Abstract
Partial differential equation (PDE) models are commonly used to model complex dynamic systems in applied sciences such as biology and finance. The forms of these PDE models are usually proposed by experts based on their prior knowledge and understanding of the dynamic system. Parameters in PDE models often have interesting scientific interpretations, but their values are often unknown, and need to be estimated from the measurements of the dynamic system in the present of measurement errors. Most PDEs used in practice have no analytic solutions, and can only be solved with numerical methods. Currently, methods for estimating PDE parameters require repeatedly solving PDEs numerically under thousands of candidate parameter values, and thus the computational load is high. In this article, we propose two methods to estimate parameters in PDE models: a parameter cascading method and a Bayesian approach. In both methods, the underlying dynamic process modeled with the PDE model is represented via basis function expansion. For the parameter cascading method, we develop two nested levels of optimization to estimate the PDE parameters. For the Bayesian method, we develop a joint model for data and the PDE, and develop a novel hierarchical model allowing us to employ Markov chain Monte Carlo (MCMC) techniques to make posterior inference. Simulation studies show that the Bayesian method and parameter cascading method are comparable, and both outperform other available methods in terms of estimation accuracy. The two methods are demonstrated by estimating parameters in a PDE model from LIDAR data.
Some Key Words: Asymptotic theory, Basis function expansion, Bayesian method, Differential equations, Measurement error, Parameter cascading
1 Introduction
Differential equations are important tools in modeling dynamic processes, and are widely used in many areas. The forward problem of solving equations or simulating state variables for given parameters that define the differential equation models has been studied extensively by mathematicians. However, the inverse problem of estimating parameters based on observed error-prone state variables has a relatively sparse statistical literature, and this is especially the case for partial differential equation (PDE) models. There is growing interest in developing efficient estimation methods for such problems.
Various statistical methods have been developed to estimate parameters in ordinary differential equation (ODE) models. There is a series of work in the study of HIV dynamics in order to understand the pathogenesis of HIV infection. For example, Ho, et al. (1995) and Wei, et al. (1995) used standard nonlinear least squares regression methods, while Wu, Ding and DeGruttola (1998) and Wu and Ding (1999) proposed a mixed-effects model approach. Refer to Wu (2005) for a comprehensive review of these methods. Furthermore, Putter, et al. (2002), Huang and Wu (2006), and Huang, Liu and Wu (2006) proposed hierarchical Bayesian approaches for this problem. These methods require repeatedly solving ODE models numerically, which could be time-consuming. Ramsay (1996) proposed a data reduction technique in functional data analysis which involved solving for coefficients of linear differential operators, see Poyton, et al. (2006) for an example of application. Li, et al. (2002) studied a pharmacokinetic model and proposed a semiparametric approach for estimating time-varying coefficients in an ODE model. Ramsay, et al. (2007) proposed a generalized smoothing approach, based on profile likelihood ideas, which they named parameter cascading, for estimating constant parameters in ODE models. Cao, Wang and Xu (2011) proposed robust estimation for ODE models when data have outliers. Cao, Huang and Wu (2012) proposed a parameter cascading method to estimate time-varying parameters in ODE models. These methods estimate parameters by optimizing certain criteria. In the optimization procedure, using gradient-based optimization techniques may have the parameter estimates converge to a local minima, otherwise global optimization is computationally intensive.
Another strategy to estimate parameters of ODE is the two-stage method, which in the first stage estimates the function and its derivatives from noisy observations using data smoothing methods without considering differential equation models, and then in the second stage estimates of ODE parameters are obtained by least squares. Liang and Wu (2008) developed a two-stage method for a general first order ODE model, using local polynomial regression in the first stage, and established asymptotic properties of the estimator. Similarly, Chen and Wu (2008) developed local estimation for time-varying coefficients. The two-stage methods are easy to implement, however, they might not be statistically efficient, due to the fact that derivatives cannot be estimated accurately from noisy data, especially higher order derivatives.
As for PDEs, there are two main approaches. The first is similar to the two-stage method in Liang and Wu (2008). For example, Bar, Hegger and Kantz (1999) modeled unknown PDEs using multivariate polynomials of sufficiently high order, and the best fit was chosen by minimizing the least squares error of the polynomial approximation. Based on the estimated functions, the PDE parameters were estimated using least squares (Muller and Timmer 2004). The issues of noise level and data resolution were addressed extensively in this approach. See also Parlitz and Merkwirth (2000) and Voss, et al. (1999) for more examples. The second approach uses numerical solutions of PDEs, thus circumventing derivative estimation. For example, Muller and Timmer (2002) solved the target least-squares type minimization problem using an extended multiple shooting method. The main idea was to solve initial value problems in sub-intervals and integrate the segments with additional continuity constraints. Global minima can be reached in this algorithm, but it requires careful parameterization of the initial condition, and the computational cost is high.
In this article, we consider a multidimensional dynamic process, g(x), where x = (x1, …, xp)T ∈ ℝp is a multi-dimensional argument. Suppose this dynamic process can be modeled with a PDE model
| (1) |
where θ = (θ1, …, θm)T is the parameter vector of primary interest, and the left hand side of (1) has a parametric form in g(x) and its partial derivatives. In practice, we do not observe g(x) but instead observe its surrogate Y (x). We assume that g(x) is observed over a meshgrid with measurement errors, so that for i = 1, …, n, we observe data (Yi, xi) satisfying
where εi, i = 1,…, n, are independent and identically distributed measurement errors and are assumed here to follow a Gaussian distribution with mean zero and variance . Our goal is to estimate the unknown θ in the PDE model (1) from noisy data, and to quantify the uncertainty of the estimates.
As mentioned before, a straightforward two-stage strategy, though easy to implement, has difficulty in estimating derivatives of the dynamic process accurately, leading to biased estimates of the PDE parameter. We propose two joint modeling schemes: (a) a parameter cascading or penalized profile likelihood approach and (b) a fully Bayesian treatment. We conjecture that joint modeling approaches are more statistically efficient than a two-stage method, a conjecture that is borne out in our simulations. For the parameter cascading approach, we make two crucial contributions besides the extension to multivariate splines. First, we develop an asymptotic theory for the model fit, along with a new approximate covariance matrix that includes the smoothing parameters. Second, we propose a new criterion for the smoothing parameter selection, which is shown to outperform available criteria used in ODE parameter estimation. Because of the nature of the penalization in the parameter cascading approach, there is no obvious direct ”Bayesianization” of it. Instead, we develop a new hierarchical model for the PDE. At the first stage of the hierarchy, the unknown function is related to the data. At the next stage, the PDE induces a prior on the parameters which is very different from the penalty used in the parameter cascading algorithm. This PDE restricted prior is new in the Bayesian literature. Further, we allow multiple smoothing parameters and perform Bayesian model mixing to obtain the whole uncertainty distribution of the smoothing parameters. Our MCMC based method is of course also very different than the parameter cascading method where we jointly draw parameters rather than using conditional optimization.
