Abstract
A recent genome-wide association meta-analysis showed a suggestive association between alcohol intake in humans and a common single nucleotide polymorphism in the ras-specific guanine nucleotide releasing factor 2 gene. Here, we tested whether this variant – associated with lower alcohol consumption – showed associations with brain structure and longitudinal ventricular expansion over time, across two independent elderly cohorts, totaling 1,032 subjects. We first examined a large sample of 738 elderly participants with neuroimaging and genetic data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI1). Then, we assessed the generalizability of the findings by testing this polymorphism in a replication sample of 294 elderly subjects from a continuation of the first ADNI project (ADNI2) to minimize the risk of reporting false positive results. The minor allele – previously linked with lower alcohol intake – was associated with larger volumes in various cortical regions, notably the medial prefrontal cortex and cingulate gyrus in both cohorts. Intriguingly, the same allele also predicted faster ventricular expansion rates in the ADNI1 cohort at 1- and 2-year follow up. Despite a lack of alcohol consumption data in this study cohort, these findings, combined with earlier functional imaging investigations of the same gene, suggest the existence of reciprocal interactions between genes, brain, and drinking behavior.
Keywords: neuroimaging genetics, ventricular expansion, aging neuroscience, rasgrf2, brain volume, structural MRI
INTRODUCTION
A recent genome-wide association meta-analysis of alcohol intake in humans showed a suggestive association with a common variant (rs26907) in the RASGRF2 gene (Schumann et al., 2011). Each copy of the minor A allele at rs26907 (MAF = 0.17), was associated with about 2.6% lower alcohol consumption (Schumann et al., 2011). Functional analysis of the RASGRF2 gene by the same group revealed that alcohol preference is associated with whole-brain RASGRF2 mRNA expression and that RASGRF2 regulates alcohol-induced reinforcement by affecting mesolimbic neuron activity and dopamine release (Stacey et al., 2012).
Ras-specific guanine nucleotide-releasing factor 2 (Ras-GRF2) is a protein encoded by the RASGRF2 gene. It is involved in signal transduction from ion channel receptors (Tian et al., 2004) and the regulation of MAPK signaling cascades, including the ERK pathway (Fasano and Brambilla, 2011; Feig, 2011), which plays a critical role in drug-induced reinforcement and synaptic plasticity (Girault et al., 2007). RASGRF2 also helps to regulate neuronal excitability, neuronal survival in response to ischemia, learning and memory formation, and carcinogenesis (Jin and Feig, 2010; Feig, 2011; Fernandez-Medarde and Santos, 2011).
A growing enigma in neurology is how alcohol intake, and related genes, affect brain aging. Moderate alcohol intake protects against cardiovascular events (Rimm et al., 1999; Mukamal et al., 2006; Hvidtfeldt et al., 2010), but it is less clear how moderate drinking affect the brain over the long term. Elderly people who drink moderately may have fewer white matter abnormalities and infarcts than both non-drinkers and heavy drinkers (Mukamal et al., 2001; den Heijer et al., 2004; Mukamal, 2004). Genetic factors affect alcohol intake (Olfson and Bierut, 2012; Blaine et al., 2013; Meyers et al., 2013) but we do not know if the same genes affect brain structure and the rate of brain atrophy.
Dynamic changes in the brain’s lateral ventricles reveal the rate of brain atrophy as we age and reflect brain tissue loss with high effect sizes (Hua et al., 2013). Lateral ventricle expansion accompanies gray and white matter degeneration globally and in nearby subcortical regions (Ferrarini et al., 2008). The ventricles do not play a role in cognition, but ventricular expansion is associated with many brain-related health factors in the elderly, including current cognitive status and future memory decline (Coffey et al., 2001).
Here, we hypothesized that this candidate variant for reduced drinking (the minor A allele at the rs26907 locus in RASGRF2) might be associated with structural differences in fronto-limbic regions relevant to alcohol reinforcement in elderly subjects. To test if the findings generalized, we examined an independent replication sample of elderly participants, reducing the risk of false positive findings. We predicted that this variant might be associated with the ventricular expansion rate, in the larger sample whose ventricular volumes were measured at baseline, and at 1- and 2-year follow ups.
