Abstract
Membrane structures and cytoskeleton dynamics are intimately inter-connected in the eukaryotic cell. Recently, the molecular mechanisms operating at this interface have been progressively addressed. Many experiments have revealed that the actin cytoskeleton can interact with membranes through various discrete membrane domains. The actin-binding protein, profilin has been proven to inhibit actin polymerization and to promote F-actin elongation. This is dependent on many factors, such as the profilin/G-actin ratio and the ionic environment of the cell. Additionally, profilin has specific domains that interact with phosphoinositides and poly-L-proline rich proteins; theoretically, this gives profilin the opportunity to interact with membranes, and a large number of experiments have confirmed this possibility. In this article, we summarize recent findings in plant cells, and discuss the evidence of the connections among actin cytoskeleton, profilin and biomembranes through direct or indirect relationships.
Keywords: actin cytoskeleton, profilin, plasma membrane, organelle, vesicle, plants
INTRODUCTION
The membrane is a lipid bilayer that functions to divide and separate the cells and organelles. It undergoes many dynamic morphological changes during cellular processes such as endocytosis, exocytosis, vesicular transport, and morphogenesis. Growing evidence has demonstrated that actin cytoskeleton dynamics are involved in these processes. However, the interactions between microfilaments and membranes vary in different cell types and locations. Some cytoskeletal elements may interact with membranes directly. Transmembrane proteins can regulate membrane-cytoskeleton interactions directly or indirectly through adaptor proteins or adaptor complexes. Furthermore, some proteins have domains that can associate with the membrane, and domains that can interact with cytoskeletal components. These are the main types of membrane-cytoskeleton interactions (Doherty and McMahon, 2008). The extracellular matrix (ECM) of animals mainly consists of proteinaceous materials. However, the plant cell wall, which deviates plant cell from spherical shapes, mainly consists of carbohydrates. This implies that there are differences in the intermolecular interactions that occur in membrane-cytoskeleton of animal and plant. In mammals, cytoskeletal proteins that can function as adaptors, such as talin (Heise et al., 1991), vinculin (Geiger et al., 1980), and filamin (Stossel et al., 2001) bind the actin cytoskeleton to membranes; homologs of these proteins are absent from plants (Hussey et al., 2002). There are many plant-specific linker molecules. For example, myosin VIII binds directly or indirectly to plasma membrane-localized callose synthase complexes (Verma and Hong, 2001; Ostergaard et al., 2002) and it also binds to actin filaments in the cytoplasm, which implies that myosin VIII associates plasma membrane with actin filaments in plants. Moreover, a plant-specific Networked (NET) superfamily of actin-binding proteins is found in Arabidopsis. Members of the NET superfamily localize to the actin cytoskeleton and specify different membrane compartments. NET1A is located at the plasma membrane and binds directly to actin filaments through a novel actin-binding domain. The NET superfamily is grouped into four phylogenetic clades, and other members have functions at the tonoplast, nuclear membrane, and pollen tube plasma membrane, which suggest that this superfamily is involved in regulating actin-membrane interactions (Deeks et al., 2012).
A large amount of literature has fostered our current understanding of the membrane, the actin cytoskeleton, and of actin-binding proteins that mediate membrane and actin cytoskeleton components. Profilins are actin-binding proteins, and have the capacity to interact with three classes of ligands. In addition to G-actin, they also associate with poly-L-proline (PLP) which can interact with the binding cleft formed from the N-terminal and C-terminal helices of profilin (Metzler et al., 1994; Mahoney et al., 1999) and phosphoinositides (Gibbon and Staiger, 2000; Jockusch et al., 2007) which offers the possibility that profilin interacts with the membrane. In recent years, much evidence has been verified that profilins can interact with membranes directly or indirectly. In this review, we will summarize recent findings and focus predominantly on the functions of profilins in the direct or indirect relationships among actin cytoskeleton, profilin and membranes in plant cells.