The main idea of our two methods is to represent the unknown dynamic process via a nonparametric function while using the PDE model to regularize the fit. In both methods, the nonparametric function is expressed as a linear combination of B-spline basis functions. In the parameter cascading method, this nonparametric function is estimated using penalized least squares, where a penalty term is defined to incorporate the PDE model. This penalizes the infidelity of the nonparametric function to the PDE model, so that the non-parametric function is forced to better represent the dynamic process modeled by the PDE. In the Bayesian method, the PDE model information is coded in the prior distribution. We recognize that there is no exact solution by substituting the nonparametric function into the PDE model (1). This PDE modeling error is then modeled as a random process, hence inducing a constraint on the basis function coefficients. We also introduce in the prior an explicit penalty on the smoothness of the nonparametric function. Our two methods avoid direct estimation of the derivative of the dynamic process, which can be obtained easily as a linear combination of the derivatives of the basis functions, and also avoids specifying boundary conditions.
In principle, the proposed methods are applicable to all PDEs, thus having potentially wide applications. As quick examples of PDEs, the heat equation and wave equation are among the most famous ones. The heat equation, also known as the diffusion equation, describes the evolution in time of the heat distribution or chemical concentration in a given region, and is defined as . The wave equation is a simplified model for description of waves, such as sound waves, light waves and water waves, and is defined as . More examples of famous PDE are the Laplace equation, the transport equation and the beam equation. See Evans (1998) for a detailed introduction to PDEs.
For illustration, we will do specific calculations based on our empirical example of LIDAR data described in Section 5 and also used in our simulations in Section 4. There we propose a PDE model for received signal g(t, z) over time t and range z given as
| (2) |
The PDE (2) is a linear PDE of parabolic type in one space dimension and is also called a (one-dimensional) linear reaction-convection-diffusion equation. If g(t, z) were observable, (2) has a closed form solution, obtained by separation of variables, but the solution is the sum of an infinite sequence. Such a solution requires a high computational load to evaluate the solution over a meshgrid of moderate size.
The rest of the article is organized as follows. The parameter cascading method is introduced in Section 2, and the asymptotic properties of the proposed estimator are established. In Section 3 we introduce the Bayesian framework and explain how to make posterior inference using a Markov chain Monte Carlo (MCMC) technique. Simulation studies are presented in Section 4 to evaluate the finite sample performance of our two methods in comparison with a two-stage method. In Section 5 we illustrate the methods using a LIDAR data. Finally, we conclude with some remarks in Section 6.
2 Parameter Cascading Method
2.1 Basis Function Approximation
When solving partial differential equations, it is possible to obtain a unique, explicit formula for certain specific examples, such as the wave equation. However, most PDEs used in practice have no explicit solutions, and can only be solved by numeric methods such as finite difference method (Morton and Mayers, 2005) and finite element method (Brenner and Scott, 2010). Instead of repeatedly solving PDEs numerically for thousands of candidate parameters, which is computationally expensive, we represent the dynamic process, g(x), modeled in (1), by a nonparametric function, which can be expressed as a linear combination of basis functions
| (3) |
where b(x) = {b1(x), …, bK(x)}T is the vector of basis functions, and β = (β1, …, βK)T is the vector of basis coefficients.
We choose B-splines as basis functions in all simulations and applications in this article, since B-splines are non-zero only in short subintervals, a feature called the compact support property (de Boor 2001), which is useful for efficient computation and numerical stability, compared with other basis (e.g. truncated power series basis). The B-spline basis functions are defined with their order, the number and locations of knots. Some work has been aimed at automatic knot placement and selection. Many of the feasible frequentist methods, for example, Friedman and Silverman (1989) and Stone, et al. (1997), are based on stepwise regression. A Bayesian framework is also available, see Denison, Mallick and Smith (1997) for example. Despite good performance, knot selection procedures are highly computationally intensive. To avoid the complicated knot selection problem, we use a large enough number of knots to make sure the basis functions are sufficiently flexible to approximate the dynamic process. To prevent the nonparametric function overfitting the data, one penalty term will be defined with the PDE model in the next subsection to penalize the roughness of the nonparametric function.
The PDE model (1) can be expressed using the same set of B-spline basis functions by substituting (3) into model (1) as follows
In the special case of linear PDEs, the above expression is also linear in β, which can be expressed as
| (4) |
where f{b(x), ∂b(x)/∂x1, … ; θ} is a linear function of the basis functions and their derivatives. In the following, we denote ℱ{x, g(x), …; θ} by the short hand notation ℱ{g(x); θ}, and f{b(x), ∂b(x)/∂x1, … ; θ} by f (x; θ). For the PDE example (2), the form of f (x; θ) is given in Appendix A.1.
2.2 Estimating β and θ
Following Section 2.1, the dynamic process, g(x), is expressed as a linear combination of basis functions. It is natural to estimate the basis function coefficients, β, using penalized splines (Eilers and Marx, 2010; Ruppert, Wand and Carroll, 2003). If we were simply interested in estimating g(·) = bT(·)β, then we would use the usual penalty λβTPTPβ, where λ is a penalty parameter and P is a matrix performing differencing on adjacent elements of β (Eilers and Marx, 2010). Such a penalty does penalize to achieve smoothness of the estimated function, however, it is not in fidelity with (1). Instead, for fixed θ, we define the roughness penalty as ∫ [ℱ{g(x); θ}]2dx. This penalty incorporates the PDE model, containing derivatives involved in the model. As a result, the penalty is able to regularize the spline fit. It also shows fidelity to the PDE model, i.e., smaller value indicates more fidelity of the spline approximation to the PDE. Hence, we propose to estimate the coefficients, β, for fixed θ by minimizing the penalized least squares
| (5) |
The integration in (5) can be approximated numerically by numerical integration methods. Burden and Douglas (2000) suggested that a composite Simpson’s rule provided an adequate approximation, a suggestion that we use.. See Appendix B.1 in the Supplementary Material for details.
The PDE parameter θ is then estimated using a higher level of optimization. Denote the estimate of the spline coefficients by β̂(θ), which is considered as a function of θ. Define ĝ(x, θ) = bT(x)β̂ (θ). Because the estimator β̂(θ) is already regularized, we propose to estimate θ by minimizing the least squares measure of fit
| (6) |
For a general nonlinear PDE model, the function β̂(θ) might have no closed form, and the estimate is thus obtained numerically. This lower level of optimization for fixed θ is embedded inside the optimization of θ. The objective functions J(β|θ) and H(θ) are minimized iteratively until convergence to a solution. In some cases, the optimization can be accelerated and made more stable by providing the gradient, whose analytic form, by the chain rule, is ∂H(θ)/∂θ = {∂β̂(θ)/∂θ}T × ∂H(θ)/∂β̂(θ). Although β̂ (θ) does not have an explicit expression, the implicit function theorem can be applied to find the analytic form of the first-order derivative of β̂ (θ) with respect to θ required in the above gradient. Because β̂ is the minimizer of J(β|θ), we have ∂J(β|θ)/∂β|β̂ = 0. By taking the total derivative with respect to θ on the left hand side, and assuming ∂2J(β|θ)/∂βT∂β|β̂ is nonsingular, the analytic expression of the first-order derivative of β̂ is
When the PDE model (1) is linear, β̂ has a close form and the algorithm can be stated as follows. By substituting in (3) and (4), the lower level criterion (5) becomes
Let B be the n × K basis matrix with ith row bT(xi), and define Y = (Y1, …, Yn)T, and the K × K penalty matrix R(θ) = ∫ f (x; θ)fT(x; θ)dx. See Appendix B.1 in the Supplementary Material for calculation of R(θ) for the PDE example (2). Then the penalized least squares criterion (5) can be expressed in the matrix notation
| (7) |
which is a quadratic function of β. By minimizing the above penalized least squares criterion, the estimate for β, for fixed θ, can be obtained in close form as
| (8) |
Then by substituting in (8), (6) becomes
| (9) |
To summarize, when estimating parameters in linear PDE models, we minimize criterion (9) to obtain an estimate, θ̂, for parameters in linear PDE models. The estimated basis coefficients, β̂, are obtained by substituting θ̂ into (8).