MATERIALS AND METHODS
SUBJECTS
Data used in preparing this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database1. ADNI, followed by ADNI-GO and ADNI-2, recruited over 1500 adults, ages 55–90, to participate in the research, consisting of cognitively normal older individuals, people with early or late mild cognitive impairment (MCI), and Alzheimer’s disease (AD). Subjects originally recruited for ADNI-1 and ADNI-GO had the option to be followed in ADNI-2. For up-to-date information, see http://www.adni-info.org.
Here, we analyzed two independent samples of elderly subjects with neuroimaging and genome-wide data from the ADNI1 and ADNI2 cohorts. We refer to ADNI-GO and ADNI-2 participants as “ADNI2,” as the only distinction was the grant funding that supported data collection, and the data collection was identical for them both. All ADNI studies are conducted according to the Good Clinical Practice guidelines, the Declaration of Helsinki, and U.S. 21 CFR Part 50 (Protection of Human Subjects), and Part 56 (Institutional Review Boards). Written informed consent was obtained from all participants before protocol-specific procedures were performed. To avoid the known effects of population stratification on genetic analysis (Lander and Schork, 1994), we only included non-Hispanic Caucasian subjects identified by self-report and confirmed by multi-dimensional scaling (MDS) analysis (Stein et al., 2010) in both ADNI cohorts.
Alzheimer’s disease neuroimaging initiative1
The ADNI1 cohort included three diagnostic groups: people with AD, MCI, and healthy elderly (cognitively normal) participants. We included participants from all diagnostic groups, as power is limited when performing any genetic association analysis. Typical effects of candidate genes on the phenotype are often around 1% of the mean value per allele (Stein et al., 2012), so we have often been able to pick up effects only when ADNI’s full sample is included. Even so, we have been able to replicate effects from ADNI in other non-overlapping samples, showing that affects found in ADNI can be robust and can generalize (Stein et al., 2011; Hibar et al., 2013; Roussotte et al., 2013). Effect sizes for individual genetic variants on brain structure in particular are expected to be small, so the genetic analysis would be underpowered if we further subdivided the sample (Stein et al., 2012). Our final analysis comprised 738 individuals (average age ± s.d. = 75.52 ± 6.78 years; 438 men/300 women) including 173 AD, 359 MCI, and 206 healthy participants (Table 1).
Table 1.
Demographic and genetic data for the ADNI1 cohort.
| ADNI1 | Males | Females | Total |
|---|---|---|---|
| Total | 438 | 300 | 738 |
| Healthy elderly | 112 | 94 | 206 (28%) |
| MCI | 231 | 128 | 359 (49%) |
| AD | 95 | 78 | 173 (23%) |
| rs26907 0 A alleles | 318 | 226 | 544 (74%) |
| rs26907 1 A alleles | 114 | 67 | 181 (24%) |
| rs26907 2 A alleles | 6 | 7 | 13 (2%) |
| Mean age (±sd) | 75.90 (±6.76) | 74.98 (±6.78) | 75.53 (±6.78) |
Alzheimer’s disease neuroimaging initiative2
The ADNI2 cohort also included people with MCI who were further subdivided into early and late MCI (EMCI, LMCI). When we conducted these analyses, just under 300 ADNI2 subjects had been genotyped and processed using tensor based morphometry (TBM; see Minimal Deformation Target and Tensor Based Morphometry). Our final analysis comprised 294 individuals (average age ± s.d. = 73.16 ± 7.33 years; 166 men/128 women) including 25 AD, 66 LMCI, 81 EMCI, and 122 healthy participants (Table 2).
Table 2.
Demographic and genetic data for the ADNI2 cohort.
| ADNI2 | Males | Females | Total |
|---|---|---|---|
| Total | 166 | 128 | 294 |
| Healthy elderly | 64 | 58 | 122 (41%) |
| EMCI | 47 | 34 | 81 (28%) |
| LMCI | 38 | 28 | 66 (22%) |
| AD | 17 | 8 | 25 (9%) |
| rs26907 0 A alleles | 117 | 88 | 205 (70%) |
| rs26907 1 A alleles | 46 | 38 | 84 (28%) |
| rs26907 2 A alleles | 3 | 2 | 5 (2%) |
| Mean age (±sd) | 74.49 (±7.14) | 71.45 (±7.25) | 73.16 (±7.33) |
GENOTYPING AND SNP SELECTION
In ADNI, genome-wide association study (GWAS) data was collected from 1252 participants. All 818 subjects (including the non-Caucasians not used in this study) from the ADNI1 sample were genotyped using the Illumina Human 610-Quad BeadChip (San Diego, CA, USA), and DNA samples were genotyped from 434 ADNI-GO/ADNI-2 participants using the Illumina OmniExpress genotyping array.