MULTIFUNCTIONAL PROFILINS
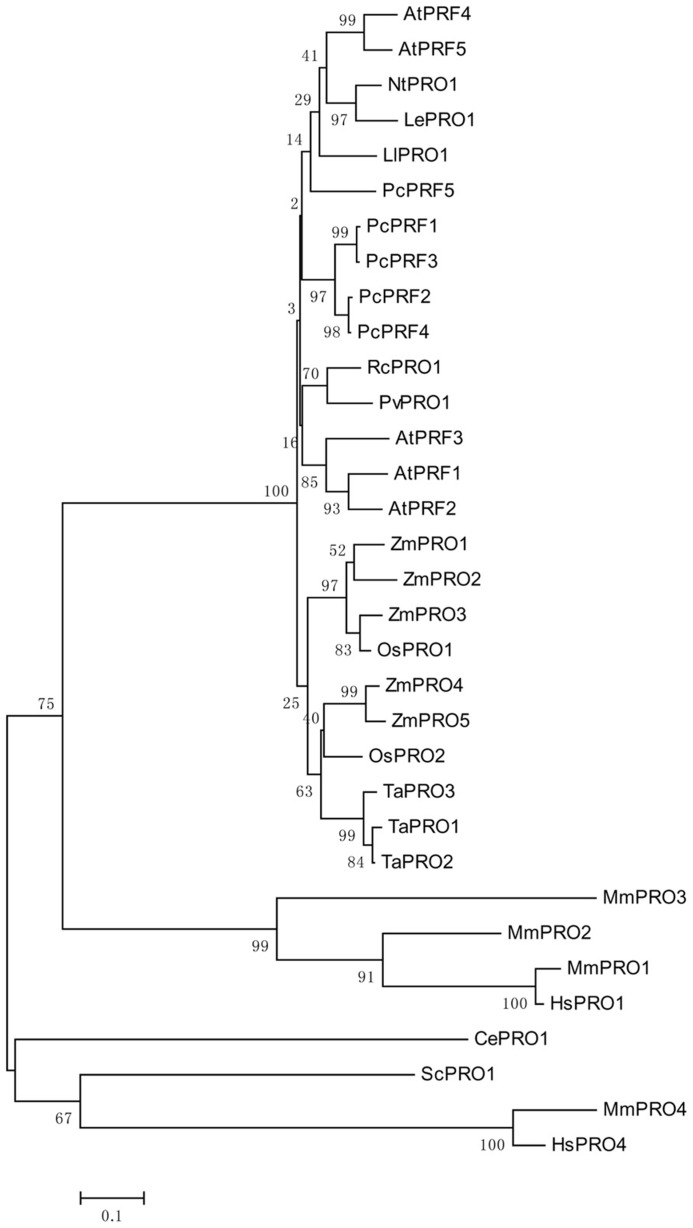
Genomic DNA sequences of putative profilins contain three exons; these may be separated by introns of different sizes (Huang et al., 1996), and are dispersed throughout the genome. Comparing the amino acid sequences of different profilins reveal that profilins have less than 25% identity across different kingdoms (Pollard and Quirk, 1994), but are highly conserved, with at least 70% identity, across various plant species (Mittermann et al., 1995; Vidali et al., 1995). This is consistent with the analysis of the phylogenetic tree shown in Figure 1. Although the secondary and tertiary structures of all profilins are well conserved (Fedorov et al., 1997; Thorn et al., 1997; Jockusch et al., 2007), the fact that many varieties of profilins isoforms exist in different species, and even in the same organism, may indicate that members of the profilin family have diverse functions. Plant profilins are from multigene families and can be divided into two major groups: the vegetative group, in which profilins exist extensively and are constitutively expressed in all plant tissues; and the reproductive group, where profilins are expressed in reproductive tissues (Kandasamy et al., 2002). The Arabidopsis profilin family includes five highly different isoforms: AtPRF1–AtPRF5; AtPRF1–AtPRF3 belong to the vegetative class, and AtPRF4 and AtPRF5 to the reproductive class (Christensen et al., 1996; Kandasamy et al., 2002). AtPRF1 has much higher affinities for both PLP and G-actin than AtPRF2 (Wang et al., 2009). The tobacco profilin gene, pronp1, is prominently expressed in mature pollen, elongating pollen tubes, and the root hairs of developing seedlings. Pronp1 represents a unique profilin as it has activities in two kinds of tip-growing cells, the pollen tubes and root hairs, which rapidly regulate the organization of the actin cytoskeleton (Swoboda et al., 2001). In tomato, LePRO1 was found to be expressed only in pollen grains, and not in other parts of the anther or in other organs using a non-radioactive labeling method (Yu et al., 1998). RcPRO1, a Ricinus communis phloem profilin, is expressed in epidermal, cortex, pith, and xylem tissue. In the sieve-tube exudates, RcPRO1 has 15-fold molar excess to actin, which suggests that actin filament formation is blocked in the assimilate stream (Schobert et al., 2000). In maize, five profilins have been identified (ZmPRO1–ZmPRO5); ZmPRO1–ZmPRO3 are major profilin isoforms of a pollen-abundant class, whereas ZmPRO4 and ZmPRO5 appear to be members of a predominantly endosperm profilin class. Furthermore, ZmPRO1 inhibits hydrolysis of membrane phosphatidylinositol-4, 5-bisphosphate (PIP2) by phospholipase C more effectively than ZmPRO5. Conversely, ZmPRO5 has higher affinity for PLP and sequesters more monomeric actin to inhibit actin polymerization better than ZmPRO1 (Staiger et al., 1993; Kovar et al., 2000). Currently, there are over 400 profilins from 100 plant species, which are effective at NCBI GenBank database (Pruitt et al., 2007; Jimenez-Lopez et al., 2012). All of the above evidences support that profilins are multifunctional proteins according to their expressions and locations.
FIGURE 1.
An unrooted phylogenetic tree of profilins. The plant genes are Arabidopsis thaliana AtPRF1–AtPRF5 (AT2G19760, AT4G29350, AT5G56600, AT4G29340, AT2G19770), Petroselinum crispum PcPRF1–PcPRF5 (AY900012-AY900016), Zea mays ZmPRO1–ZmPRO5 (X73279, X73280, X73281, AF032370, AF201459), Oryza sativa OsPRO1–OsPRO2 (LOC_Os10g17680, LOC_Os06g05880), Triticum aestivum TaPRO1–TaPRO3 (X89825-X89827), Nicotiana tabacum NtPRO1 (pronp1 AJ130969), tomato LePRO1 (U50195), Ricinus communis RcPRO1 (AF092547), Phaseolus vulgaris PvPRO1 (CAA57508), Lilium longiflorum LlPRO1 (AF200184). Selected fungal and metazoan sequences are included: Mus musculus MmPRO1–MmPRO4 (NP_035202, NP_062283, NP_083579, AK013595), Homo sapiens HsPRO1 and HsPRO4 (BC057828, BC029523), Caenorhabditis elegans CePRO1 (PFN-1, NP_493258) Saccharomyces cerevisiae ScPRO1 (PFY1, NP_014765). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches (Felsenstein, 1985). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree.
