Abstract
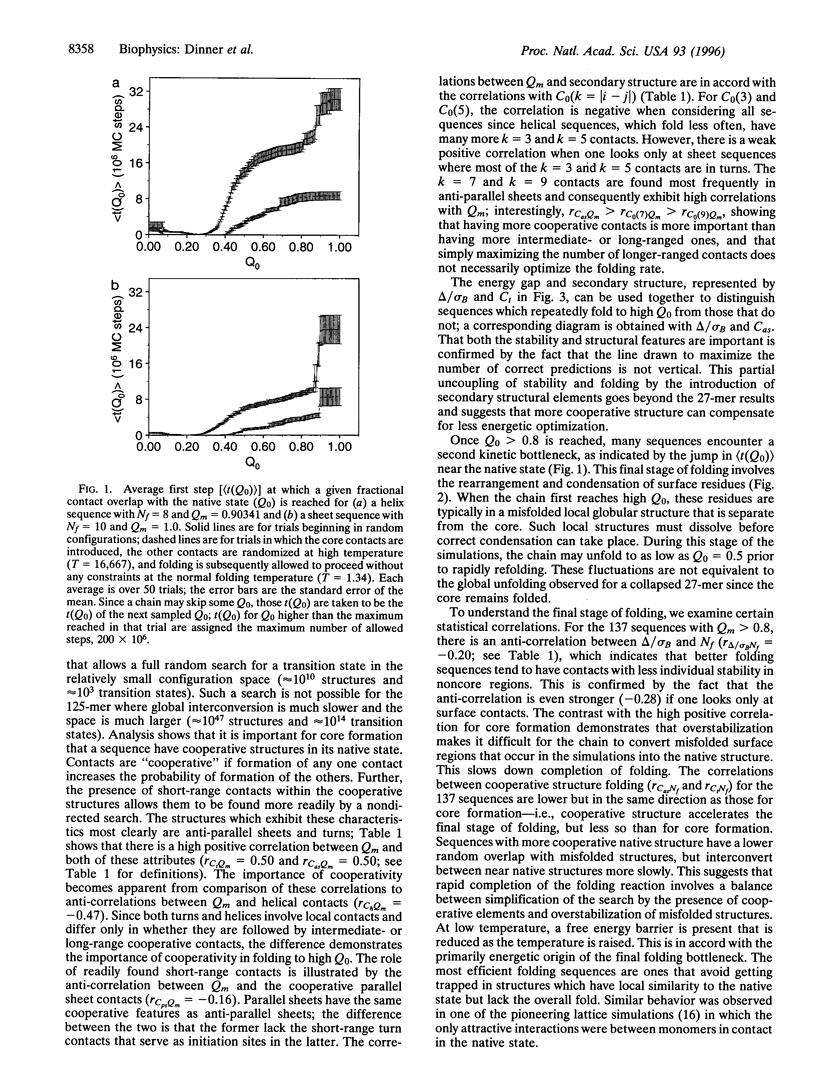
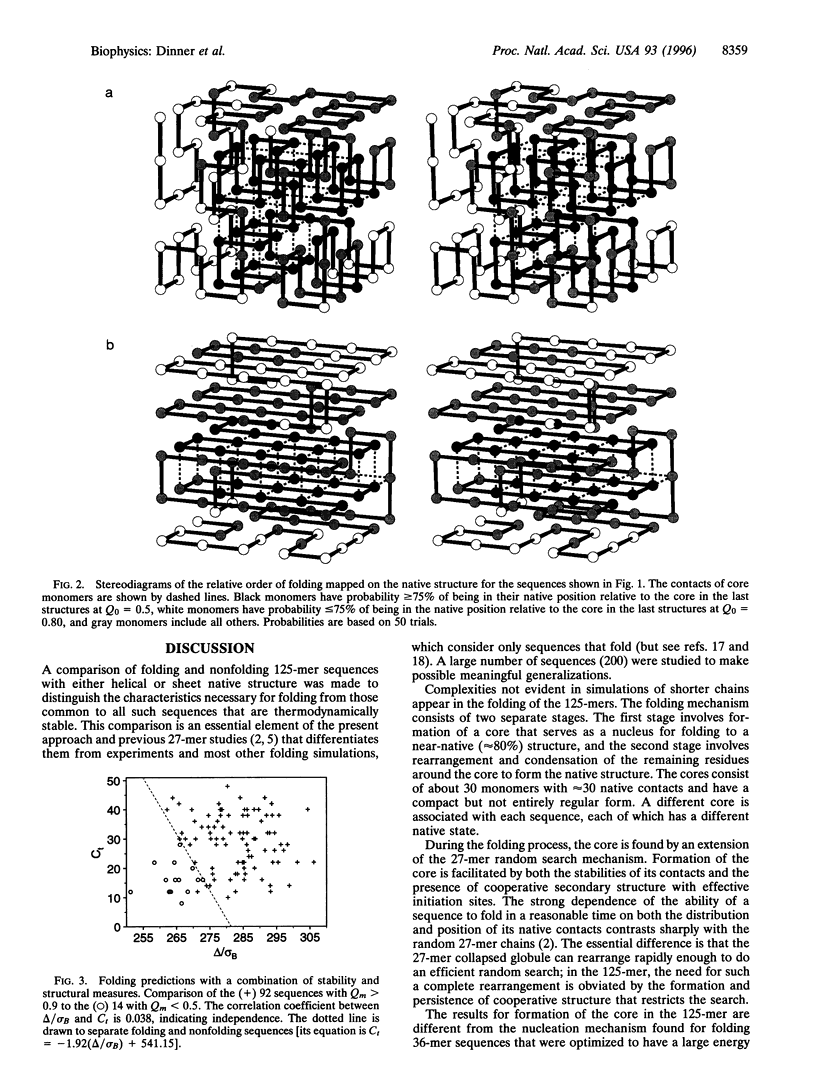
The folding mechanism of a 125-bead heteropolymer model for proteins is investigated with Monte Carlo simulations on a cubic lattice. Sequences that do and do not fold in a reasonable time are compared. The overall folding behavior is found to be more complex than that of models for smaller proteins. Folding begins with a rapid collapse followed by a slow search through the semi-compact globule for a sequence-dependent stable core with about 30 out of 176 native contacts which serves as the transition state for folding to a near-native structure. Efficient search for the core is dependent on structural features of the native state. Sequences that fold have large amounts of stable, cooperative structure that is accessible through short-range initiation sites, such as those in anti-parallel sheets connected by turns. Before folding is completed, the system can encounter a second bottleneck, involving the condensation and rearrangement of surface residues. Overly stable local structure of the surface residues slows this stage of the folding process. The relation of the results from the 125-mer model studies to the folding of real proteins is discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abkevich V. I., Gutin A. M., Shakhnovich E. I. Impact of local and non-local interactions on thermodynamics and kinetics of protein folding. J Mol Biol. 1995 Sep 29;252(4):460–471. doi: 10.1006/jmbi.1995.0511. [DOI] [PubMed] [Google Scholar]
- Abkevich V. I., Gutin A. M., Shakhnovich E. I. Specific nucleus as the transition state for protein folding: evidence from the lattice model. Biochemistry. 1994 Aug 23;33(33):10026–10036. doi: 10.1021/bi00199a029. [DOI] [PubMed] [Google Scholar]
- Avbelj F., Moult J. Determination of the conformation of folding initiation sites in proteins by computer simulation. Proteins. 1995 Oct;23(2):129–141. doi: 10.1002/prot.340230203. [DOI] [PubMed] [Google Scholar]
- Bai Y., Sosnick T. R., Mayne L., Englander S. W. Protein folding intermediates: native-state hydrogen exchange. Science. 1995 Jul 14;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie J. U., Reidhaar-Olson J. F., Lim W. A., Sauer R. T. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990 Mar 16;247(4948):1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- Brocchieri L., Karlin S. How are close residues of protein structures distributed in primary sequence? Proc Natl Acad Sci U S A. 1995 Dec 19;92(26):12136–12140. doi: 10.1073/pnas.92.26.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C., Levitt M., Richardson D. Structure of proteins: packing of alpha-helices and pleated sheets. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4130–4134. doi: 10.1073/pnas.74.10.4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais J. R., Handel T. M. De novo design of the hydrophobic cores of proteins. Protein Sci. 1995 Oct;4(10):2006–2018. doi: 10.1002/pro.5560041006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Bromberg S., Yue K., Fiebig K. M., Yee D. P., Thomas P. D., Chan H. S. Principles of protein folding--a perspective from simple exact models. Protein Sci. 1995 Apr;4(4):561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill K. A., Fiebig K. M., Chan H. S. Cooperativity in protein-folding kinetics. Proc Natl Acad Sci U S A. 1993 Mar 1;90(5):1942–1946. doi: 10.1073/pnas.90.5.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J., Merutka G., Waltho J. P., Lerner R. A., Wright P. E. Folding of peptide fragments comprising the complete sequence of proteins. Models for initiation of protein folding. I. Myohemerythrin. J Mol Biol. 1992 Aug 5;226(3):795–817. doi: 10.1016/0022-2836(92)90633-u. [DOI] [PubMed] [Google Scholar]
- Elöve G. A., Chaffotte A. F., Roder H., Goldberg M. E. Early steps in cytochrome c folding probed by time-resolved circular dichroism and fluorescence spectroscopy. Biochemistry. 1992 Aug 4;31(30):6876–6883. doi: 10.1021/bi00145a003. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. The sixth Datta Lecture. Protein folding and stability: the pathway of folding of barnase. FEBS Lett. 1993 Jun 28;325(1-2):5–16. doi: 10.1016/0014-5793(93)81405-o. [DOI] [PubMed] [Google Scholar]
- Gutin A. M., Abkevich V. I., Shakhnovich E. I. Is burst hydrophobic collapse necessary for protein folding? Biochemistry. 1995 Mar 7;34(9):3066–3076. doi: 10.1021/bi00009a038. [DOI] [PubMed] [Google Scholar]
- Hooke S. D., Radford S. E., Dobson C. M. The refolding of human lysozyme: a comparison with the structurally homologous hen lysozyme. Biochemistry. 1994 May 17;33(19):5867–5876. doi: 10.1021/bi00185a026. [DOI] [PubMed] [Google Scholar]
- Karplus M., Sali A. Theoretical studies of protein folding and unfolding. Curr Opin Struct Biol. 1995 Feb;5(1):58–73. doi: 10.1016/0959-440x(95)80010-x. [DOI] [PubMed] [Google Scholar]
- Karplus M., Weaver D. L. Protein folding dynamics: the diffusion-collision model and experimental data. Protein Sci. 1994 Apr;3(4):650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemer C. M., Rooman M. J., Wodak S. J. Protein structure prediction by threading methods: evaluation of current techniques. Proteins. 1995 Nov;23(3):337–355. doi: 10.1002/prot.340230308. [DOI] [PubMed] [Google Scholar]