2.3 Smoothing Parameter Selection
Our ultimate goal is to obtain an estimate for the PDE parameter θ such that the solution of the PDE is close to the observed data. For any given value of the smoothing parameter, λ, we obtain the PDE parameter estimate, θ̂, and the basis coefficient estimate, β̂(θ̂). Both can be treated as functions of λ, which are denoted as θ̂(λ) and β̂{θ̂(λ), λ}. Define ei(λ) = Yi − ĝ{xi, θ̂(λ), λ} and ηi(λ) = ℱ{ĝ(xi); θ̂(λ)}, the latter of which is f̂T{xi; θ̂(λ)}β̂{θ̂(λ), λ} for linear PDE models. Fidelity to the PDE can be measured by , while fidelity to the data can be measured by . Clearly, minimizing just leads to λ = 0, and gives far too undersmoothed data fits, while simultaneously not taking the PDE into account. On the other hand, our experience shows that minimizing always results in the largest candidate value for λ.
Hence, we propose the following criterion, which considers data fitting and PDE model fitting simultaneously. To choose an optimal λ, we minimize
The first summation term in G(λ), which measures the fit of the estimated dynamic process to the data, tends to choose a small value of the smoothing parameter. The second summation term in G(λ), which measures the fidelity of the estimated dynamic process to the PDE model, tends to choose a large value of the smoothing parameter. Adding these two terms together allows a choice of the value for the smoothing parameter that makes the best trade-off between fitting to data and fidelity to the PDE model. For the sake of completeness, we tried cross-validation and generalized cross-validation to estimate the smoothing parameter. The result was to greatly undersmooth the function fit, while leading to biased and quite variable estimates of the PDE parameters.
2.4 Limit Distribution and Variance Estimation of Parameters
We analyze the limiting distribution of θ̂ following the same line of argument as in Yu and Ruppert (2002), under Assumptions 1–4 in Appendix A.2. As in their work, we assume that the spline approximation is exact so that g(x) = bT(x)β0 for a unique β0 = β0(θ0), our Assumption 2. Let θ0 be the true value of θ, and define λ̃ = λ/n, , Gn(θ) = Sn + λ̃R(θ), Rjθ(θ) = ∂R(θ)/∂θj, and . Define Λn(θ) to have (j, k)th element
Define , where 𝒞n(θ0) has (j, k)th element . Let be the inverse of the symmetric square root of Σn,prop.
Following the same basic outline of Yu and Ruppert (2002), and essentially their assumptions although the technical details are considerably different, we show in Appendix A.2 that under Assumptions 1–4 stated there, and assuming homoscedasticity,
| (10) |
Estimating Σn,prop, is easy by replacing θ0 by θ̂ and β0 by β̂ = β̂ (θ̂), and estimating by fitting a standard spline regression and then forming the residual variance. In the case of heteroscedastic errors, the term in 𝒞n,jk(θ0) can be replaced by its consistent estimate , where p is the number of parameters in the B-spline. We use this sandwich-type method in our numerical work.
3 Bayesian Estimation and Inference
3.1 Basic Methodology
In this section we introduce a Bayesian approach for estimating parameters in PDE models. In this Bayesian approach, the dynamic process modeled by the PDE model is represented by a linear combination of B-spline basis functions, which is estimated with Bayesian P-splines. The coefficients of the basis functions are regularized through the prior, which contains the PDE model information. Therefore, data fitting and PDE fitting are incorporated into a joint model. As described in the paragraph after equation (1), our approach is not a direct “Bayesianization” of the methodology described in Section 2.
We use the same notation as before. With the basis function representation given in (3), the basis function model for data fitting is Yi = bT(xi)β+εi, where the εi are independent and identically distributed measurement errors and are assumed to follow a Gaussian distribution with mean zero and variance . The basis functions are chosen with the same rule introduced in the previous section.
In conventional Bayesian P-splines, which will be introduced in Section 3.2, the penalty term penalizes the smoothness of the estimated function. Rather than using a single optimal smoothing parameter as in frequentist methods, the Bayesian approach performs model mixing with respect to this quantity. In other words, many different spline models provide plausible representations of the data, and the Bayesian approach treats such model uncertainty through the prior distribution of the smoothing parameter.
In our problem, we know further that the underlying function satisfies a given PDE model. Naturally, this information should be coded into the prior distribution to regularize the fit. Because we recognize that there may be no basis function representation that exactly satisfies the PDE model (1), for the purposes of Bayesian computation, we will treat the approximation error as random, and the PDE modeling errors are
| (11) |
where the random modeling errors, ζ(xi), are assumed to be independent and identically distributed with a prior distribution , where the precision parameter, γ0, should be large enough so that the approximation error in solving (1) with a basis function representation is small. Similarly, instead of using a single optimal value for the precision parameter, γ0, a prior distribution is assigned to γ0. The modeling error distribution assumption (11) and a roughness penalty constraint together induce a prior distribution on the basis function coefficients β. The choice of roughness penalty depends on the dimension of x. For simplicity, we state the Bayesian approach with the specific penalty shown in Section 3.2. The prior distribution of β is
| (12) |
where, as before, K denotes the number of basis functions, γ0 is the precision parameter, ζ(β, θ) = [ℱ{bT(x1)β; θ}, …, ℱ{bT(xn)β; θ}]T, γ1 and γ2 control the amount of penalty on smoothness, and the penalty matrices H1, H2, H3 are the same as in the usual Bayesian P-splines, given in (14). We assume conjugate priors for and γℓ as , γℓ ~ Gamma(aℓ, bℓ), for ℓ = 0, 1, 2, where IG(a,b) denotes the Inverse-Gamma distribution with mean (a − 1)−1b. For the PDE parameter, θ, we assign a prior, with variance large enough to remain noninformative.
Denote γ = (γ0, γ1, γ2)T and . Based on the above model and prior specification, the joint posterior distribution of all unknown parameters is
| (13) |
The posterior distribution (13) is not analytically tractable, hence we use a Markov chain Monte Carlo (MCMC) based computation method (Gilks, Richardson and Spiegelhalter, 1996) or more precisely Gibbs sampling (Gelfand and Smith, 1990) to simulate the parameters from the posterior distribution. To implement the Gibbs sampler, we need the full conditional distributions of all unknown parameters. Due to the choice of conjugate priors, the full conditional distributions of and γℓ’s are easily obtained as Inverse-Gamma and Gamma distributions, respectively. The full conditional distributions of β and θ are not of standard form, and hence we employ Metropolis-Hastings algorithm to sample them.
