutant to multiple pathogens, and implicate the ZmLOX3 gene as a target for SM1-mediated ISR elicitation.
Methods
Plant and fungal materials
The lox3-4 mutants were identified using a reverse genetics resource generated by Mu-transposable element-insertional mutagenesis as described previously (Gao et al., 2008). Both the mutant and wild type plants (WT) are near-isogenic lines (NILs) at the backcross 7 (BC7) genetic stage in the B73 background. In addition to B73, the lox3-4 mutant allele has been backcrossed into FR2128 four times (BC4), and the increased resistance of lox3-4 in this genetic background is presented in Figure 1A. For the remaining experiments, the mutant in the B73 genetic background was used so that it can be directly compared to the B73 line (hereafter designated WT). The mutant and WT seeds were grown at approximately 25–28°C in commercial soil (Metro-Mix 366; Scotts-Sierra Horticultural Products, Marysville, OH, U.S.A.) under 14 h of daylight with 150 μmol m−2 s−1 (Quantum Meter; Apogee Instruments, Logan, UT, U.S.A.). The seedlings were grown in long conical tubes (20.5 by 4 cm) for 2–3 weeks until they had three fully expanded leaves (V3 developmental stage). To study the effect of root colonization by T. virens on C. graminicola disease development, WT and lox3-4 seed were treated with T. virens GV29-8 strain according to Djonovic et al. (2007) and sown in conical pots (15 by 3 cm) filled with steam-sterilized coarse sand/peat mix (6:1, vol/vol).
To study the effects of T. virens colonization of roots, we used a hydroponic system as described previously (Djonovic et al., 2007). This system was chosen to prevent contamination with microbes and allow root sampling with minimal mechanical damage (Supplementary Figure 1A). Briefly, disinfected seeds were germinated over 2 days on LDB medium and microbe-free germinated seedlings were transferred to hydroponic vessels containing half-strength Murashige and Skoog (1/2 MS) medium amended with Gamborg's vitamins (pH = 5.6) (Sigma-Aldrich, St. Louis, MO, U.S.A). Strains of T. virens were grown and applied to the hydroponic media as described by Djonovic et al. (2007) 24 h after the seedling transfer.
C. graminicola strain 1.001 was cultured on potato dextrose agar, and the conidial suspension for infections was prepared and infection performed as described by (Gao et al., 2007).
Xylem sap collection and application to seedling stems
WT and lox3-4 plants were grown to V4 developmental stage (4th leaf fully developed) in a greenhouse under natural light (during months of April until July). One day before sap collection, the plants were transferred to lighted shelves (150 μmol m−2 s−1 (Quantum Meter; Apogee Instruments, Logan, UT, U.S.A.) with 14 h of daylight and were immediately watered. 24 h later, plants were re-watered until the soil was saturated, and then plants were carefully decapitated with a sharp scalpel above the first leaf (Supplemental Figure 1B). The first droplet of sap was discarded and subsequent droplets were collected for 8 h and stored on ice. The plants were periodically recut when cut site sealed, the next droplets were discarded as above. Sap collected from plants of the same genotype was mixed thoroughly before freezing in liquid N2 and stored at -80°C until needed for the experiments. B73 plants were grown to the V4 stage to match the developmental stage of the collected sap. WT and lox3-4 sap were thawed and diluted 1:1 with sterile distilled water to reduce viscosity. Plants were placed horizontally in a sterilized tray and two 1 cm long incisions were made between the first and second leaves (Supplemental Figure 1C). The incisions were made halfway through the depth of the stem so as not to puncture the backside, and then 50 μl of diluted sap was added to each incision. Masking tape was used to seal the wound sites and to ensure that the sap did not leak out.
RNA extraction, semi-Q-RT-PCR and Q-RT-PCR analysis
Leaves from the mutant and WT plants were harvested at designated times after inoculation and were immediately frozen in liquid N2, and stored at −80°C. Roots of hydroponically grown plants were harvested at designated incubation intervals, excised with a sharp scalpel, immediately frozen in liquid N2, and stored at −80°C. Total RNA was extracted from tissues using TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH, U.S.A) and treated with DNase according to the manufacturer's instructions (RNase-free DNase kit, Qiagen, Valencia, CA, U.S.A). For semi-quantitative PCR (semi-Q-RT-PCR) experiments, 1 μg of RNA was used for cDNA synthesis as described previously (Gao et al., 2007) with GAPc used as a reference gene.
For quantitative reverse transcription-PCR (Q-RT-PCR) assays, a one-step qPCR procedure was performed using Thermo Scientific Verso One-Step RT-qPCR Kits (Thermo Scientific, Waltha, MA, U.S.A). Reactions were optimized for RNA and primer concentrations with each 10 μ l reaction consisting of 40 ng of DNase-free RNA and 200 nM primers. Q-RT-PCR analysis was performed with an Applied Biosystem StepOne Plus Real-Time PCR instrument. Primers used in this study are described in Supplementary Table 1. The PCR program consisted of a 15 min cDNA synthesis step at 50°C, followed by polymerase activation step of 15 min at 95°C, followed by 40 cycles of 15 s at 95°C, 30 s at 56°C, and 30 s at 72°C followed by a melt curve analysis. Primers were designed using Primer3Plus software in accordance with the criteria required for quantitative PCR primer design (Udvardi et al., 2008). The specificity of primers, lack of primer-dimer formation, and the absence of contaminating genomic DNA was verified, respectively, using amplicon dissociation curves, PCR in the absence of cDNA, and by PCR analysis of RNA samples before reverse transcription. The amplification efficiency of primers was calculated using LinReg (11.0) was ≥ 80%. Initially, the GAPc and Cullin genes were tested for reference gene suitability, and Csullin was chosen based upon its more stable expression across samples. Expression levels were normalized using Cullin as a reference gene, and relative expressions of genes compared to control (time 0) were calculated using the method of Ruijter et al. (2009).
Bioinformatics
ZmLOX3 RNAseq expression data was compiled from publically available gene expression data through qteller (http://qteller.com/qteller3/bar_chart.php?name=GRMZM2G109130
Hydrogen peroxide detection in response to infection and treatment of maize leaf with alamethicin
The leaf segments (approximately 5 cm length) of lox3-4 mutant and WT plants at V4-stage were treated with alamethicin (ALA, Sigma-Aldrich, St Louis, MO, U.S.A) at 10 μg/mL by applying 10 μl of solution to the midrib vain of the leaf segment and incubating for 48 h. H2O2 production in WT and lox3-4 mutants upon ALA treatment was examined according to the DAB staining method (Gao et al., 2008) with modifications. Briefly, leaves were excised and subsequently immersed in DAB (1mg/ml; pH 3.8; 3, 3′-diaminobenzidine, Sigma-Aldrich, St Louis, MO, U.S.A) solution with low vacuum pressure for 30 min, followed by an overnight incubation at room temperature in the dark. The stained leaves were fixed and cleared in alcoholic lactophenol (95% ethanol: lactic acid: phenol = 2: 1: 1) at 65°C, rinsed once with 50% ethanol, and twice with H2O. The destained leaves were stored in 50% glycerol or subjected to microscopic observation.