- Otzen D. E., Itzhaki L. S., elMasry N. F., Jackson S. E., Fersht A. R. Structure of the transition state for the folding/unfolding of the barley chymotrypsin inhibitor 2 and its implications for mechanisms of protein folding. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10422–10425. doi: 10.1073/pnas.91.22.10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford S. E., Dobson C. M., Evans P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992 Jul 23;358(6384):302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- Rooman M. J., Kocher J. P., Wodak S. J. Extracting information on folding from the amino acid sequence: accurate predictions for protein regions with preferred conformation in the absence of tertiary interactions. Biochemistry. 1992 Oct 27;31(42):10226–10238. doi: 10.1021/bi00157a009. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. How does a protein fold? Nature. 1994 May 19;369(6477):248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- Sali A., Shakhnovich E., Karplus M. Kinetics of protein folding. A lattice model study of the requirements for folding to the native state. J Mol Biol. 1994 Feb 4;235(5):1614–1636. doi: 10.1006/jmbi.1994.1110. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Engineering of stable and fast-folding sequences of model proteins. Proc Natl Acad Sci U S A. 1993 Aug 1;90(15):7195–7199. doi: 10.1073/pnas.90.15.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Implications of thermodynamics of protein folding for evolution of primary sequences. Nature. 1990 Aug 23;346(6286):773–775. doi: 10.1038/346773a0. [DOI] [PubMed] [Google Scholar]
- Shakhnovich EI. Proteins with selected sequences fold into unique native conformation. Phys Rev Lett. 1994 Jun 13;72(24):3907–3910. doi: 10.1103/PhysRevLett.72.3907. [DOI] [PubMed] [Google Scholar]
- Sippl M. J. Knowledge-based potentials for proteins. Curr Opin Struct Biol. 1995 Apr;5(2):229–235. doi: 10.1016/0959-440x(95)80081-6. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Englander S. W. Molecular collapse: the rate-limiting step in two-state cytochrome c folding. Proteins. 1996 Apr;24(4):413–426. doi: 10.1002/(SICI)1097-0134(199604)24:4<413::AID-PROT1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kim P. S. A kinetic explanation for the rearrangement pathway of BPTI folding. Nat Struct Biol. 1995 Dec;2(12):1123–1130. doi: 10.1038/nsb1295-1123. [DOI] [PubMed] [Google Scholar]
- Zwanzig R., Szabo A., Bagchi B. Levinthal's paradox. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):20–22. doi: 10.1073/pnas.89.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]