In the special case of a linear PDE, simplifications arise. With approximation (4), the PDE modeling errors are represented as ζ(xi) = fT(xi; θ)β, for i = 1, …, n. Define the matrix F(θ) = {f (x1; θ), …, f (xn; θ)}T. Then the prior distribution of β given in (12) becomes
where the exponent is quadratic in β. Then the joint posterior distribution of all unknown parameters given in (13) becomes
Under linear PDE models, the full conditional of β is easily seen to be a Normal distribution. This reduces the computational cost significantly compared with sampling under nonlinear cases, because the length of the vector β increases quickly as dimension increases. Computational details of both nonlinear and linear PDEs are shown in Appendix A.3.
3.2 Bayesian P-Splines
Here we describe briefly the implementation of Bayesian penalized splines, or P-splines. Eilers and Marx (2003) and Marx and Eilers (2005) deal specifically with bivariate penalized B-splines. In the simulation studies and the application of this article, we use the bivariate B-spline basis, which is formed by the tensor product of one-dimensional B-spline basis.
Following Marx and Eilers (2005), we use the difference penalty to penalize the interaction of one-dimensional coefficients as well as each dimension individually. Denote the number of basis functions in each dimension by kℓ, the one-dimensional basis function matrices by Bℓ, and the order difference matrix of size (kℓ − mℓ) × kℓ by Dℓ, for ℓ = 1, 2. The prior density of the basis function coefficient β of length K = k1k2 is assumed to be [β|γ1, γ2] ∝ (γ1γ2)K/2 exp{−βT(γ1H1 + γ2H2 + γ1γ2H3)β/2}, where γ1 and γ2 are hyper-parameters, and the matrices are
| (14) |
When assuming conjugate prior distributions as , [γ1] = Gamma(a1, b1), and [γ2] = Gamma(a2, b2), the posterior distribution can be derived easily and sampled using the Gibbs sampler. Though the prior distribution of β is improper, the posterior distribution is proper (Berry, Carroll and Ruppert, 2002).
4 Simulations
4.1 Background
In this section, the finite sample performances of our methods are investigated via Monte Carlo simulations, which are also compared with a two-stage method described below.
The two-stage method is constructed for PDE parameter estimation as follows. In the first stage, g(x) and the partial derivatives of g(x) are estimated by the multidimensional penalized signal regression (MPSR) method (Marx and Eilers 2005). Marx and Eilers (2005) showed that their MPSR method was competitive with other popular methods and had several advantages such as taking full advantage of the natural spatial information of the signals and being intuitive to understand and use. Let β̂ denote the estimated coefficients of the basis functions in the first stage. In the second stage, we plug the estimated function and partial derivatives into the PDE model, ℱ{g(x); θ} = 0, for each observation, i.e., we calculate ℱ̂{ĝ(xi); θ} for i = 1, …, n. Then, a least-squares type estimator for the PDE parameter, θ, is obtained by minimizing . For comparison purposes, the standard errors of two-stage estimates of the PDE parameters are estimated using a parametric bootstrap, which is implemented as follows. Let θ̂ denote the estimated PDE parameter using the two-stage method, and S(x|θ̂) denote the numerical solution of PDE (2) using θ̂ as the parameter value. New simulated data are generated by adding independent and identically distributed Gaussian noises with the same standard deviation as the data to the PDE solutions at every 1 time unit and every 1 range unit. The PDE parameter is then estimated from the simulated data using the two-stage method, and the PDE parameter estimate is denoted as θ̃(j), where j = 1, …, 100, is the index of replicates in the parametric bootstrap procedure. The experimental standard deviation of θ̃(j) is set as the standard error of two-stage estimates.
4.2 Data Generating Mechanism
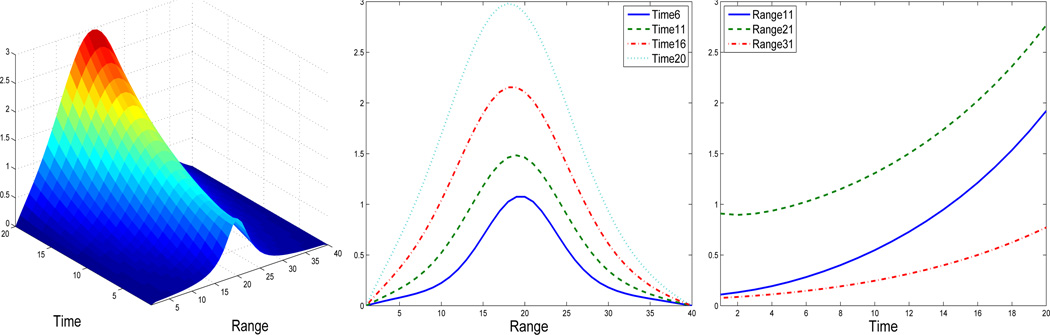
The PDE model (2) is used to simulate data. The PDE model (2) is numerically solved by setting the true parameter values as θD = 1, θS = 0.1, and θA = 0.1, the boundary condition as g(t, 0) = 0, and the initial condition as g(0, z) = {1 + 0.1 × (20 − z)2}ℒ1 over a meshgrid in the time domain t ∈ [1, 20] and the range domain z ∈ [1, 40]. In order to obtain a precise numerical solution, we take grid of size 0.0005 in the time domain and of size 0.001 in the range domain. The numerical solution is shown in Figure 1, together with cross sectional views along time and range axis. Then the observed error-prone data is simulated by adding independent and identically distributed Gaussian noises with standard deviation σ = 0.02 to the PDE solutions at every 1 time unit and every 1 range unit. In other words, our data is on a 20-by-40 meshgrid in the domain [1, 20] × [1, 40]. This value of σ is close to that of our data example in Section 5. In order to investigate the effect of data noise on the parameter estimation, we do another simulation study in which the simulated data are generated in the exact same setting except that the standard deviation of noises is set as σ = 0.05.
Figure 1.
Snapshots of the numerical solution, g(t, z), for the PDE model (2). Left: 3-D plot of the surface g(t, z). Middle: plot of g(ti, z) for time values ti over range, with ti = 6, 11, 16, 20. Right: plot of g(t, zj) for range values zj over time, with zj = 11, 21, 31.
4.3 Performance of the Proposed Methods
The parameter cascading method, the Bayesian method, and the two-stage method were applied to estimate the three parameters in the PDE model (2) from the simulated data. The simulation is implemented with 1000 replicates. This section summarizes the performance of these three methods in this simulation study.
The PDE model (2) indicates that the second partial derivative with respect to z is continuously differentiable, and thus we choose quartic basis functions in the range domain. Therefore, for representing the dynamic process, g(t, z), we use a tensor product of one-dimensional quartic B-splines to form the basis functions, with 5 and 17 equally spaced knots in time domain and range domain, respectively, in all three methods.