Nitric oxide (NO) detection in the leaves
To visualize NO production, the leaf segments were excised from 3-week-old lox3-4 mutant and WT seedlings at the V4 stage and injected with 1ug/mL ALA on the middle site of main vein of leaf. NO production was examined by diaminofluorescein (DAF) as described by Delledonne et al. (1998).
Distribution of materials
Novel materials described in this publication may be available for non-commercial research purposes upon acceptance and signing of a material transfer agreement. In some cases such materials may contain or be derived from materials obtained from a third party. In such cases, distribution of material will be subject to the requisite permission from any third-party owners, licensors or controllers of all or parts of the material. Obtaining any permission will be the sole responsibility of the requestor.
Results
ZmLOX3 is preferentially expressed in roots but not in infected leaves
Our previous studies showed that elimination of the ZmLOX3 gene in B73 inbred line background conferred increased resistance to C. graminicola infection in leaves (Gao et al., 2007). Since that study, we have created near-isogenic WT and mutant lines in FR2128 genetic background. As shown in Figure 1A, similar to what was observed in the B73 background in the previous study, disruption of ZmLOX3 in the FR2128 genetic background resulted in easily visible and strong reduction of disease severity to southern corn leaf blight under natural field conditions. Surprisingly, no ZmLOX3 transcripts could be detected by semi-Q-RT-PCR analyses even at an unusually high number of PCR cycles during the entire time-course of infection by C. graminicola (Figure 1B). Importantly, the time-course tested was extended for 96 h, a time point by which most infected tissue is macerated by the fungus. As a positive control for RT-PCR analyses, ZmLOX3 transcripts accumulated in the roots. A survey of ZmLOX3 gene expression from several RNAseq sources assembled through qTeller revealed that ZmLOX3 is preferentially expressed in maize roots and shows low expression in seedling shoots (Figure 1C). These results prompted a hypothesis that disruption of ZmLOX3, which is normally expressed in roots, results in a constitutive production of a systemic signal(s) that renders the entire plant, including distal above-ground tissues, resistant to C. graminicola and other pathogens (Gao et al., 2007).
Expression of defense related genes in lox3-4 mutant upon C. graminicola infection
To explore the molecular mechanism of the enhanced resistance in lox3-4 mutants, we examined the expression of a set of defense-related genes in the leaves of both the mutant and near-isogenic WT upon infection with C. graminicola. Genes used for this study were pathogenesis related protein 1 (PR1), hydroperoxide lyase (HPL), phenylalanine ammonia lyase (PAL1), glutathione S-transferase (GST), and maize proteinase inhibitor (MPI). GAPc was used as the reference gene.
Interestingly, we did not detect any difference in the level of PR1, the gene most commonly used as SAR molecular marker, in leaves of lox3-4 or WT plants in response to C. graminicola, indicating that the observed resistance of the mutant may not be due to SAR. PAL1 and MPI were induced earlier in the mutant leaves in response to inoculation with C. graminicola and GST was induced earlier and stronger in mutant than in WT (Figure 2). PAL1 showed higher expression in the leaves of the mutant even at time 0, and was further enhanced in lox3-4 mutants compared to WT. The striking similarity of these results with those previously reported in maize colonized with T. virens (Djonovic et al., 2007) prompted us to postulate that lox3-4 mutant plants may constitutively express an ISR-like phenomenon that induces wide-spectrum systemic resistance to foliar leaf blight and other diseases reported previously (Gao et al., 2007).
Figure 2.
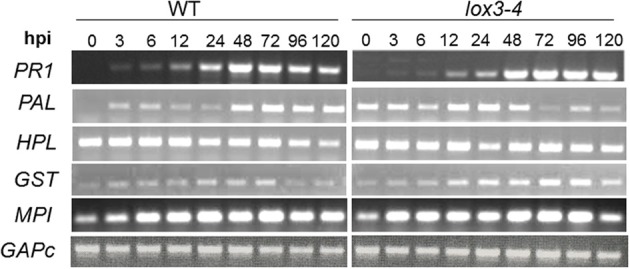
Expression of defense-related genes in leaves of lox3-4 mutant vs B73 NIL (WT) in response to C. graminicola infection. Approximately 3-week-old lox3-4 mutant and WT seedlings at 4 true leaf stage were infected with C. graminicola at 1 × 106 spores.ml−1.mlnicolaola with with at 4 tTrizol reagent and cDNA synthesized with Ambion 1st strand cDNA synthesis kit. PCR was conducted using standard procedures and the expression of GAPc was used as an internal control. HPL, hydroperoxide lyase; PAL, phenylalanine ammonia lyase; GST, glutathione S-transferase; MPI, maize proteinase inhibitor; PR1, pathogenesis related protein 1; GAPc, glyceraldehyde-3-phosphate dehydrogenase.
Xylem sap extracted from lox3-4 mutant increased resistance of wt plants to C. graminicola
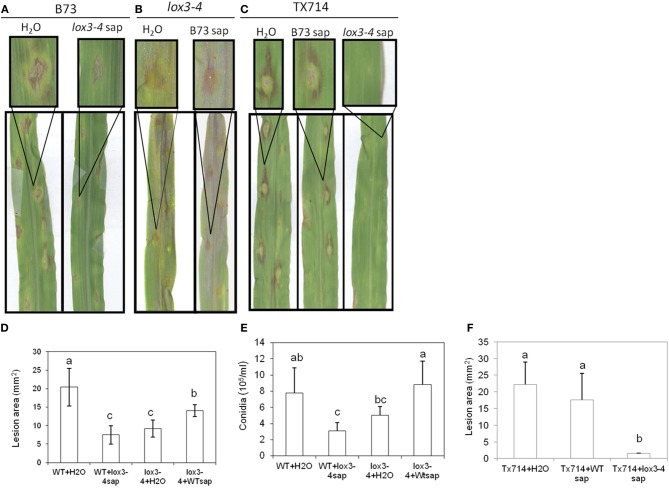
To address whether the induced resistance to C. graminicola in the aboveground organs of lox3-4 mutant resulted from constitutive production of long-distance signal(s) by its roots, xylem sap from WT and the lox3-4 mutant seedlings were collected and applied to lox3-4 and WT plants and followed by inoculation with C. graminicola. Consistent with our previous findings (Gao et al., 2007), lox3-4 mutant showed increased resistance to C. graminicola compared to WT (Figures 3A,B,D,E). Application of xylem sap collected from WT plants on lox3-4 mutant seedlings compromised the resistance level observed in lox3-4 mutants as evidenced by greater lesion area and increased conidial production (Figures 3B,D,E). Remarkably, application of lox3-4 mutant sap to WT plants significantly increased resistance as compared with that of WT supplemented with either H2O or WT sap (Figures 3A,D,E), evident by significant reduction in lesion size and conidial formation (Figures 3D,E). In addition to the mutant sap-conferred resistance in B73, the application of lox3-4 sap resulted in even more dramatic decrease of disease symptoms in leaves of Tx714 inbred line (Figures 3C,F). These results suggest that the long-distance signal(s) in xylem sap of lox3-4 is capable of inducing disease resistance in diverse genetic backgrounds. Taken together, these results strongly support the hypothesis that lox3-4 mutant roots constitutively produce an ISR-like long-distance signal that is transported to above-ground organs via xylem sap.