PROFILIN IS INVOLVED IN PLASMA MEMBRANE-ACTIN CYTOSKELETON INTERACTIONS
Binding interactions between the plasma membrane and the actin cytoskeleton define cell functions such as cytoplasmic streaming, cytokinesis, and endocytosis. Profilin is one of the crucial linkers of the membrane-cytoskeleton interaction. The inherent interaction of the actin cytoskeleton with the plasma membrane is through the relationship between actin-binding proteins and PIP2, which itself localizes to the inner side of the plasma membrane (Nebl et al., 2000; Caroni, 2001). PIP2 can bind to transmembrane adhesion protein, and also interacts with several actin-binding proteins including profilin (Goldschmidt-Clermont et al., 1990; Heiska et al., 1998; Couchman et al., 2002). Immunofluorescence analysis revealed that at the plasma membrane of maize root cells, PIP2 is targeted to discrete domains that resemble profilin-enriched domains. PIP2redistributes and the actin cytoskeleton remodels following treatment with phospholipase C activator mastoparan (Baluska et al., 2001). Therefore, profilin may be a linker between the plasma membrane and actin cytoskeleton through PIP2. Furthermore, profilins can interact with the proteins that contain PLP stretches of at least eight to ten prolines in continuous or discontinuous sequences (Schluter et al., 1997). In eukaryotes, formins are a group of actin-binding proteins that contain the FH1 domain with different numbers of PLP stretches; they are considered to act as morphological regulation proteins that direct the assembly of unbranched actin filaments (Paul and Pollard, 2009). Profilins or actin/profilin complexes can interact with the PLP stretches of different formins to promote actin filament polymerization (Chang et al., 1997; Pruyne et al., 2002; Kovar et al., 2006; Paul and Pollard, 2009). Additionally, type I formins contain an N-terminal transmembrane domains; this is the region of formin association with the plasma membrane in plants (Cvrckova et al., 2004). For example, in Arabidopsis, formin homology 6 (AtFH6) interacting with profilin locates at the plasma membrane and is uniformly distributed (Favery et al., 2004). Furthermore, AtFH1 and AtFH5 are reported to associate with the cell membrane (Banno and Chua, 2000; Cheung and Wu, 2004; Ingouff et al., 2005). This verifies that plant type I formins are likely to be membrane-bound, with AtFH8 being the exception, as it is targeted to the nuclear envelope (Xue et al., 2011). The site of the profilin binding FH1 PLP tracks is on the opposite face of the actin binding site of profilin (Schutt et al., 1993), and this explains why profilin can bind PLP and actin simultaneously without mutual influence (Tanaka and Shibata, 1985; Perelroizen et al., 1994). Profilin has an indirect connection and possibly acts as a regulator in the linkage of the plasma membrane and the actin cytoskeleton.
The plant cell is able to defend itself from infection by exogenous fungi. During this process, the cytoskeleton reorganizes and the papilla localizes at penetration sites, this leads to a thick cell wall being formed to prevent pathogen ingress (Schmelzer, 2002). Material is site-directed to arrive at positions around the fungal infection structure beneath the cell wall, and the actin filament and microtubule re-orientate their structures toward the penetration site (Schmelzer, 2002; Takemoto et al., 2003). In cultured parsley cells, undergoing attack from infection with the oomycetous plant pathogen Phytophthora infestans, profilin is expressed and accumulates at the site of infection on the plasma membrane, and the actin cables focus at the penetration site where Rop GTPases also accumulate (Schutz et al., 2006). In addition, in developing microspores and mature pollen of Zea mays, profilin is associated with the plasma membrane (von Witsch et al., 1998). Profilin accumulates in the tip zone near the plasma membrane in root hairs of Arabidopsis (Braun et al., 1999; Baluska et al., 2000). These results suggest that profilins play a role in both signal transduction and linkage between the plasma membrane and actin cytoskeleton
PROFILIN IS INVOLVED IN ORGANELLE LOCATION WITH THE ACTIN CYTOSKELETON
There is much evidence, that in various eukaryotic cells the cytoskeleton is involved in organelle movements. In plant cells, the role of the actin cytoskeleton in organelle movements has been reported for movements of chloroplasts (Kandasamy and Meagher, 1999), the endoplasmic reticulum (ER; Boevink et al., 1998), and the Golgi apparatus (Boevink et al., 1998; Nebenfuhr et al., 1999).
In Arabidopsis, CHUP1 (Chloroplast unusual positioning 1) which is a 112 kDa protein that is closely related with chloroplast movement (Kasahara et al., 2002; Oikawa et al., 2003) is directly targeted to the outer envelope of the chloroplast; this is dependent on its N-terminus domain (Oikawa et al., 2003). In addition to the N-terminus domain, the CHUP1 protein has four other domains, including two leucine-zippers, an actinin-type actin binding domain (Gimona et al., 2002), and a proline-rich motif (PRM) that is similar to PRM1 identified as a profilin binding motif (Holt and Koffer, 2001). A fusion protein which includes GST and the actin binding domain of CHUP1 can bind F-actin in vitro (Oikawa et al., 2003). The in vitro biochemical analyzes revealed that CHUP1 interacts with profilin as a modulator of actin polymerization through the PRM of C-terminal part of CHUP1 (CHUP1-CT). The experiment of CHUP1-CT titrated to a mixture of profilin and actin confirmed that the trimeric complex of actin, profilin, and CHUP1-CT is more stable than the individual binary complex. Though CHUP1 can bind F-actin directly, profilin has been reported to enhance the connection between chloroplasts and actin filaments (Schmidt von Braun and Schleiff, 2008).