In the two-stage method for estimating PDE parameters, the Bayesian P-Spline method is used to estimate the dynamic process and its derivatives by setting the hyper-parameters defined in Section 3.1 as aε = bε = a1 = b1 = a2 = b2 = 0.01, and taking the third order difference matrix to penalize the roughness of the second derivative in each dimension. In the Bayesian method for estimating PDE parameters, we take the same smoothness penalty as in the two-stage method, and the hyper-parameters defined in Section 3 are set to be aε = bε = aℓ = bℓ = 0.01 for ℓ = 0, 1, 2, and . In the MCMC sampling procedure, we collect every 5th sample after a burn-in stage of length 5000, until 3000 posterior samples are obtained.
We summarize the simulation results in Table 1, including the biases, standard deviations, square root of average squared errors, and coverage probabilities of 95% confidence intervals for each method. We see that the Bayesian method and the parameter cascading method are comparable, and both have smaller biases, standard deviations and root average squared errors than the two-stage method. The improvement in θD is substantial, which is associated with the second partial derivative, ∂2g(t, z)/∂z2. This is consistent with our conjecture that the two-stage strategy is not statistically efficient because of the inaccurate estimation of derivatives, especially higher order derivatives.
Table 1.
The biases, standard deviations (SD), square roots of average squared errors (RASE) of the parameter estimates for the PDE model (2) using the Bayesian method (BM), the parameter cascading method (PC), and the two-stage method (TS) in the 1,000 simulated data sets when the data noise has the standard deviation σ = 0.02, 0.05. The actual coverage probabilities (CP) of nominal 95% credible/confidence intervals are also shown. The true parameter values are also given the second row.
| Noise | σ = 0.02 | σ = 0.05 | |||||
|---|---|---|---|---|---|---|---|
| Parameters | θD | θS | θA | θD | θS | θA | |
| True | 1.0 | 0.1 | 0.1 | 1.0 | 0.1 | 0.1 | |
| Bias × 103 | BM | −16.5 | −0.4 | −0.2 | −35.6 | 1.0 | 0.6 |
| PC | −29.7 | −0.1 | −0.3 | −55.9 | −0.2 | −0.5 | |
| TS | −225.2 | −0.7 | −1.8 | −337.8 | 0.5 | 0.6 | |
| SD × 103 | BM | 9.1 | 1.6 | 0.2 | 22.2 | 3.8 | 0.5 |
| PC | 24.9 | 3.8 | 0.5 | 40.5 | 6.2 | 0.8 | |
| TS | 91.0 | 5.9 | 1.1 | 140.7 | 10.2 | 2.1 | |
| RASE × 103 | BM | 18.81 | 1.66 | 0.27 | 42.0 | 3.9 | 0.8 |
| PC | 38.96 | 3.75 | 0.54 | 69.1 | 6.2 | 1.0 | |
| TS | 243.21 | 5.91 | 20.66 | 365.9 | 10.2 | 2.2 | |
| CP | BM | 93.9% | 99.9% | 98.8% | 74.0% | 97.8% | 86.4% |
| PC | 84.3% | 96.7% | 94.9% | 78.1% | 96.5% | 93.5% | |
| TS | 41.8% | 93.6% | 72.1% | 37.6% | 94.0% | 93.8% | |
To validate numerically the proposed sandwich estimator of variance in the parameter cascading method, we applied a parametric bootstrap of size 200 to each of the same 1,000 simulated data sets, and obtain the bootstrap estimator for standard errors of parameter estimates in each of the 1000 data sets. Table 2 displays the means of sandwich and bootstrap standard error estimators, which are highly consistent to each other. Both are also close to the sample standard deviations of parameter estimates obtained from the same 1000 simulated data sets.
Table 2.
Numerical validation of the proposed sandwich estimator in the parameter cascading method when the data noise has the standard deviation σ = 0.02, 0.05. Under each scenario, the first two rows are means of 1000 sandwich and bootstrap standard error estimators obtained from the same 1000 simulated data sets, respectively; the last row is the sample standard deviation of 1000 parameter estimates obtained from the same 1000 simulated data sets.
| Parameters | θD | θS | θA | |||
|---|---|---|---|---|---|---|
| σ = 0.02 | Mean of Sandwich Estimators | 0.0246 | 0.00375 | 0.000467 | ||
| Mean of Bootstrap Estimators | 0.0257 | 0.00374 | 0.000474 | |||
| Sample Standard Deviation | 0.0249 | 0.00375 | 0.000465 | |||
| σ = 0.05 | Mean of Sandwich Estimators | 0.0392 | 0.00599 | 0.000783 | ||
| Mean of Bootstrap Estimators | 0.0404 | 0.00597 | 0.000791 | |||
| Sample Standard Deviation | 0.0405 | 0.00617 | 0.000795 | |||
The modeling error for the PDE (2) is estimated as ℱ̂{ĝ(t, z); θ̂} = ∂ĝ(t, z)/∂t − θ̂D∂2ĝ(t, z)/∂z2 − θ̂S∂ĝ(t, z)/∂z − θ̂Aĝ(t, z). To assess the accuracy of the estimated dynamic process, ĝ(t, z), and the estimated PDE modeling errors, ℱ̂{ĝ(t, z); θ̂}, we use the square root of the average squared errors (RASEs), which are defined as
| (15) |
| (16) |
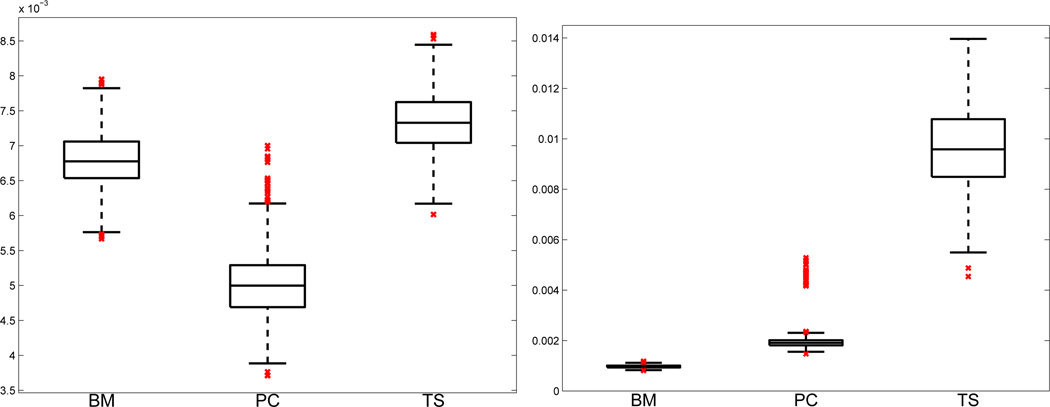
where mtgrid and mzgrid are the number of grid points in each dimension, tj, zk are grid points for j = 1, …, mtgrid, and k = 1, …, mzgrid. Figure 2 presents the boxplots of RASEs for the estimated dynamic process, ĝ(t, z), and PDE modeling errors, ℱ̂{ĝ̂(t, z); θ̂}, from the simulated data sets. The Bayesian method and the parameter cascading method have much smaller RASEs for the estimated PDE modeling errors, ℱ̂{ĝ(t, z); θ̂}, than the two-stage method, because the two-stage method produces inaccurate estimation of derivatives, especially higher order derivatives.