Figure 3.
Effect of xylem sap from lox3-4 mutant and B73 NIL plants on the pathogenicity of C. graminicola infected on the leaves. The xylem sap were collected from approximately 3-week-old lox3-4 mutant and B73 seedlings at 4 true leaf stage by cutting the stem from the position above the soil. Twenty microliter of sap per plant was injected into the wounding site on the stem of lox3-4 mutant and B73 or TX714 NIL, respectively, subsequently followed by the infection with C. graminicola at 1 × 106 spores/ml. The disease symptoms were recorded and photographed at 3 days post infection on (A) B73 plants treated with H2O or lox3-4 xylem sap, (B) lox3-4 plants treated with H2O or B73 sap, and (C)TX714 plants treated with H2O, B73 sap, or lox3-4 xylem sap. The insert panels show the close-up of representative lesions showing on the corresponding leaf. The (D) lesion size and (E) number of conidia on lox3-4 and B73 leaves infected with C. graminicola upon treatment H2O, and lox3-4 or B73 sap and (F) lesion size on TX714 leaves infected with C. graminicola upon treatment H2O, and lox3-4 or B73 sap. The data are shown as mean ± SE (n > 15 for lesion size and n = 3 for conidia number experiments, respectively). The different letters above the bar indicate the significant difference between different treatments analyzed by SPSS software One-Way ANOVA (p < 0.05).
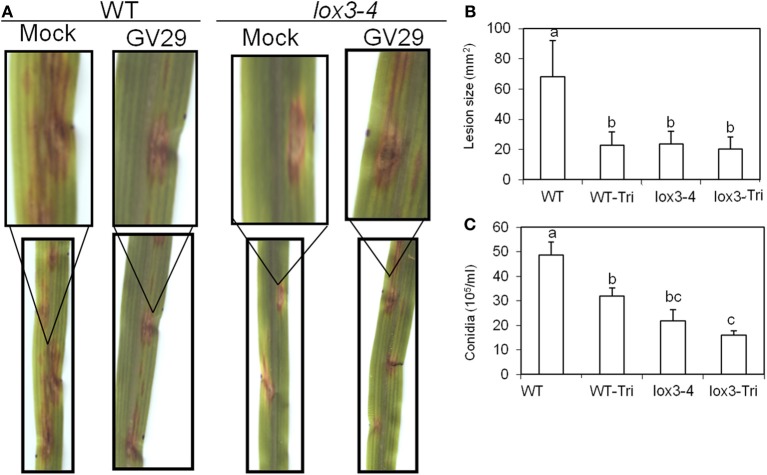
Interestingly, the level of reduction of lesion size triggered by lox3-4 xylem sap in WT was identical to treatment of WT plants with T. virens (Figures 4A,B,C). Furthermore, root colonization by T. virens did not enhance resistance of lox3-4 mutants, indicating that lox3-4 mutants constitutively express an ISR-like signal transported through xylem sap that induces resistance in the leaves.
Figure 4.
Effect of colonization of roots of lox3-4 mutant and B73 plants on the pathogenicity of C. graminicola infected on the leaves. Approximately 3-week-old lox3-4 mutant and B73 seedlings at 4 true leaf stage were treated with T. virens in the roots. Then the leaves of treated plants were infected with 10 μ l of C. graminicola conidial suspension at 1 × 106 spores/ml at 3 days post treatment. (A) The disease symptoms were recorded and photographed at 3 days post infection. The insert panels show the close-up of representative lesions showing on the corresponding leaf. (B) The lesion size on lox3-4 and B73 leaves infected with C. graminicola upon treatment with or without T. virens. The data are shown as mean ± SE (n > 15) of lesions examined. (C) The conidia number of C. graminicola from the infected leaves of lox3-4 mutant and WT leaf segment upon treatment with or without T. virens. The data are shown as mean ± SE (n = 3) of leaves examined. The different letters above the bar indicate the significant difference between different treatments analyzed by SPSS software One-Way ANOVA (p < 0.05).
ZmLOX3 is down-regulated by T. virens
The results presented above provide strong evidence that ZmLOX3 acts as a potent negative regulator of ISR. If this hypothesis is true, then we reasoned that ISR-inducing microorganisms must suppress this gene to facilitate ISR activation. To test this hypothesis, we compared ZmLOX3 expression in roots of WT maize in response to colonization by ISR-inducing T. virens GV29 and the Δsm1 mutant strain. In our previous study, we demonstrated that this mutant strain is incapable of inducing ISR against C. graminicola in maize (Djonovic et al., 2007). Interestingly, root colonization by GV29 suppressed ZmLOX3 expression in the roots as early as 24 hpi in the hydroponic system, a time point at which there is no observable root colonization occurring, indicating that the secretion of SM1 from hyphae of T. virens has an effect on ZmLOX3 expression in the hydroponic system used (Figure 5). The expression of this gene remained suppressed compared to control at 48 and 72 hpi when fungal colonization of roots typically occurs, although the difference with control was only significant at 24 and 72 hpi. Importantly, no change in the expression of ZmLOX3 was detected in maize roots colonized by Δsm1 (Figure 5) suggesting that ZmLOX3 is one of the target genes by which SM1 induces ISR.
Figure 5.