Although profilin can bind to formin, the type II formins do not contain the transmembrane domains present in type I formins (Cvrckova et al., 2004). In rice, like other plant type II formins, formin homology 5 (FH5) has a characteristic N-terminal phosphatase tensin (PTEN)-related domain that may interact with membranes (Cvrckova et al., 2004). The experiments of transiently expressing the PTEN-RFP fusion protein in tobacco (Nicotiana tabacum) cells and immunostaining analysis using rice leaf cells revealed that the PTEN-like domain of FH5 is sufficient to confer localization of the protein to the chloroplast surface. This suggests that the PTEN domain of FH5 may be a bridge between chloroplasts and the actin cytoskeleton (Zhang et al., 2011). Furthermore, FH5 was capable of nucleating actin assembly from the actin/profilin complex in vitro biochemical analyzes (Yang et al., 2011; Zhang et al., 2011). Therefore, profilin is indirectly involved in the localization of chloroplast to the actin filaments. In Arabidopsis, observations of living cells in stable transgenic plants revealed that 35S:: GFP-AtPRF1 forms a filamentous network likely associated with actin filaments; this was verified by treatment with latrunculin A, and through a recovery experiment involving the removal of latrunculin A. Whereas, 35S:: GFP-AtPRF2 forms polygonal meshes resembling ER in the same latrunculin A treatment conditions (Wang et al., 2009). Furthermore, in plants, profilins possibly participate in the linkage of the nuclear envelope and the actin cytoskeleton during the interphase of Arabidopsis; this is because AtFH8 locates primarily to the nuclear envelope at this stage (Xue et al., 2011).
PROFILIN IS INVOLVED IN VESICLE TRAFFICKING
Profilins are known to play an important role in endocytosis and membrane trafficking in lower eukaryotes (Wolven et al., 2000; Pearson et al., 2003). In mammalian cells, profilins may also be involved in membrane trafficking. It has been reported that profilin 1 exists in budding Golgi vesicles, and that dynamin 2 recruitment to the Golgi is dependent on profilin 1 (Dong et al., 2000). Moreover, in mammalian cells, there are multiple phosphoinositide 3-kinases (PI3Ks), and these can be grouped into three main classes. Class I and II PI3Ks can induce receptor-dependent trafficking processes, such as phagocytosis. Class III PI3Ks, which represent the most ancient form of PI3Ks, and are the only ones conserved in lower eukaryotes, mammals, and plants. Class III PI3Ks mainly regulate receptor-independent trafficking events, such as endocytic membrane traffic (Lindmo and Stenmark, 2006). In animal cells, PI3Ks have been reported to play many different roles in vesicle trafficking, and inhibition of PI3Ks induces the inhibition of clathrin-dependent endocytosis (Martys et al., 1996; Spiro et al., 1996). In plant cells, Class III PI3K protein complexes may have a regulatory function during vesicle trafficking (Matsuoka et al., 1995; Kim et al., 2001; Jung et al., 2002). In Phaseolus vulgaris, in addition to the N- and C-terminal PLP-binding domain, profilin has a domain around Tyr72; this can recognize and bind PLP and PI3K (Aparicio-Fabre et al., 2006). Profilin can bind directly to Class III PI3Ks in a manner reliant upon the tyrosine phosphorylation status of the PLP domain in profilin. This interaction between profilin and Class III PI3Ks suggests that profilin may participate in membrane trafficking, and may act as a linker between the endocytic pathway and the actin reorganization dynamics (Aparicio-Fabre et al., 2006).
With advances in biotechnology, diverse pharmaceutical drugs have been used to study the interaction between vesicular trafficking and cytoskeleton. Brefeldin A (BFA) is a drug that inhibits the recycling of vesicular trafficking, and disrupts secretion in yeast, mammalian, and plant cells (Vogel et al., 1993; Samaj et al., 2004; Citterio et al., 2008; Robinson et al., 2008). In Arabidopsis roots, BFA-compartments can be formed due to the accumulation of trans-Golgi network (TGN) secretory and recycling vesicles, which gather together following BFA treatment (Geldner et al., 2003). During this process, profilin 2 is up-regulated and accumulates in the BFA-compartments, which then interacts with the actin to remodel the actin cytoskeleton. This study suggested that profilin 2 may bridge vesicular trafficking to the actin cytoskeleton in a BFA-dependent manner (Takac et al., 2011). Table 1 lists the profilins cited in the present article and emphasizes some of their cellular functions. Therefore, the recently investigated interactions between membranes and the actin cytoskeleton have revealed profilins to be of particular interest, this is because they may act as linkers and regulate communication and cooperation between the two cellular members in plants. Currently available studies suggest that diverse interaction mechanisms are required to satisfy the different structural and dynamic requirements of particular systems. Future research is required to unravel how membrane-actin cytoskeleton interactions are regulated through profilins and their different ligands.
Table 1.