Figure 2.
Boxplots of the square roots of average squared errors (RASE) for the estimated dynamic process, ĝ(t, z), and the PDE modeling errors, ℱ̂{ĝ(t, z); θ̂}, using the Bayesian method (BM), the parameter cascading method (PC), and the two-stage method (TS) from 1000 data sets in the simulation study. Left: boxplots of RASE(ĝ), defined in (15), by all three methods. Right: boxplots of RASE(ℱ̂), defined in (16), by all three methods.
5 Application
5.1 Background and Illustration
We have access to a small subset of long range infrared light detection and ranging (LIDAR) data described by Warren, et al. (2008, 2009, 2010). A comic describing the LIDAR data is given in Figure 3. Our data set consists of samples collected for 28 aerosol clouds, 14 of them biological and the other 14 being non-biological. Briefly, for each sample, there is a transmitted signal that is sent into the aerosol cloud at 19 laser wavelengths, and for t = 1, …, T time points. For each wavelength and time point, received LIDAR data were observed at equally spaced ranges z = 1, .., Z. The experiment also included background data, i.e., before the aerosol cloud was released, and the received data were then background corrected.
Figure 3.
A comic describing the LIDAR data. A point source laser is transmitted into an aerosol cloud at multiple wavelengths and over multiple time points. There is scattering of the signal and reflected back to a receiver over multiple range values. See Figure 4 for an example of the received data over bursts and time for a single wavelength and a single sample.
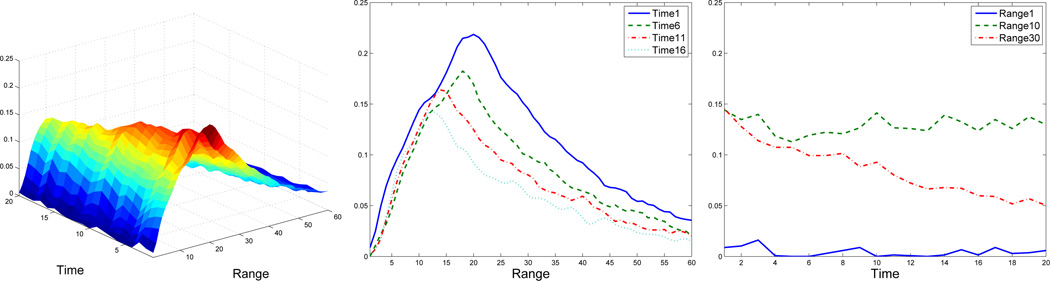
An example of the background-corrected received data for a single sample and a single wavelength are given in Figure 4. Data such as this are well-described by the PDE (2). This equation is a linear PDE of parabolic type in one space dimension and is also called a (one-dimensional) linear reaction-convection-diffusion equation. If we describe this equation as g(t, z), the parameters θD, θS and θA describe the diffusion rate, the drift rate/shift and the reaction rate, respectively.
Figure 4.
Snapshots of the empirical data. Left: 3D plot of the received signal. Middle: the received signal at a few time values, ti = 1, 6, 11, 16, over the range. Right: the received signal at a few range values, zj = 1, 10, 30, over the time.
In fitting model (2) to the real data, we take T = 20 time points and Z = 60 range values, so that the sample size n is 20 × 60 = 1, 200. To illustrate what happens with the data in Figure 4, the parameter cascading method, Bayesian method, and the two-stage method were applied to estimate the three parameters in the PDE model (2) from the above LIDAR data set. All three methods use bivariate quartic B-spline basis functions constructed with 5 inner knots in the time domain and 20 inner knots in the range domain.
Table 3 displays the estimates for the three parameters in the PDE model (2). While the three methods produce similar estimates for parameters θS and θA, the parameter cascading estimate and Bayesian estimate for θD are more consistent with each other than with the two-stage estimate. This phenomenon is consistent with what was seen in our simulations. Moreover, in this application, the three methods produce almost identical smooth curves, but not derivatives. This fact is also found in our simulation studies, where all three methods lead to similar estimates for the dynamic process, g(t, z), but the two-stage method performs poorly for estimating its derivatives.
Table 3.
Estimated parameters for the PDE model (2) from the LIDAR data set using the Bayesian method (BM), the parameter cascading method (PC), and the two-stage method (TS).
| θD | θS | θA | ||
|---|---|---|---|---|
| Estimates | BM | −0.4470 | 0.2563 | −0.0414 |
| PC | −0.3771 | 0.2492 | −0.0407 | |
| TS | −0.1165 | 0.2404 | −0.0436 |
5.2 Differences Among the Types of Samples
To understand if there are differences between the received signals for biological and non-biological samples, we performed the following simple analysis. For each sample, and for each wavelength, we fit the PDE model (2) to obtain estimates of (θD, θS, θA), and then performed t-tests to compare them across aerosol types. Strikingly, there was no evidence that the diffusion rate θD differed between the aerosol types at any wavelength, with a minimum p-value being of 0.12 across all wavelengths and both the parameter cascade and Bayesian methods. For the drift rate/shift θS, all but 1 wavelength had a p-value < 0.05 for both methods, and multiple wavelengths reached Bonferroni significance. For the reaction rate θA the results are somewhat intermediate. While for both methods, all but 1 wavelength had a p-value < 0.05, none reached Bonferroni significance. In summary, the differences between the two types of aerosol clouds is clearly expressed by the drift rate/shift, with some evidence of differences in the reaction rate, but no differences in the diffusion rate. In almost all cases, the drift rate is larger in the non-biological samples, while the reaction rate is larger in the biological samples.
6 Concluding Remarks
Differential equation models are widely used to model dynamic processes in many fields such as engineering and biomedical sciences. The forward problem of solving equations or simulating state variables for given parameters that define the models have been extensively studied in the past. However, the inverse problem of estimating parameters based on observed state variables is relatively sparse in the statistical literature, and this is especially the case for partial differential equation models.
We have proposed a parameter cascading method and a fully Bayesian treatment for this problem, which are compared with a two-stage method. The parameter cascading method and Bayesian method are joint estimation procedures which consider the data fitting and PDE fitting simultaneously. Our simulation studies show that the proposed two methods are more statistically efficient than a two-stage method, especially for parameters associated with higher order derivatives. Basis function expansion plays an important role in our new methods, in the sense that it makes joint modeling possible and links together fidelity to the PDE model and fidelity to data through the coefficients of basis functions. A potential extension of this work would be to estimate time-varying parameters in PDE models from error-prone data.
Supplementary Material
Acknowledgments
The research of Mallick, Carroll and Xun was supported by grants from the National Cancer Institute (R37-CA057030) and the National Science Foundation DMS grants 0914951. This publication is based in part on work supported by Award Number KUS-CI-016-04, made by King Abdullah University of Science and Technology (KAUST). Cao’s research is supported by a discovery grant (PIN: 328256) from the Natural Science and Engineering Research Council of Canada (NSERC). Maity’s research was performed while visiting Department of Statistics, Texas A&M University, and was partially supported by Award Number R00ES017744 from National Institute of Environmental Health Sciences.