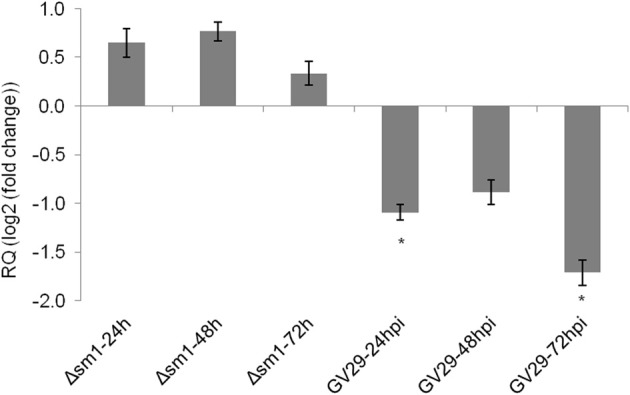
Relative expression of ZmLOX3 in response to root colonization by T. virens strain GV-29-8 and Δ sm1, a mutant that is unable to induce ISR in maize. Two days old disease free seedlings were transferred to hydroponic media with half-strength Murashige and Skooge media and were inoculated with the hyphae of the fungus. At each time point, three biological replicates, each consisting of three plants were sampled. The experiment was repeated twice and results were pooled and analyzed together. The data are shown as mean log2 (fold change) ± SE and only those means those sample with statistically significant change in gene expression (α = 5%) are designated with an asterisk.
lox3-4 plants respond to C. graminicola or alamethicin treatment by increased production of ROS and NO
To understand the biochemical mechanisms behind increased resistance observed in lox3-4 mutants, we measured accumulation of H2O2in leaves of WT and lox3-4 mutants in response to C. graminicola infection. Noticeable change was observed in hydrogen peroxide (H2O2) accumulation. ROS produced by maize is implicated in defense against C. graminicola (Vargas et al., 2012). Using DAB staining, we observed that both WT and lox3-4 mutants accumulate hydrogen peroxide around the site of C. graminicola inoculation. However, lox3-4 mutants tend to respond faster and accumulate higher hydrogen peroxide levels compared to WT (Figure 6A), which may limit the duration of biotrophic stage of the fungal life cycle during disease development.
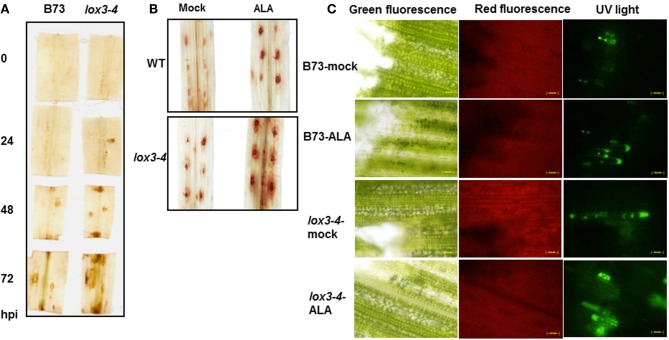
Figure 6.
Effect of (A) C. graminicola and (B) alamethincin (ALA) elicitor on accumulation of H2O2 or (C) Nitric Oxide in lox3-4 mutant and B73 leaves. Leaf segment from lox3-4 mutant and B73 (A) inoculated with C. graminicola or (B) treated with ALA for 24 h were stained according to DAB staining procedure to elucidate the ROS accumulation. (C) Nitric oxide accumulation was visualized by DAF fluorecence 4 h after ALA treatment.
To further test if the lox3-4 mutant is primed for faster ROS accumulation in response to general defense elicitor and not specifically by C. graminicola, we tested if such a primed response occured upon treatment with the elicitor alamethicin. Treatment with ALA only slightly increased accumulation of ROS in WT leaf segments, whereas much greater ROS production was detected in ALA-treated lox3-4 mutant (Figure 6B). Similarly, ALA triggers more NO accumulation in lox3-4 mutant leaf compared to WT (Figure 6C). ALA increases membrane permeability, a hallmark of lipid peroxidation and hypersensitive response (HR). These results indicate that defense responses were expressed earlier and to a greater extent in the lox3-4 mutant plants compared to WT, consistent with the previous reported observations of boosted responses associated to a “primed” state in the ISR- expressing plants.
Xylem sap from lox3-4 mutant altered response of wt leaves to C. graminicola at molecular level
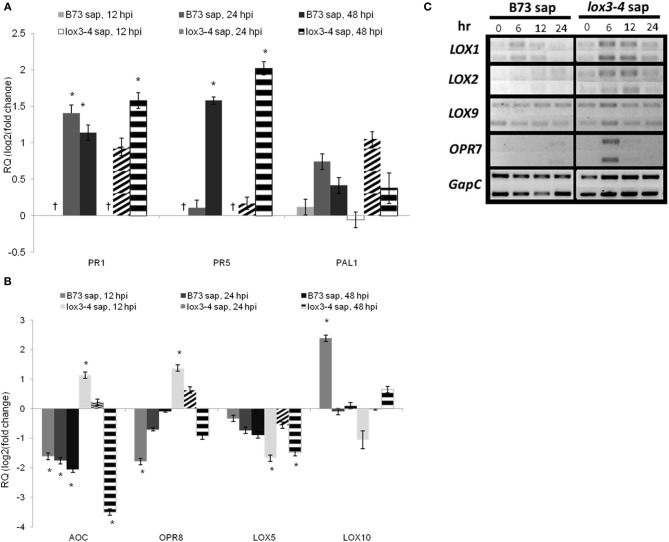
The fourth leaf of WT plants treated with xylem sap extracted from either WT or lox3-4 mutants were inoculated with C. graminicola. Samples were taken at time 0 (the control) and designated times after inoculation to analyze changes in expression of genes involved in basal defense, JA, and other oxylipins biosynthesis. The gene expression analysis was performed using Q-RT-PCR, except for ZmLOX1, ZmLOX2, ZmLOX5, and OPR7 for which we used semi-Q-RT-PCR. The relative gene expression was determined as fold change compared to the control (time 0).
The relative expression of PR1 and PR5 were similar in both treatments, but at 48 hpi leaves of plants treated with lox3-4 sap showed higher relative expression of both genes than those treated with WT sap. PAL1 relative expression increased only in leaves of plants treated with lox3-4 sap at 24 hpi.
The relative expression of genes involved in JA biosynthesis pathway, allene oxide cyclase (AOC), 12-oxophytodienoate reductase 7 and 8 (OPR7 and OPR8), were enhanced at earlier time points after inoculation with C. graminicola in plants treated with lox3-4 mutant sap. However, their relative expression levels were reduced or remained unchanged when treated with WT sap.