Data from both cohorts were imputed to a common reference space; the 1000 genomes CEU (Caucasian) reference set following freely available imputation protocols (ENIGMA2, 2012). The imputed data were filtered for standard imputation quality criteria (imputation quality: Rsq < 0.3) and minor allele frequency (MAF < 0.05). The final, filtered genetic datasets were used for our genetic analyses.
We analyzed a common (G/A, minor allele frequency: A = 0.170) single nucleotide polymorphism (rs26907) in the ras-specific guanine nucleotide releasing factor 2 (RASGRF2) gene previously implicated in alcohol consumption (Schumann et al., 2011; Stacey et al., 2012) for association with regional brain volumes in both ADNI cohorts, and longitudinal ventricular expansion in the ADNI1 subjects who had been included in the regional brain volumes association study. The allele distribution in both samples did not deviate from the Hardy-Weinberg equilibrium (p = 0.6118 for ADNI1 and p = 0.1912 for ADNI2).
IMAGE ACQUISITION
ADNI1 subjects were scanned with a standardized MRI protocol developed for this cohort (Leow et al., 2006; Jack et al., 2008). Briefly, high-resolution structural brain MRI scans were acquired at 58 sites across North America, using 1.5 T MRI scanners. A sagittal 3D MP-RAGE sequence was used, optimized for consistency across sites (Jack et al., 2008; TR/TE = 2400/1000 ms; flip angle = 8°; FOV = 24 cm; final reconstructed voxel resolution = 0.9375 × 0.9375 × 1.2 mm3). Each ADNI2 subject received a 3 T accelerated T1-weighted MRI scan. By vendor, General Electric (GE) scanners use IR-SPGR sequences and Philips and Siemens use MP-RAGE sequences. Scan vendors and sequences for ADNI2 are available online.
REGIONAL BRAIN VOLUMES ASSOCIATION STUDIES (ADNI1 AND ADNI2)
Image correction and pre-processing
For both ADNI samples, image corrections were applied using a processing pipeline at the Mayo Clinic, consisting of: (1) a procedure termed GradWarp to correct geometric distortion due to gradient non-linearity (Jovicich et al., 2006), (2) a “B1-correction”, to adjust for image intensity inhomogeneity due to B1 non-uniformity using calibration scans (Jack et al., 2008), (3) “N3” bias field correction, for reducing residual intensity inhomogeneity (Sled et al., 1998), and (4) geometrical scaling, according to a phantom scan acquired for each subject (Jack et al., 2008) to adjust for scanner- and session-specific calibration errors2. To adjust for global differences in brain positioning and scale, all subjects’ scans were linearly registered to the stereotaxic space defined by the International Consortium for Brain Mapping (ICBM-53; Mazziotta et al., 2001), using a 9-parameter (9P) transformation (three translations, three rotations, three scales (Collins et al., 1994). For both ADNI cohorts, we used standard trilinear interpolation and resampled the resulting aligned scans to have 1mm isotropic voxels. Subjects’ brain images were not skull-stripped during pre-processing.
Minimal deformation target and tensor based morphometry
For ADNI1, we created a minimal deformation target (MDT), which serves as an unbiased average template image for automated image registration, and to reduce statistical bias. The MDT was created using the MRI scans of 40 randomly selected healthy elderly subjects, as detailed elsewhere (Hua et al., 2008a,b). The MDT image was calculated as a geometrically centered mean anatomical image, using a method called sKL-MI to align data to an average affine registered target image; this procedure leads to fairly “sharp” average brain image for a group and follows a procedure we developed and tested elsewhere (Hua et al., 2008a,b).
To quantify 3D patterns of volumetric tissue variations, all individual T1-weighted images (N = 1,032) were non-linearly aligned to the MDT template created for ADNI1 with an inverse-consistent 3D elastic warping technique using a mutual information cost function (Leow et al., 2005). For each subject, a separate Jacobian matrix field was derived from the gradients of the deformation field that aligned that individual brain to the MDT template. The determinant of the local Jacobian matrix was derived from the forward deformation field to characterize local volume differences. Color-coded Jacobian determinants were used to illustrate regions of volume expansion, i.e., those with det J(r) >1, or contraction, i.e., det J(r) <1 (Freeborough and Fox, 1998; Thompson et al., 2000; Chung et al., 2001; Riddle et al., 2004) relative to the template. All images were registered to the same template, so these Jacobian maps shared common anatomical coordinates, defined by the normal template. Individual Jacobian maps were retained for further statistical analyses.