Appendix
A.1 Calculation of f (x; θ) and F (θ)
Here we show the form of f (x; θ) and F(θ) for the PDE example (2). The vector f (x; θ) is a linear combination of basis functions and their derivatives involved in model (2). We have that f (x; θ) = ∂b(x)/∂t − θD∂2b(x)/∂z2 − θS∂b(x)/∂z − θAb(x). Similar to the basis function matrix B = {b(x1), …, b(xn)}T, we define the following n × K matrices consisting of derivatives of the basis functions
Then the matrix F(θ) = {f (x1; θ), …, f (xn; θ)}T = Bt − θDBzz − θSBz − θAB.
A.2 Sketch of the Asymptotic Theory
A.2.1 Assumptions and Notation
Asymptotic theory for our estimators follows in a fashion very similar to that of Yu and Ruppert (2002). Let λ̃ = λ/n, denote the true value of θ as θ0 and define
The parameter θ is estimated by minimizing
| (A.1) |
Assumption 1 The sequence λ̃ is fixed and satisfies λ̃ = o(n−1/2).
Assumption 2 The function g(x) = bT(x)β0 for a unique β0, i.e., the spline approximation is exact, and hence .
Assumption 3 The parameter θ0 is in the interior of a compact set and, for j = 1, …, n, is the unique solution to .
Assumption 4 Assumptions (1)–(4) of Yu and Ruppert (2002) hold with their m(υ, θ) being our bT(x)βn(θ).
A.2.2 Characterization of the Solution to (A.1)
Remember the matrix fact that for any nonsingular symmetric matrix A(z) for scalar z, ∂A−1(z)/∂z = −A−1(z){∂A(z)/∂z}A−1(z). This means that for j = 1, …, m,
| (A.2) |
Minimizing ℒn(θ) is equivalent to solving for j = 1, …, m for the system of equations
where we define Ψij(θ) = {Yi − bT(xi)β̂n(θ)}bT(xi){∂β̂n(θ)∂θj}. From now on, we define the score for θj as and define 𝒯n(θ) = 𝒯{n1(θ), …, 𝒯nm(θ)}T.
There are some further simplifications of 𝒯n(θ). Because of (A.2),
However,
Thus for any θ,
| (A.3) |
Hence, θ̂ is the solution to the system of equations .
A.2.3 Further Calculations
Yu and Ruppert show that if λ̃ → 0 as n → ∞, then uniformly in θ, β̂n(θ) = β0 + op(1) and that if λ̃ = o(n−1/2) as n → ∞, then . Define the Hessian matrix as ℳn(θ) = ∂𝒯n(θ)/∂θT. Because of these facts and Assumption 3, it follows that θ̂ = θ0 + op(1), i.e., consistency. It then follows that
where θ* = θ0 + op(1) is between θ̂ and θ0, and hence that
| (A.4) |
Define Λn(θ) to have (j, k)th element
In what follows, as in Yu and Ruppert (2002), we continue to assume that λ̃ = o(n−1/2). However, with a slight abuse of notation we will write Gn(θ0) → Ω2(θ0) rather that Gn(θ0) → Ω1, because we have found that implementing the covariance matrix estimator for θ̂ is more accurate if this is retained: a similar calculation is done in Yu and Ruppert’s Section 3.2. Now using Assumption 3, we see that
Define and . Then we have that
| (A.5) |
Now recall that Sn → Ω1 and Gn(θ0) → Ω2(θ0) in probability. Hence we have that
in distribution. So using (A.5) the (j, k)th element of the covariance matrix of 𝒯n is given by
We now analyze the term n−1/2ℳn(θ*). Because of consistency of θ̂,
| (A.6) |
The (j, k)th element of ℳn(θ) is
We see that by (A.2),
Now using the fact that β̂n(θ) = βn(θ) + op(1) for any θ, and recalling the definition of Λn(θ), we have at θ0 that
Similarly for the remaining term of the Hessian matrix we have
By Assumption 3, and since ε(x) has mean zero, we see that
| (A.7) |
Hence using (A.4) and (A.6, it follows that
| (A.8) |
Hence using (A.8) we obtain (10), but with Ω1 and Ω2(θ) replaced by their consistent estimates Sn and Gn(θ).
A.3 Full Conditional Distributions
To sample from the posterior distribution (13) using Gibbs sampler, we need full conditional distributions of all the unknowns. Due to conjugacy, parameters and the γ terms have close form full conditionals. Define SSE = (Y−Bβ)T(Y−Bβ). If we define ”rest” to mean conditional on everything else, we have
The parameters β and θ do not have closed form full conditionals, which are instead
To draw samples from these full conditionals, a Metropolis-Hastings (MH) update within the Gibbs sampler is applied for each component of θi. The proposal distribution for the ith component is a normal distribution Normal(θi,curr, σi,prop), where the mean θi,curr is the current value and the standard deviation σi,prop is a constant.
In the special case of a linear PDE, the model error is also linear in β, represented by ζ(β, θ) = F(θ)β. Then the term ζT(β, θ)ζ(β, θ) is a quadratic function in β. Define H = H(θ) = γ0FT(θ)F(θ) + γ1H1 + γ2H2 + γ1γ2H3, and . By completing the square in [β|rest], the full conditional of β under linear PDE models is in the explicit form
Contributor Information
Xiaolei Xun, Email: xiaolei.xun@novartis.com, Beijing Novartis Pharma Co. Ltd., Pudong New District, Shanghai, 201203, China.
Jiguo Cao, Email: cao@stats.uwo.ca, Department of Statistical and Actuarial Sciences, University of Western Ontario, London, ON, N6A5B7, Canada.
Bani Mallick, Email: bmallick@stat.tamu.edu, Department of Statistics, Texas A&M University, 3143 TAMU, College Station, TX, 77843-3143.
Raymond J. Carroll, Email: carroll@stat.tamu.edu, Department of Statistics, Texas A&M University, 3143 TAMU, College Station, TX, 77843-3143.
Arnab Maity, Email: amaity@ncsu.edu, Department of Statistics, North Carolina State University, Raleigh, North Carolina 27695.