ZmLOX5 and ZmLOX10 were shown to be required for normal colonization of leaves and stalks by C. graminicola in maize (Christensen, 2009; Park, 2011). Interestingly, we found that both genes were suppressed in maize leaves treated with sap from lox3-4 mutants. ZmLOX5 was not suppressed in leaves of plants treated with WT sap and ZmLOX10 was actually induced at 12 hpi in these leaves. The expression of ZmLOX9, another 13-lipoxygenase that is likely involved in JA biosynthesis (Christensen et al., 2013) was enhanced in leaves of plants treated with lox3-4 sap at 6 hpi, but not in those treated with WT sap.
Discussion
Oxylipins, an important class of fatty acid-derived signaling molecules, are implicated in plant defense responses to pathogens, insect herbivory, and abiotic stresses. However, we have previously shown that 9-oxylipins, produced by a maize 9-lipoxygenase, ZmLOX3, facilitate pathogenicity of several root, stem, leaf, and seed fungal pathogens (Gao et al., 2007; Isakeit et al., 2007). Maize mutant plants unable to express this gene were shown to gain a remarkable level of increased resistance to a number of diverse fungal pathogens; however, the mechanism of such broad-spectrum resistance was not clear at the time of those publications. Gao et al. (2008) showed that disrupting ZmLOX3 changes expression of other defense-related genes including other 9- and 13-LOXes and ACC oxidase in root of unstressed and disease-free maize tissue. As a result of this inappropriate defense activation, lox3-4 mutants are delayed in germination and development of below- and above-ground organs and premature senescence (Gao et al., 2008).
This report provides compelling evidence that the disruption of the ZmLOX3 gene, normally expressed in roots, results in constitutive production of an ISR-inducing root-to-shoot signal that renders distal tissues resistant to a number of plant diseases. Greater foliage resistance of mutant plants to C. graminicola and the potent ability of the mutant xylem sap to increase this resistance in WT plants, clearly indicate that xylem sap of lox3-4 plants contains a long-distance ISR-inducing signal synthesized in the mutant root. The identity of this signal(s) is currently unknown due to the lack of appropriate analytical and genetic tools. However, we are in the process of creating several combinations of double and triple mutants with JA, green leaf volatile, and ethylene producing enzymes in the background of lox3-4 mutant to disrupt the constitutive signaling and to enable the identification of the candidate signal(s).
Comparative gene expression showed that the treatment of WT plants with lox3-4 sap resulted in a significant change of the transcript accumulation pattern in these plants when infected with C. graminicola compared to WT plants treated with WT sap (Figure 7). Specifically, WT plants treated with lox3-4 sap responded to the inoculation with faster and stronger induction of genes involved in JA biosynthesis, i.e., 13- or 9/13-LOXs, LOX1, LOX2, LOX9, AOC, OPR7, and OPR8. Other defense-related genes including PR1, PR5, and PAL1 were also differentially expressed in lox3-4-sap treated WT plants. PAL1 was only induced in WT plants treated with lox3-4 sap at 24 hpi, and PR1 and PR5 were expressed at higher level in WT plants treated with the lox3-4 sap. Interestingly, PAL1 gene was constitutively expressed in uninfected lox3-4 mutants as well as in infected plants throughout the entire time course examined, but was induced later and to lower levels in WT plants infected with the pathogen (Figure 2). PAL enzymes are involved in the biosynthesis of SA and other phenolic compounds, which are implicated in induction of defense in systemic tissue, and are required for cell wall strengthening via lignification processes (Mauch-Mani and Slusarenko, 1996; Chen et al., 2009) Therefore, we speculate that these PAL-derived defense metabolites may be one potential mechanism behind increased resistance of the lox3-4 mutant (Figure 8).
Figure 7.
Effect of B73 and lox3-4 sap on the expression of defense genes in B73 maize inoculated with C. graminicola. (A) relative expression of SAR-related genes PR1 (pathogenesis related 1), PR5 (pathogenesis related 5), and PAL1 (phennyl alanine amonia lyase), (B) relative expression of genes involved in octadecanoid pathway, AOC (allene oxide synthase), OPR8 (12-oxo-phytodienoic acid 8), LOX5, and LOX10 (lipoxygenase 5 and 10, respectively) and (C) semi-Q-RT-PCR analysis of expression of LOX1, LOX2, LOX9, and OPR7. B73 maize plants at V4 stage were treated with xylem sap from B73 or lox3-4 plants (as described in materials and methods) and were inoculated with C. graminicola 3 h later. Control sample was taken prior to sap treatment and the rest of the samples were taken at designated times after inoculation. For (A) and (B) quantitative real-time PCR (Q-RT-PCR) was used to examine the gene expression and Cullin was used as reference gene. (C) Wherever efficiency of q-RT-PCR reaction was bellow 80% semi-Q-RT-PCR analysis was used to study the gene expression and GAPc was used as internal reference. The data are presented as mean ± SE. The †represents the sample that no amplification was detected for the gene expression. The sample with statistically significant change in gene expression (α = 5%) are designated with an asterisk.
Figure 8.
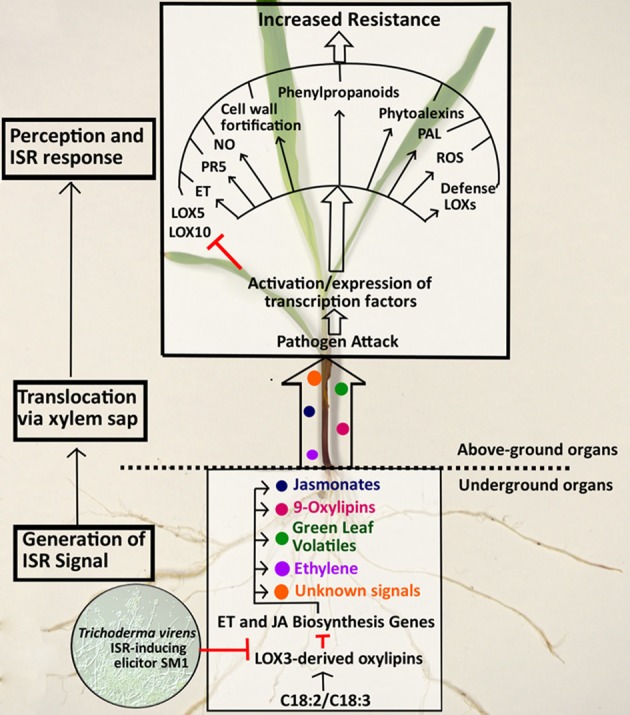
Working model of ZmLOX3 involvement in suppression of ISR Responses. In unperturbed maize roots, LOX3-derived oxylipins down-regulate defense-related genes and maintain levels of the defensive and developmental hormones, jasmonate (JA) and ethylene (ET) at healthy levels required for normal seed germination, plant growth, and normal life span (Gao et al., 2008). Upon colonization of roots, Trichoderma virens produces small molecules that possesses signaling activity and modulate gene expression in the roots to allow for mutualistic interactions with the host. One of such signal effecting expression of the host genes is a secreted proteinaceous elicitor, SM1 (Djonovic et al., 2007). One of the functions of SM1-mediated signaling is the down-regulation of expression of the host gene, ZmLOX3, which, in turn, results in de-repression of several defense and signaling host molecules including 9/13-LOX products (such as JA, traumatin, green leaf volatiles, or other oxylipins derived either from C18:3 or C18:2 polyunsaturated fatty acids) and ET. Some of these metabolites, their derivatives or their precursors are transported via xylem into aboveground organs where they signal changes in transcription of a number of defense-related genes including, upregulation of pathogenesis-related genes (e.g., PR5), phenylalanine ammonia lyase (PAL), several defense related LOXs such as LOX1, LOX2, LOX9, and other genes required for biosynthesis of JA, ROS, NO, and cell-wall fortification (e.g., lignin), phytoalexins, and other defense-related metabolites. In addition to up-regulation of defense genes, the host susceptibility genes that promote colonization of tissues by diverse pathogens are down-regulated. This includes LOX10 and LOX5 genes shown to promote disease (Christensen, 2009; Park, 2011). Collectively, these reprogramming in metabolome and transcriptome results in heightened defense status known as ISR in systemic tissues leading to increased resistance upon pathogen attack.