Regression of structural brain differences with the candidate SNP
In both ADNI cohorts, we investigated how the rs26907 variant affected regional brain volumes using univariate linear regression to associate the number of minor A alleles (0,1, or 2) with the Jacobian values (describing the amount of brain tissue deficit or excess relative to the standard template) at each voxel in the brain, after covarying for age, sex, and diagnosis (i.e., AD, MCI, and healthy elderly for ADNI1 and AD, LMCI, EMCI, and healthy elderly for ADNI2).
Multiple comparisons correction
Computing thousands of association tests across the brain can introduce a high Type I (false positive) error rate in neuroimaging studies, if not controlled. We used a searchlight method for false discovery rate (FDR) correction (Langers et al., 2007), which controls the FDR in any reported statistical map. This corrected the map of statistical associations between the image phenotype (morphometry) and genotype at the rs26907 locus. All maps shown are thresholded at the appropriate corrected p-value, after performing searchlight FDR (q = 0.05), to show only regions of significance that passed the multiple comparisons correction.
LONGITUDINAL VENTRICULAR EXPANSION STUDY (ADNI1)
Image correction and pre-processing
For lateral ventricle segmentation, we analyzed baseline (N = 738), 1-year (N = 623), and 2-year (N = 481) follow-up brain MRI scans from all the ADNI1 subjects included in the regional brain volumes association study described above. Raw MRI scans were pre-processed to reduce signal inhomogeneity and linearly registered to a template (using 9 parameter registration). Segmentations were assessed visually for defects from multiple views. All subjects were quality controlled for ventricular segmentation, and one baseline ADNI1 subject was removed from the longitudinal ventricular expansion study after quality control of the ventricular surfaces. Our final analysis thus included 737 ADNI1 subjects at baseline, 623 at 12-month follow-up, and 481 at 24-month follow-up.
Segmentation of the lateral ventricles
Prior methods for ventricular segmentation have used semi-automated, automated (Chou et al., 2008), and single-atlas or multi-atlas methods (Chou et al., 2009). Here we segmented the ventricles with our modified multi-atlas approach (Gutman et al., 2013; Madsen et al., 2013). Our segmentation approach uses group-wise surface registration to existing templates in addition to surface-based template blending to yield more accurate results. The lateral ventricles were segmented in each subject using a validated method (Chou et al., 2008). Ventricular surfaces were then extracted from these segmentations and an inverse-consistent fluid registration with a mutual information fidelity term aligned a set of hand-labeled ventricular templates to each scan (Leow et al., 2007). The template surfaces were registered into homologous point-to-point correspondence as a group using medial-spherical registration (Gutman et al., 2012).
To construct a surface boundary of the new subject, a normalized similarity measure between each template image and the new image was computed in a neighborhood around each vertex point of each deformed template surface. The position of each point of the new boundary was defined by the template, which showed the best similarity score, here normalized mutual information. The final surface was then constrained to be a smooth approximation of this winner-takes-all construction. This approach is very similar to that of (Yushkevich et al., 2010), except ours is based on surface geometry rather than voxels in an image. The approach is advantageous compared to whole-template approaches typically used in multi-atlas segmentation, allowing more flexible segmentation, particularly for outliers.
Statistical analyses: associations of rs26907 genotype with ventricular volumes
Statistical tests using the number of minor A alleles at the rs26907 locus to predict left and right ventricular volumes were conducted with SPSS 21.0. This assumes an additive model of allele effects – a common assumption. We tested general linear models (GLMs) with outcome variables of ventricular volume at baseline [N = 737, one subject was excluded as mentioned in Section “Image Correction and Pre-Processing” under Longitudinal Ventricular Expansion Study (ADNI1)], difference between ventricular volume at baseline and volume after 1 year (in cubic mm, N = 623), difference between ventricular volume at baseline and volume after 2 years (in cubic mm, N = 481), and covarying for age, sex, and diagnosis (i.e., healthy elderly control, MCI, or AD). As the volume images were already normalized for overall brain size during the 9-parameter affine alignment, additional volume normalization was unnecessary. It would only have obscured the effect of faster relative expansion rates. As our expansion rates are computed over some snapshot in time - here only 1 or 2 years – the variation in rate of expansion, after being normalized for overall brain size, would have already had an effect on total volume prior to baseline image acquisition. In all analyses, the left and right ventricular volumes were tested separately and combined.