Profilin and its cellular functions in plant cells.
| Profilin involving in the cellular pathway | Profilins | Cells or ligands | Reference |
|---|---|---|---|
| Plasma membrane-actin cytoskeleton interaction | ZmPRO3 | Root cells of maize PIP2 | Baluska et al. (2001) |
| AtPRF1 etc | Arabidopsis seed endosperm, root cells etc AtFH1, AtFH5, and AtFH6 | Banno and Chua (2000); Cheung and Wu (2004), Ingouff et al. (2005), Favery et al. (2004) | |
| PcPRF1 | Cultured parsley cells | Schutz et al. (2006) | |
| Organelles location with the actin cytoskeleton | Profilins from Arabidopsis | Arabidopsis mesophyll protoplasts CHUP1 | Oikawa et al. (2003); Schmidt von Braun and Schleiff (2008) |
| NA | Rice leaf cells OsFH5 | Zhang et al. (2011); Yang et al. (2011) | |
| AtPRF2 | Arabidopsis epidermal cells, trichomes, stem epidermal cells ER | Wang et al. (2009) | |
| NA | Arabidopsis root cells AtFH8 | Xue et al. (2011) | |
| Vesicle trafficking with the actin cytoskeleton | PvPRO1 | Phaseolus vulgaris root nodules Class III PI3Ks | Aparicio-Fabre et al. (2006) |
| AtPRF2 | Arabidopsis roots TGN | Takac et al. (2011) |
NA, not available.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31130005) and the National Basic Research Program of China (2013CB126902) to Haiyun Ren.
REFERENCES
- Aparicio-Fabre R., Guillen G., Estrada G., Olivares-Grajales J., Gurrola G., Sanchez F. (2006). Profilin tyrosine phosphorylation in poly-L-proline-binding regions inhibits binding to phosphoinositide 3-kinase in Phaseolus vulgaris. Plant J. 47 491–500 10.1111/j.1365-313X.2006.02787.x [DOI] [PubMed] [Google Scholar]
- Baluska F., Salaj J., Mathur J., Braun M., Jasper F., Samaj J., et al. (2000). Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 227 618–632 10.1006/dbio.2000.9908 [DOI] [PubMed] [Google Scholar]
- Baluska F., von Witsch M., Peters M., Hlavacka A., Volkmann D. (2001). Mastoparan alters subcellular distribution of profilin and remodels F-actin cytoskeleton in cells of maize root apices. Plant Cell Physiol. 42 912–922 10.1093/pcp/pce116 [DOI] [PubMed] [Google Scholar]
- Banno H., Chua N. H. (2000). Characterization of the arabidopsis formin-like protein AFH1 and its interacting protein. Plant Cell Physiol. 41 617–626 10.1093/pcp/41.5.617 [DOI] [PubMed] [Google Scholar]
- Boevink P., Oparka K., Santa Cruz S., Martin B., Betteridge A., Hawes C. (1998). Stacks on tracks: the plant Golgi apparatus traffics on an actin/ER network. Plant J. 15 441–447 10.1046/j.1365-313X.1998.00208.x [DOI] [PubMed] [Google Scholar]
- Braun M., Baluska F., von Witsch M., Menzel D. (1999). Redistribution of actin, profilin and phosphatidylinositol-4, 5-bisphosphate in growing and maturing root hairs. Planta 209 435–443 10.1007/s004250050746 [DOI] [PubMed] [Google Scholar]
- Caroni P. (2001). Actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 20 4332–4336 10.1093/emboj/20.16.4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Drubin D., Nurse P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137 169–182 10.1083/jcb.137.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A. Y., Wu H. M. (2004). Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16 257–269 10.1105/tpc.016550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen H. E., Ramachandran S., Tan C. T., Surana U., Dong C. H., Chua N. H. (1996). Arabidopsis profilins are functionally similar to yeast profilins: identification of a vascular bundle-specific profilin and a pollen-specific profilin. Plant J. 10 269–279 10.1046/j.1365-313X.1996.10020269.x [DOI] [PubMed] [Google Scholar]
- Citterio C., Vichi A., Pacheco-Rodriguez G., Aponte A. M., Moss J., Vaughan M. (2008). Unfolded protein response and cell death after depletion of brefeldin A-inhibited guanine nucleotide-exchange protein GBF1. Proc. Natl. Acad. Sci. U.S.A. 105 2877–2882 10.1073/pnas.0712224105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couchman J. R., Vogt S., Lim S. T., Lim Y., Oh E. S., Prestwich G. D., et al. (2002). Regulation of inositol phospholipid binding and signaling through syndecan-4. J. Biol. Chem. 277 49296–49303 10.1074/jbc.M209679200 [DOI] [PubMed] [Google Scholar]
- Cvrckova F., Novotny M., Pickova D., Zarsky V. (2004). Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genomics 5:44 10.1186/1471-2164-5-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks M. J., Calcutt J. R., Ingle E. K., Hawkins T. J., Chapman S., Richardson A. C., et al. (2012). A superfamily of actin-binding proteins at the actin-membrane nexus of higher plants. Curr. Biol. 22 1595–1600 10.1016/j.cub.2012.06.041 [DOI] [PubMed] [Google Scholar]
- Doherty G. J., McMahon H. T. (2008). Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu. Rev. Biophys. 37 65–95 10.1146/annurev.biophys.37.032807.125912 [DOI] [PubMed] [Google Scholar]