Interestingly, both ZmLOX5 and ZmLOX10 were down-regulated in response to C. graminicola in leaves of WT plants treated with lox3-4 sap, while in plants treated with WT sap, ZmLOX5 expression was similar to control and ZmLOX10 was up-regulated. Our previous work showed that both lox5 (Park, 2011) and lox10 (Christensen, 2009) mutants were more resistant to C. graminicola, suggesting that these two genes are susceptibility factors in maize. Suppression of these two genes in the lox3-4-sap treated plants is a logical explanation of yet another mechanism behind increased resistance in lox3-4 mutant (Figure 8). The role of these genes in the lox3 mutant background is currently under investigation through generation of double mutants in our laboratory.
Previous analyses showed that JA, ET, SA, and genes involved in JA biosynthesis pathway including LOX8, AOS, AOC, and ethylene biosynthesis gene ACO31, are up-regulated in lox3-4 maize roots (Gao et al., 2007, 2008). These findings suggest that JA and/or ethylene may themselves be mobile signals present in the lox3-4 sap that activate systemic resistance. Alternatively, increased JA/ethylene levels result in transcriptional activation of other pathways that produce the systemic mobile signal (Figure 8).
H2O2 and NO are synergistic in their signaling of defense responses, especially to biotrophs and hemibiotrophs (Mittler et al., 2004; Besson-Bard et al., 2008). Earlier and higher accumulation of H2O2 and NO in response to C. graminicola and/or ALA treatment together with earlier induction and greater expression of diverse set of defense related genes, PR1, PAL, GST, and MPI, in lox3-4 leaves may explain the increased resistance of this mutant. H2O2 was shown to limit growth of hemibiotroph Septoria tritici (Shetty et al., 2007) and the ability to scavenge and detoxify ROS is directly related to growth and virulence of hemibiotroph M. oryzea (Huang et al., 2011) and necrotroph Sclerotinia sclerotiorum (Veluchamy et al., 2012). In C. graminicola infected B73, H2O2 started to accumulate at ~36 hpi, which is similar to previous reports (Vargas et al., 2012). However, lox3-4 mutants started to accumulate H2O2 earlier (~24 hpi). In addition to direct effect on fungal growth, H2O2 is associated with cell wall lignification and cross-linking of cell wall proteins that further limits fungal growth. Continued increase in H2O2 when fungi enter the necrotrophic stage is suggested to contribute to plant cell death and disease development. lox3-4 leaves continued to accumulate H2O2 even at ~72 hpi, during the necrotrophic stage. Significant increase of GST expression in the leaves of mutant plants compared to wild type may help explain the lesser extent of necrotrophic lesion in these plants compared to WT, since GST is associated with quantitative disease resistance in maize and is implicated in resistance to multiple maize diseases (Wisser et al., 2011).
A recent study has implicated an Arabidopsis 9-lipoxygenase, AtLOX5, and its 9-oxylipins in susceptibility to foliar green peach aphid (GPA) (Nalam et al., 2012), and showed that the movement of GPA inducible 9-oxylipins via xylem sap facilitate GPA growth and host colonization. These studies and our previous reports (Gao et al., 2007; Christensen, 2009; Park, 2011) identified AtLOX5, ZmLOX3, ZmLOX5, and ZmLOX10 as “susceptibility factors” and possible targets for pathogen or insects manipulation. This raises the question of why these genes have perpetuated throughout plant evolution. The unprecedented observation that ZmLOX3 is downregulated by a beneficial root-colonizing fungus provides a possible explanation. Ubiquitous Trichoderma spp. with a wide host range may naturally downregulate this and/or other LOX genes in the host leading to de-repression/activation of ISR only when needed. The ecological cost of expressing secondary defense mechanisms such as SAR and ISR justifies the presence of their negative regulators at one or multiple layers, whose down-regulation in response to environmental cues allows for initiation and activation of the secondary defense processes. Besides Trichoderma spp., other ISR inducing microorganisms interact with maize roots in the rhizosphere and it is yet to be seen whether suppression of 9-LOXs by mutualistic rhizosphere microbes such as plant growth promoting rhizobacteria is conserved. Whether this gene is permanently suppressed in colonized plants remains to be determined.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Seth Murray, Texas A&M University and his graduate and undergraduate students for the continuous help with field-related part of our research program. We are grateful to Dr. Nasser Yalpani from Pioneer - a DuPont Company for thoughtful comments on this manuscript and his continuous support of our research. Thanks to the undergraduate student interns, Pierce Jamieson and Zack Gorman, who helped with setting up the hydroponic study. The Q-RT-PCR studies were performed at the Jo lab and his assistance and the assistance of his lab members especially Andrew Cromwell is appreciated. This work was supported by Texas A&M AgriLife Research Monocot Improvement grant and by National Science Foundation grants NSF IOB-0544428 and NSF IOS-0951272 to Michael V. Kolomiets, and by the Start-up funding and the Fundamental Research Funds for the Central Universities from Nanjing Agricultural University to Xiquan Gao. We wish to thank the Hispanic Leaders in Agriculture and the Environment Program and the Alfred P. Sloan Foundation Minority PhD Program who provided financial support to Eli J. Borrego, the Graduate Diversity Fellowship program that provided financial support to Ramadhika Damarwinasis and funding to Nasie N. Constantino from Texas A&M University's Mentoring for Success in Research and First Year Experience program.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fpls.2013.00510/abstract
(A) Image depicting hydroponic system. Setup allows for plant roots to be submerged in liquid media allowing root inoculation. (B) Image depicts xylem sap collected from V4 maize plants forming as droplets at cut sites. Cuts were made at a diagonal angle with a razor blade. (C) Illustrated image depicting dimensions of incision sites.