References
- Bar M, Hegger R, Kantz H. Fitting differential equations to space-time dynamics. Physical Review, E. 1999;59:337–342. [Google Scholar]
- Berry SM, Carroll RJ, Ruppert D. Bayesian smoothing and regression splines for measurement error problems. Journal of the American Statistical Association. 2002;97:160–169. [Google Scholar]
- Brenner SC, Scott R. The Mathematical Theory of Finite Element Methods. Springer; 2010. [Google Scholar]
- Burden RL, Douglas FJ. Numerical Analysis. Ninth edition. California: Brooks/Cole; 2010. [Google Scholar]
- Cao J, Wang L, Xu J. Robust estimation for ordinary differential equation models. Biometrics. 2011;67:1305–1313. doi: 10.1111/j.1541-0420.2011.01577.x. [DOI] [PubMed] [Google Scholar]
- Cao J, Huang JZ, Wu H. Penalized nonlinear least squares estimation of time-varying parameters in ordinary differential equations. Journal of Computational and Graphical Statistics. 2012;21:42–56. doi: 10.1198/jcgs.2011.10021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Wu H. Efficient local estimation for time-varying coeffients in deterministic dynamic models with applications to HIV-1 dynamics. Journal of the American Statistical Association. 2008;103:369–384. [Google Scholar]
- de Boor C. Applied Mathematical Sciences. Revised edition. Vol. 27. New York: Springer-p; 2001. A Practical Guide to Splines. [Google Scholar]
- Denison DGT, Mallick BK, Smith AFM. Automatic Bayesian curve fitting. Journal of the Royal Statistical Society, Series B. 1997;60:333–350. [Google Scholar]
- Eilers P, Marx B. Multidimensional calibration with temperature interaction using two-dimensional penalized signal regression. Chemometrics and Intelligent Laboratory Systems. 2003;66:159–174. [Google Scholar]
- Eilers P, Marx B. Splines, knots and penalties. Wiley Interdisciplinary Reviews: Computational Statistics. 2010;2:637–653. [Google Scholar]
- Evans LC. Graduate Studies in Mathematics. Vol. 19. American Mathematical Society, US.; 1998. Partial Differential Equations. [Google Scholar]
- Friedman JH, Silverman BW. Flexible parsimonious smoothing and additive modeling. Technometrics. 1989;31:3–21. [Google Scholar]
- Gelfand AE, Smith AFM. Sampling-based approaches to calculating marginal densities. Journal of the American Statistical Association. 1990;85:398–409. [Google Scholar]
- Gilks WR, Richardson S, Spiegelhalter DJ. Markov Chain Monte Carlo in Practice: Interdisciplinary Statistics. Chapman & Hall; 1996. [Google Scholar]
- Ho DD, Neumann AS, Perelson AS, Chen W, Leonard JM, Markowitz M. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature. 1995;373:123–126. doi: 10.1038/373123a0. [DOI] [PubMed] [Google Scholar]
- Huang Y, Liu D, Wu H. Hierarchical Bayesian methods for estimation of parameters in a longitudinal HIV dynamic system. Biometrics. 2006;62:413–423. doi: 10.1111/j.1541-0420.2005.00447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wu H. A Bayesian approach for estimating antiviral efficacy in HIV dynamic models. Journal of Applied Statistics. 2006;33:155–174. [Google Scholar]
- Li L, Brown MB, Lee KH, Gupta S. Estimation and inference for a spline-enhanced population pharmacokinetic model. Biometrics. 2002;58:601–611. doi: 10.1111/j.0006-341x.2002.00601.x. [DOI] [PubMed] [Google Scholar]
- Liang H, Wu H. Parameter estimation for differential equation models using a framework of measurement error in regression models. Journal of the American Statistical Association. 2008;103:1570–1583. doi: 10.1198/016214508000000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx B, Eilers P. Multidimensional penalized signal regression. Technometrics. 2005;47:13–22. [Google Scholar]
- Morton KW, Mayers DF. Numerical Solution of Partial Differential Equations, An Introduction. Cambridge University Press; 2005. [Google Scholar]
- Muller T, Timmer J. Fitting parameters in partial differential equations from partially observed noisy data. Physical Review, D. 2002;171:1–7. [Google Scholar]
- Muller T, Timmer J. Parameter identification techniques for partial differential equations. International Journal of Bifurcation and Chaos. 2004;14:2053–2060. [Google Scholar]
- Parlitz U, Merkwirth C. Prediction of spatiotemporal time series based on reconstructed local states. Physical Review Letters. 2000;84:1890–1893. doi: 10.1103/PhysRevLett.84.1890. [DOI] [PubMed] [Google Scholar]
- Poyton AA, Varziri MS, McAuley KB, McLellan PJ, Ramsay JO. Parameter estimation in continuous-time dynamic models using principal differential analysis. Computer and Chemical Engineering. 2006;30:698–708. [Google Scholar]
- Putter H, Heisterkamp SH, Lange JMA, De Wolf F. A Bayesian approach to parameter estimation in HIV dynamical models. Statistics in Medicine. 2002;21:2199–2214. doi: 10.1002/sim.1211. [DOI] [PubMed] [Google Scholar]
- Ramsay JO. Principal differential analysis: data reduction by differential operators. Journal of the Royal Statistical Society, Series B. 1996;58:495–508. [Google Scholar]
- Ramsay JO, Hooker G, Campbell D, Cao J. Parameter estimation for differential equations: a generalized smoothing approach (with discussion) Journal of the Royal Statistical Society, Series B. 2007;69:741–796. [Google Scholar]
- Ruppert D, Wand MP, Carroll RJ. Semiparametric Regression. Cambridge University Press; 2003. [Google Scholar]
- Stone CJ, Hansen MH, Kooperberg C, Truong YK. Polynomial splines and their tensor products in extended linear modeling. Annals of Statistics. 1997;25:1371–1425. [Google Scholar]
- Voss HU, Kolodner P, Abel M, Kurths J. Amplitude equations from spatiotemporal binary-fluid convection data. Physical Review Letters. 1999;83:3422–3425. [Google Scholar]
- Warren RE, Vanderbeek RG, Ben-David A, Ahl JL. Simultaneous estimation of aerosol cloud concentration and spectral backscatter from multiple-wavelength lidar data. Applied Optics. 2008;47(24):4309–4320. doi: 10.1364/ao.47.004309. [DOI] [PubMed] [Google Scholar]
- Warren RE, Vanderbeek RG, Ahl JL. Detection and classification of atmospheric aerosols using multi-wavelength LWIR lidar. Proceedings of SPIE. 2009;7304:73040E. [Google Scholar]
- Warren RE, Vanderbeek RG, Ahl JL. Estimation and discrimination of aerosols using multiple wavelength LIWR lidar. Proceedings of SPIE. 2010;7665:766504-1. [Google Scholar]
- Wei X, Ghosh SK, Taylor ME, Johnson VA, Emini EA, Deutsch P, Lifson JD, Bonhoeer S, Nowak MA, Hahn BH, Saag MS, Shaw GM. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–123. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- Wu H. Statistical methods for HIV dynamic studies in AIDS clinical trials. Statistical Methods in Medical Research. 2005;14:171–192. doi: 10.1191/0962280205sm390oa. [DOI] [PubMed] [Google Scholar]
- Wu H, Ding A. Population HIV-1 dynamics in vivo: applicable models and inferential tools for virological data from AIDS clinical trials. Biometrics. 1999;55:410–418. doi: 10.1111/j.0006-341x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- Wu H, Ding A, DeGruttola V. Estimation of HIV dynamic parameters. Statistics in Medicine. 1998;17:2463–2485. doi: 10.1002/(sici)1097-0258(19981115)17:21<2463::aid-sim939>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Yu Y, Ruppert D. Penalized spline estimation for partially linear single-index models. Journal of the American Statistical Association. 2002;97:1042–1054. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.