- Dong J., Radau B., Otto A., Muller E., Lindschau C., Westermann P. (2000). Profilin I attached to the Golgi is required for the formation of constitutive transport vesicles at the trans-Golgi network. Biochim. Biophys. Acta. 1497 253–260 10.1016/S0167-4889(00)00056-2 [DOI] [PubMed] [Google Scholar]
- Favery B., Chelysheva L. A., Lebris M., Jammes F., Marmagne A., De Almeida-Engler J., et al. (2004). Arabidopsis formin AtFH6 is a plasma membrane-associated protein upregulated in giant cells induced by parasitic nematodes. Plant Cell 16 2529–2540 10.1105/tpc.104.024372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A. A., Ball T., Mahoney N. M., Valenta R., Almo S. C. (1997). The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin. Structure 5 33–45 10.1016/S0969-2126(97)00164-0 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39 783–791 10.2307/2408678 [DOI] [PubMed] [Google Scholar]
- Geiger B., Tokuyasu K. T., Dutton A. H., Singer S. J. (1980). Vinculin, an intracellular protein localized at specialized sites where microfilament bundles terminate at cell membranes. Proc. Natl. Acad. Sci. U.S.A. 77 4127–4131 10.1073/pnas.77.7.4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., et al. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112 219–230 10.1016/S0092-8674(03)00003-5 [DOI] [PubMed] [Google Scholar]
- Gibbon B. C., Staiger C. J. (2000). “Profilin,” in Actin: a Dynamic Framework for Multiple Plant Cell Functions eds Staiger C. J., Baluska F., Volkmann D., Barlow P. (Dordrecht, The Netherlands: Kluwer Academic Publishers; ) 45–65 [Google Scholar]
- Gimona M., Djinovic-Carugo K., Kranewitter W. J., Winder S. J. (2002). Functional plasticity of CH domains. FEBS Lett. 513 98–106 10.1016/S0014-5793(01)03240-9 [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont P. J., Machesky L. M., Baldassare J. J., Pollard T. D. (1990). The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science 247 1575–1578 10.1126/science.2157283 [DOI] [PubMed] [Google Scholar]
- Heise H., Bayerl T., Isenberg G., Sackmann E. (1991). Human platelet P-235, a talin-like actin binding protein, binds selectively to mixed lipid bilayers. Biochim. Biophys. Acta. 1061 121–131 10.1016/0005-2736(91)90276-E [DOI] [PubMed] [Google Scholar]
- Heiska L., Alfthan K., Gronholm M., Vilja P., Vaheri A., Carpen O. (1998). Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2). Regulation by phosphatidylinositol 4, 5-bisphosphate. J. Biol. Chem. 273 21893–21900 10.1074/jbc.273.34.21893 [DOI] [PubMed] [Google Scholar]
- Holt M. R., Koffer A. (2001). Cell motility: proline-rich proteins promote protrusions. Trends Cell Biol. 11 38–46 10.1016/S0962-8924(00)01876-6 [DOI] [PubMed] [Google Scholar]
- Huang S., McDowell J. M., Weise M. J., Meagher R. B. (1996). The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 111 115–126 10.1104/pp.111.1.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey P. J., Allwood E. G., Smertenko A. P. (2002). Actin-binding proteins in the Arabidopsis genome database: properties of functionally distinct plant actin-depolymerizing factors/cofilins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357 791–798 10.1098/rstb.2002.1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingouff M., Fitz Gerald J. N., Guerin C., Robert H., Sorensen M. B., Van Damme D., et al. (2005). Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 7 374–380 10.1038/ncb1238 [DOI] [PubMed] [Google Scholar]
- Jimenez-Lopez J. C., Morales S., Castro A. J., Volkmann D., Rodriguez-Garcia M. I., Alche J. D. (2012). Characterization of profilin polymorphism in pollen with a focus on multifunctionality. PLoS ONE 7:e30878 10.1371/journal.pone.0030878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jockusch B. M., Murk K., Rothkegel M. (2007). The profile of profilins. Rev. Physiol. Biochem. Pharmacol. 159 131–149 [DOI] [PubMed] [Google Scholar]
- Jung J. Y., Kim Y. W., Kwak J. M., Hwang J. U., Young J., Schroeder J. I., et al. (2002). Phosphatidylinositol 3- and 4-phosphate are required for normal stomatal movements. Plant Cell 14 2399–2412 10.1105/tpc.004143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy M. K., McKinney E. C., Meagher R. B. (2002). Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil. Cytoskeleton 52 22–32 10.1002/cm.10029 [DOI] [PubMed] [Google Scholar]
- Kandasamy M. K., Meagher R. B. (1999). Actin-organelle interaction: association with chloroplast in arabidopsis leaf mesophyll cells. Cell Motil. Cytoskeleton 44 110–118 [DOI] [PubMed] [Google Scholar]
- Kasahara M., Kagawa T., Oikawa K., Suetsugu N., Miyao M., Wada M. (2002). Chloroplast avoidance movement reduces photodamage in plants. Nature 420 829–832 10.1038/nature01213 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Eu Y. J., Yoo C. M., Kim Y. W., Pih K. T., Jin J. B., et al. (2001). Trafficking of phosphatidylinositol 3-phosphate from the trans-Golgi network to the lumen of the central vacuole in plant cells. Plant Cell 13 287–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Drobak B. K., Staiger C. J. (2000). Maize profilin isoforms are functionally distinct. Plant Cell 12 583–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124 423–435 10.1016/j.cell.2005.11.038 [DOI] [PubMed] [Google Scholar]