Primers used in this study for RT-PCR and Q-RT-PCR.
References
- Balmer D., Planchamp C., Mauch-Mani B. (2013a). On the move: induced resistance in monocots. J. Exp. Bot. 64, 1249–1261 10.1093/jxb/ers248 [DOI] [PubMed] [Google Scholar]
- Balmer D., de Papajewski D. V., Planchamp C., Glauser G., Mauch-Mani B. (2013b). Induced resistance in maize is based on organ-specific defence responses. Plant J. 74, 213–225 10.1111/tpj.12114 [DOI] [PubMed] [Google Scholar]
- Baltruschat H., Fodor J., Harrach B. D., Niemczyk E., Barna B., Gullner G., et al. (2008). Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytol. 180, 501–510 10.1111/j.1469-8137.2008.02583.x [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Pugin A., Wendehenne D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39 10.1146/annurev.arplant.59.032607.092830 [DOI] [PubMed] [Google Scholar]
- Bostock R. M. (2005). Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu. Rev. Phytopathol. 43, 545–580 10.1146/annurev.phyto.41.052002.095505 [DOI] [PubMed] [Google Scholar]
- Chen Z., Zheng Z., Huang J., Lai Z., Fan B. (2009). Biosynthesis of salicylic acid in plants. Plant Signal. Behav. 4, 493–496 10.4161/psb.4.6.8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A., Fonseca S., Fernandez G., Adie B., Chico J. M., Lorenzo O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, U666–U671 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- Choudhary D. K., Prakash A., Johri B. N. (2007). Induced systemic resistance (ISR) in plants: mechanism of action. Indian J. Microbiol. 47, 289–297 10.1007/s12088-007-0054-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen S. A. (2009). The Function of The Lipoxygenase ZmLOX10 in Maize Interactions With Insects and Pathogens. Ph.D. dissertation Texas A&M University [Google Scholar]
- Christensen S. A., Nemchenko A., Borrego E., Murray I., Sobhy I. S., Bosak L., et al. (2013). The maize lipoxygenase, ZmLOX10, mediates green leaf volatile, jasmonate and herbivore-induced plant volatile production for defense against insect attack. Plant J. 74, 59–73 10.1111/tpj.12101 [DOI] [PubMed] [Google Scholar]
- Conrath U., Beckers G. J., Flors V., Garcia-Agustin P., Jakab G., Mauch F., et al. (2006). Priming: getting ready for battle. Mol. Plant Microbe. Interact. 19, 1062–1071 10.1094/MPMI-19-1062 [DOI] [PubMed] [Google Scholar]
- Delledonne M., Xia Y. J., Dixon R. A., Lamb C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588 10.1038/29087 [DOI] [PubMed] [Google Scholar]
- Djonovic S., Vargas W. A., Kolomiets M. V., Horndeski M., Wiest A., Kenerley C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889 10.1104/pp.107.103689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant W. E., Dong X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209 10.1146/annurev.phyto.42.040803.140421 [DOI] [PubMed] [Google Scholar]
- Fu Z. Q., Dong X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863 10.1146/annurev-arplant-042811-105606 [DOI] [PubMed] [Google Scholar]
- Gao X., Shim W. B., Gobel C., Kunze S., Feussner I., Meeley R., et al. (2007). Disruption of a maize 9-lipoxygenase results in increased resistance to fungal pathogens and reduced levels of contamination with mycotoxin fumonisin. Mol. Plant Microbe. Interact. 20, 922–933 10.1094/MPMI-20-8-0922 [DOI] [PubMed] [Google Scholar]
- Gao X., Starr J., Gobel C., Engelberth J., Feussner I., Tumlinson J., et al. (2008). Maize 9-lipoxygenase ZmLOX3 controls development, root-specific expression of defense genes, and resistance to root-knot nematodes. Mol. Plant Microbe. Interact. 21, 98–109 10.1094/MPMI-21-1-0098 [DOI] [PubMed] [Google Scholar]
- Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M. (2004). Trichoderma species—opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56 10.1038/nrmicro797 [DOI] [PubMed] [Google Scholar]
- Heil M. (2001). The ecological concept of costs of induced systemic resistance (ISR). Eur. J. Plant Pathol. 107, 137–146 10.1023/A:1008793009517 [DOI] [Google Scholar]
- Heil M., Baldwin I. T. (2002). Fitness costs of induced resistance: emerging experimental support for a slippery concept. Trends Plant Sci. 7, 61–67 10.1016/S1360-1385(01)02186-0 [DOI] [PubMed] [Google Scholar]
- Hu P., Zhou W., Cheng Z., Fan M., Wang L., Xie D. (2013). JAV1 controls jasmonate-regulated plant defense. Mol. Cell 50, 504–515 10.1016/j.molcel.2013.04.027 [DOI] [PubMed] [Google Scholar]
- Huang K., Czymmek K. J., Caplan J. L., Sweigard J. A., Donofrio N. M. (2011). HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathogens 7: e1001335 10.1371/journal.ppat.1001335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakeit T., Gao X., Kolomiets M. (2007). Increased resistance of a maize mutant lacking the 9−Lipoxygenase gene, ZmLOX3, to root rot caused by exserohilum pedicellatum. J. Phytopathol. 155, 758–760 10.1111/j.1439-0434.2007.01301.x [DOI] [Google Scholar]
- Mastouri F., Bjorkman T., Harman G. E. (2012). Trichoderma harzianum enhances antioxidant defense of tomato seedlings and resistance to water deficit. Mol. Plant Microbe. Interact. 25, 1264–1271 10.1094/MPMI-09-11-0240 [DOI] [PubMed] [Google Scholar]
- Mathys J., de Cremer K., Timmermans P., van Kerckhove S., Lievens B., Vanhaecke M., et al. (2012). Genome-wide characterization of ISR induced in arabidopsis thaliana by trichoderma hamatum T382 against botrytis cinerea infection. Front. Plant Sci. 3:108 10.3389/fpls.2012.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauch-Mani B., Slusarenko A. J. (1996). Production of salicylic acid precursors is a major function of phenylalanine ammonia-lyase in the resistance of arabidopsis to peronospora parasitica. Plant Cell 8, 203–212 10.2307/3870265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R., Vanderauwera S., Gollery M., van Breusegem F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498 10.1016/j.tplants.2004.08.009 [DOI] [PubMed] [Google Scholar]