RESULTS
REGIONAL BRAIN VOLUMES ASSOCIATION STUDIES
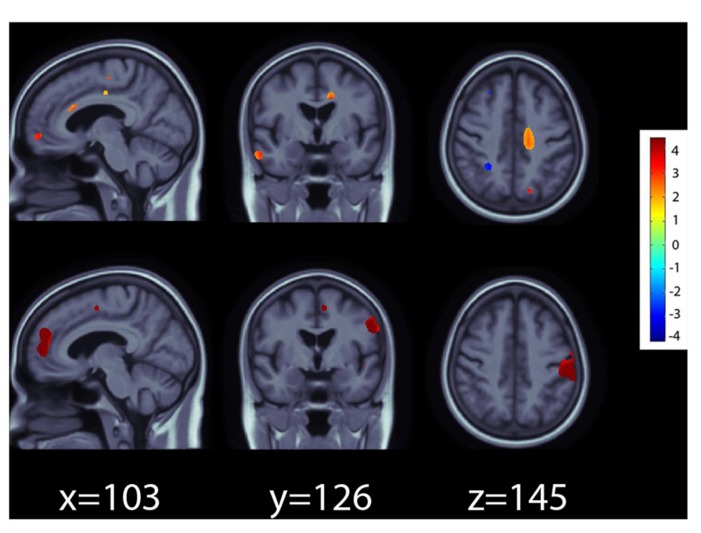
In the ADNI1 sample, the RASGRF2 polymorphism rs26907 predicted differences in regional brain volumes, after covarying for sex, age, and diagnosis, and after multiple comparisons correction at q = 0.05 (Figure 1, top panel). Larger volumes in the medial prefrontal cortices, cingulate gyrus, and right temporal lobe, were statistically related to carrying the minor A allele at the rs26907 locus. Regional volume differences associated with the minor allele ranged from 2 to 4%.
FIGURE 1.
Effects of the A allele at the rs26907 locus in the RASGRF2 gene, on regional brain volumes in the ADNI1 (top panel) and ADNI2 (bottom panel) cohorts. Positive beta values (warm colors) show regions where the minor A allele was associated with greater tissue volumes. Negative beta values (cool colors) show regions where the minor A allele was associated with lower tissue volumes. The color bar encodes the average percentage of volume difference relative to the template associated with the minor allele. Tests for associations are adjusted for age, sex, and diagnosis; maps are corrected for multiple comparisons with the searchlight false discovery rate (FDR) method at q = 0.05. Images are in radiological convention (left side of the brain shown on the right).
The RASGRF2 polymorphism rs26907 also predicted differences in regional brain volumes in the ADNI2 cohort, after covarying for sex, age, and diagnosis, and after multiple comparisons correction at q = 0.05 (Figure 1, bottom panel). As in ADNI1, the minor A allele was associated with larger volumes in the medial prefrontal cortices and cingulate gyrus. It also predicted larger volumes in the postcentral gyrus. Regional volume differences associated with the minor allele ranged from 3 to 5%.
In the ADNI1 cohort, the minor A allele was also associated with smaller volumes in the cerebellum, but this was detectable in the ADNI2 cohort only when a less conservative threshold for multiple comparisons correction at q = 0.10 was used (data not shown) possibly because the ADNI2 sample was smaller and afforded less statistical power to detect small gene effects on the brain.