- Lindmo K., Stenmark H. (2006). Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119 605–614 10.1242/jcs.02855 [DOI] [PubMed] [Google Scholar]
- Mahoney N. M., Rozwarski D. A., Fedorov E., Fedorov A. A., Almo S. C. (1999). Profilin binds proline-rich ligands in two distinct amide backbone orientations. Nat. Struct. Biol. 6 666–671 10.1038/10722 [DOI] [PubMed] [Google Scholar]
- Martys J. L., Wjasow C., Gangi D. M., Kielian M. C., McGraw T. E., Backer J. M. (1996). Wortmannin-sensitive trafficking pathways in Chinese hamster ovary cells. Differential effects on endocytosis and lysosomal sorting. J. Biol. Chem. 271 10953–10962 10.1074/jbc.271.18.10953 [DOI] [PubMed] [Google Scholar]
- Matsuoka K., Bassham D. C., Raikhel N. V., Nakamura K. (1995). Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J. Cell Biol. 130 1307–1318 10.1083/jcb.130.6.1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler W. J., Bell A. J., Ernst E., Lavoie T. B., Mueller L. (1994). Identification of the poly-L-proline-binding site on human profilin. J. Biol. Chem. 269 4620–4625 [PubMed] [Google Scholar]
- Mittermann I., Swoboda I., Pierson E., Eller N., Kraft D., Valenta R., et al. (1995). Molecular cloning and characterization of profilin from tobacco (Nicotiana tabacum): increased profilin expression during pollen maturation. Plant Mol. Biol. 27 137–146 10.1007/BF00019185 [DOI] [PubMed] [Google Scholar]
- Nebenfuhr A., Gallagher L. A., Dunahay T. G., Frohlick J. A., Mazurkiewicz A. M., Meehl J. B., et al. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1142 10.1104/pp.121.4.1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebl T., Oh S. W., Luna E. J. (2000). Membrane cytoskeleton: PIP(2) pulls the strings. Curr. Biol. 10 R351–R354 10.1016/S0960-9822(00)00465-6 [DOI] [PubMed] [Google Scholar]
- Oikawa K., Kasahara M., Kiyosue T., Kagawa T., Suetsugu N., Takahashi F., et al. (2003). Chloroplast unusual positioning1 is essential for proper chloroplast positioning. Plant Cell 15 2805–2815 10.1105/tpc.016428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L., Petersen M., Mattsson O., Mundy J. (2002). An Arabidopsis callose synthase. Plant Mol. Biol. 49 559–566 10.1023/A:1015558231400 [DOI] [PubMed] [Google Scholar]
- Paul A. S., Pollard T. D. (2009). Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton 66 606–617 10.1002/cm.20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson A. M., Baksa K., Ramet M., Protas M., McKee M., Brown D., et al. (2003). Identification of cytoskeletal regulatory proteins required for efficient phagocytosis in Drosophila. Microbes Infect. 5 815–824 10.1016/S1286-4579(03)00157-6 [DOI] [PubMed] [Google Scholar]
- Perelroizen I., Marchand J. B., Blanchoin L., Didry D., Carlier M. F. (1994). Interaction of profilin with G-actin and poly(L-proline). Biochemistry 33 8472–8478 10.1021/bi00194a011 [DOI] [PubMed] [Google Scholar]
- Pollard T. D., Quirk S. (1994). Profilins, ancient actin binding proteins with highly divergent primary structures. Soc. Gen. Physiol. Ser. 49 117–128 [PubMed] [Google Scholar]
- Pruitt K. D., Tatusova T., Maglott D. R. (2007). NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 35 D61–D65 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D., Evangelista M., Yang C., Bi E., Zigmond S., Bretscher A., et al. (2002). Role of formins in actin assembly: nucleation and barbed-end association. Science 297 612–615 10.1126/science.1072309 [DOI] [PubMed] [Google Scholar]
- Robinson D. G., Langhans M., Saint-Jore-Dupas C., Hawes C. (2008). BFA effects are tissue and not just plant specific. Trends Plant Sci. 13 405–408 10.1016/j.tplants.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Samaj J., Baluska F., Voigt B., Schlicht M., Volkmann D., Menzel D. (2004). Endocytosis, actin cytoskeleton, and signaling. Plant Physiol. 135 1150–1161 10.1104/pp.104.040683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluter K., Jockusch B. M., Rothkegel M. (1997). Profilins as regulators of actin dynamics. Biochim. Biophys. Acta 1359 97–109 10.1016/S0167-4889(97)00100-6 [DOI] [PubMed] [Google Scholar]
- Schmelzer E. (2002). Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 7 411–415 10.1016/S1360-1385(02)02307-5 [DOI] [PubMed] [Google Scholar]
- Schmidt von Braun S., Schleiff E. (2008). The chloroplast outer membrane protein CHUP1 interacts with actin and profilin. Planta 227 1151–1159 10.1007/s00425-007-0688-7 [DOI] [PubMed] [Google Scholar]