- Nalam V. J., Keeretaweep J., Sarowar S., Shah J. (2012). Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell 24, 1643–1653 10.1105/tpc.111.094110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.-S. (2011). Diverse Functions of the Two Segmentally Duplicated 9-Lipoxygenases ZmLOX4 and ZmLOX5 of Maize. Ph.D. dissertation, Texas A&M University [Google Scholar]
- Pieterse C. M., Leon-Reyes A., Van der Ent S., Van Wees S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 10.1038/nchembio.164 [DOI] [PubMed] [Google Scholar]
- Pieterse C. M. J., Vanwees S. C. M., Hoffland E., Vanpelt J. A., Vanloon L. C. (1996). Systemic resistance in Arabidopsis induced by biocontrol bacteria is independent of salicylic acid accumulation and pathogenesis-related gene expression. Plant Cell 8, 1225–1237 10.2307/3870297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozo M. J., Azcon-Aguilar C. (2007). Unraveling mycorrhiza-induced resistance. Curr. Opin. Plant Biol. 10, 393–398 10.1016/j.pbi.2007.05.004 [DOI] [PubMed] [Google Scholar]
- Ruijter J. M., Ramakers C., Hoogaars W. M., Karlen Y., Bakker O., Van den Hoff M. J., et al. (2009). Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 37, e45 10.1093/nar/gkp045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu C. M., Farag M. A., Hu C. H., Reddy M. S., Kloepper J. W., Pare P. W. (2004). Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 134, 1017–1026 10.1104/pp.103.026583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty N. P., Mehrabi R., Lütken H., Haldrup A., Kema G. H., Collinge D. B., et al. (2007). Role of hydrogen peroxide during the interaction between the hemibiotrophic fungal pathogen Septoria tritici and wheat. New Phytol. 174, 637–647 10.1111/j.1469-8137.2007.02026.x [DOI] [PubMed] [Google Scholar]
- Shoresh M., Harman G. E., Mastouri F. (2010). Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 48, 21–43 10.1146/annurev-phyto-073009-114450 [DOI] [PubMed] [Google Scholar]
- Spoel S. H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100 10.1038/nri3141 [DOI] [PubMed] [Google Scholar]
- Thaler J. S., Humphrey P. T., Whiteman N. K. (2012). Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17, 260–270 10.1016/j.tplants.2012.02.010 [DOI] [PubMed] [Google Scholar]
- Ton J., D'Alessandro M., Jourdie V., Jakab G., Karlen D., Held M., et al. (2007). Priming by airborne signals boosts direct and indirect resistance in maize. Plant J. 49, 16–26 10.1111/j.1365-313X.2006.02935.x [DOI] [PubMed] [Google Scholar]
- Torres M. A., Dangl J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403 10.1016/j.pbi.2005.05.014 [DOI] [PubMed] [Google Scholar]
- Udvardi M. K., Czechowski T., Scheible W. R. (2008). Eleven golden rules of quantitative RT-PCR. Plant Cell 20, 1736–1737 10.1105/tpc.108.061143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent S., Koornneef A., Ton J., Pieterse C. M. J. (2009a). Induced resistance—orchestrating defence mechanisms through crosstalk and priming, in Annual Plant Reviews, ed Parker J. (Oxford: Wiley-Blackwell; ). 34, 334–370 10.1002/9781444301441.ch11 [DOI] [Google Scholar]
- Van der Ent S., Van Wees S. C., Pieterse C. M. (2009b). Jasmonate signaling in plant interactions with resistance-inducing beneficial microbes. Phytochemistry 70, 1581–1588 10.1016/j.phytochem.2009.06.009 [DOI] [PubMed] [Google Scholar]
- van Hulten M., Pelser M., van Loon L. C., Pieterse C. M., Ton J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 5602–5607 10.1073/pnas.0510213103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wees S. C., Luijendijk M., Smoorenburg I., Van Loon L. C., Pieterse C. M. (1999). Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 41, 537–549 10.1023/A:1006319216982 [DOI] [PubMed] [Google Scholar]
- Van Wees S. C., Van der Ent S., Pieterse C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448 10.1016/j.pbi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Vargas W. A., Martin J. M., Rech G. E., Rivera L. P., Benito E. P., Diaz-Minguez J. M., et al. (2012). Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 158, 1342–1358 10.1104/pp.111.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veluchamy S., Williams B., Kim K., Dickman M. B. (2012). The CuZn superoxide dismutase from Sclerotinia sclerotiorum is involved with oxidative stress tolerance, virulence, and oxalate production. Physiol. Mol. Plant Pathol. 78, 14–23 10.1016/j.pmpp.2011.12.005 [DOI] [Google Scholar]
- Vlot A. C., Klessig D. F., Park S. W. (2008). Systemic acquired resistance: the elusive signal(s). Curr. Opin. Plant Biol. 11, 436–442 10.1016/j.pbi.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Waller F., Achatz B., Baltruschat H., Fodor J., Becker K., Fischer M., et al. (2005). The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proc. Natl. Acad. Sci. U.S.A. 102, 13386–13391 10.1073/pnas.0504423102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D. R., Ratsep J., Havis N. D. (2013). Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280 10.1093/jxb/ert026 [DOI] [PubMed] [Google Scholar]
- Weller D. M., Mavrodi D. V., Van Pelt J. A., Pieterse C. M., Van Loon L. C., Bakker P. A. (2012). Induced systemic resistance in arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens. Phytopathology 102, 403–412 10.1094/PHYTO-08-11-0222 [DOI] [PubMed] [Google Scholar]
- Wisser R. J., Kolkman J. M., Patzoldt M. E., Holland J. B., Yu J., Krakowsky M., et al. (2011). Multivariate analysis of maize disease resistances suggests a pleiotropic genetic basis implicates a GST gene. Proc. Natl. Acad. Sci. U.S.A. 108, 7339–7344 10.1073/pnas.1011739108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Image depicting hydroponic system. Setup allows for plant roots to be submerged in liquid media allowing root inoculation. (B) Image depicts xylem sap collected from V4 maize plants forming as droplets at cut sites. Cuts were made at a diagonal angle with a razor blade. (C) Illustrated image depicting dimensions of incision sites.
Primers used in this study for RT-PCR and Q-RT-PCR.