- Schobert C., Gottschalk M., Kovar D. R., Staiger C. J., Yoo B. C., Lucas W. J. (2000). Characterization of Ricinus communis phloem profilin, RcPRO1. Plant Mol. Biol. 42 719–730 10.1023/A:1006391508429 [DOI] [PubMed] [Google Scholar]
- Schutt C. E., Myslik J. C., Rozycki M. D., Goonesekere N. C., Lindberg U. (1993). The structure of crystalline profilin-beta-actin. Nature 365 810–816 10.1038/365810a0 [DOI] [PubMed] [Google Scholar]
- Schutz I., Gus-Mayer S., Schmelzer E. (2006). Profilin and Rop GTPases are localized at infection sites of plant cells. Protoplasma 227 229–235 10.1007/s00709-005-0151-1 [DOI] [PubMed] [Google Scholar]
- Spiro D. J., Boll W., Kirchhausen T., Wessling-Resnick M. (1996). Wortmannin alters the transferrin receptor endocytic pathway in vivo and in vitro. Mol. Biol. Cell 7 355–367 10.1091/mbc.7.3.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger C. J., Goodbody K. C., Hussey P. J., Valenta R., Drobak B. K., Lloyd C. W. (1993). The profilin multigene family of maize: differential expression of three isoforms. Plant J. 4 631–641 10.1046/j.1365-313X.1993.04040631.x [DOI] [PubMed] [Google Scholar]
- Stossel T. P., Condeelis J., Cooley L., Hartwig J. H., Noegel A., Schleicher M., et al. (2001). Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2 138–145 10.1038/35052082 [DOI] [PubMed] [Google Scholar]
- Swoboda I., Bhalla P. L., Xu H., Zhang Y., Mittermann I., Valenta R., et al. (2001). Identification of pronp1, a tobacco profilin gene activated in tip-growing cells. Plant Mol. Biol. 46 531–538 10.1023/A:1010641229366 [DOI] [PubMed] [Google Scholar]
- Takac T., Pechan T., Richter H., Muller J., Eck C., Bohm N., et al. (2011). Proteomics on brefeldin A-treated Arabidopsis roots reveals profilin 2 as a new protein involved in the cross-talk between vesicular trafficking and the actin cytoskeleton. J. Proteome Res. 10 488–501 10.1021/pr100690f [DOI] [PubMed] [Google Scholar]
- Takemoto D., Jones D. A., Hardham A. R. (2003). GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 33 775–792 10.1046/j.1365-313X.2003.01673.x [DOI] [PubMed] [Google Scholar]
- Tanaka M., Shibata H. (1985). Poly (L-proline)-binding proteins from chick embryos are a profilin and profilactin. Eur. J. Biochem. 151 291–297 10.1111/j.1432-1033.1985.tb09099.x [DOI] [PubMed] [Google Scholar]
- Thorn K. S., Christensen H. E., Shigeta R., Huddler D., Shalaby L., Lindberg U., et al. (1997). The crystal structure of a major allergen from plants. Structure 5 19–32 10.1016/S0969-2126(97)00163-9 [DOI] [PubMed] [Google Scholar]
- Verma D. P., Hong Z. (2001). Plant callose synthase complexes. Plant Mol. Biol. 47 693–701 10.1023/A:1013679111111 [DOI] [PubMed] [Google Scholar]
- Vidali L., Perez H. E., Valdes Lopez V., Noguez R., Zamudio F., Sanchez F. (1995). Purification, characterization, and cDNA cloning of profilin from Phaseolus vulgaris. Plant Physiol. 108 115–123 10.1104/pp.108.1.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. P., Lee J. N., Kirsch D. R., Rose M. D., Sztul E. S. (1993). Brefeldin A causes a defect in secretion in Saccharomyces cerevisiae. J. Biol. Chem. 268 3040–3043 [PubMed] [Google Scholar]
- von Witsch M., Baluska F., Staiger C. J., Volkmann D. (1998). Profilin is associated with the plasma membrane in microspores and pollen. Eur. J. Cell Biol. 77 303–312 10.1016/S0171-9335(98)80089-7 [DOI] [PubMed] [Google Scholar]
- Wang F., Jing Y., Wang Z., Mao T., Samaj J., Yuan M., et al. (2009). Arabidopsis profilin isoforms, PRF1 and PRF2 show distinctive binding activities and subcellular distributions. J. Integr. Plant Biol. 51 113–121 10.1111/j.1744-7909.2008.00781.x [DOI] [PubMed] [Google Scholar]
- Wolven A. K., Belmont L. D., Mahoney N. M., Almo S. C., Drubin D. G. (2000). In vivo importance of actin nucleotide exchange catalyzed by profilin. J. Cell Biol. 150 895–904 10.1083/jcb.150.4.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X. H., Guo C. Q., Du F., Lu Q. L., Zhang C. M., Ren H. Y. (2011). AtFH8 is involved in root development under effect of low-dose latrunculin B in dividing cells. Mol. Plant 4 264–278 10.1093/mp/ssq085 [DOI] [PubMed] [Google Scholar]
- Yang W., Ren S., Zhang X., Gao M., Ye S., Qi Y., et al. (2011). BENT UPPERMOST INTERNODE1 encodes the class II formin FH5 crucial for actin organization and rice development. Plant Cell 23 661–680 10.1105/tpc.110.081802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L. X., Nasrallah J., Valenta R., Parthasarathy M. V. (1998). Molecular cloning and mRNA localization of tomato pollen profilin. Plant Mol. Bio. 36 699–707 10.1023/A:1005971327353 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang Y., Tan H., Wang Y., Li G., Liang W., et al. (2011). RICE MORPHOLOGY DETERMINANT encodes the type II formin FH5 and regulates rice morphogenesis. Plant Cell 23 681–700 10.1105/tpc.110.081349 [DOI] [PMC free article] [PubMed] [Google Scholar]