LONGITUDINAL VENTRICULAR EXPANSION STUDY
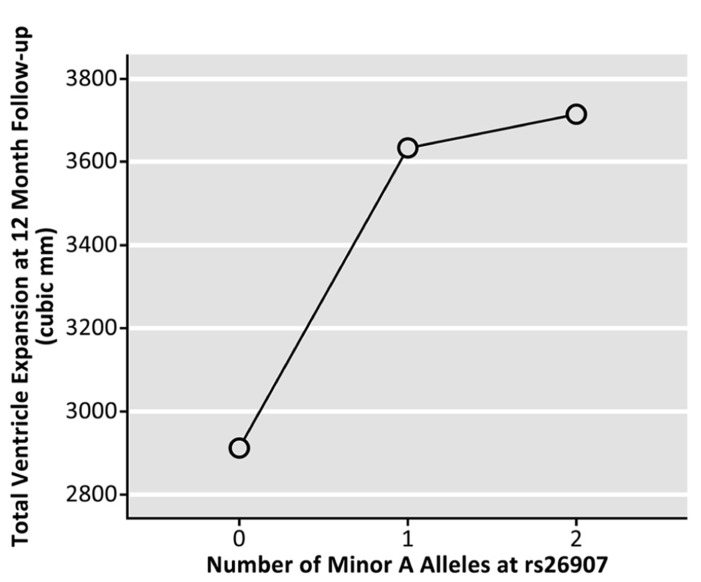
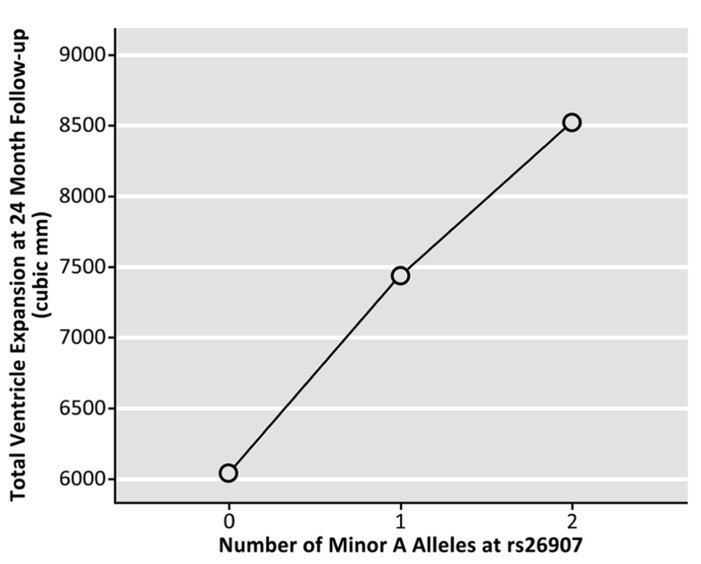
Genotype at the rs26907 locus was not significantly related to baseline volumes of the left (p = 0.173) or right (p = 0.629) lateral ventricles, after sex, age, and diagnosis were regressed out (N = 737). To determine if the allele was related to rates of brain tissue loss, we then examined the differences between ventricular volumes at baseline and volumes after 1 year (in cubic mm, N = 623), and 2 years (in cubic mm, N = 481), after covarying for sex, age, and diagnosis. Ventricular expansion rates relative to overall brain size showed a significant correlation with the number of A alleles at rs26907. After 1 year, carrying more minor A alleles was associated with greater rates of expansion in the left (p = 0.040) and right ventricle (p = 0.010), and with a greater overall rate of ventricular expansion (p = 0.017, Figure 2; N = 623). A similar pattern was observed at 2-year follow-up. Carrying more minor A alleles at the rs26907 locus was associated with faster ventricle expansion in the left (p = 0.030) and right ventricle (p = 0.028), and with greater total ventricle expansion (p = 0.024, Figure 3; N = 481).
FIGURE 2.
Effects of the A allele at the rs26907 locus in the RASGRF2 gene, on total longitudinal ventricular expansion in the ADNI1 cohort at 12-month follow-up (N = 623).
FIGURE 3.
Effects of the A allele at the rs26907 locus in the RASGRF2 gene, on total longitudinal ventricular expansion in the ADNI1 cohort at 24-month follow-up (N = 481).
DISCUSSION
This study is the first to report an association between brain structure, longitudinal ventricular expansion, and a common single nucleotide polymorphism (rs26907) associated with alcohol intake (Schumann et al., 2011; Stacey et al., 2012) in the gene encoding ras-specific guanine nucleotide releasing factor 2. The variant previously associated with lower alcohol consumption predicted larger cortical volumes in the elderly at baseline. We replicated these findings in an independent elderly cohort despite a smaller sample size with lower statistical power, suggesting that the association of this variant with larger cortical volumes in the elderly may be independent of age, sex, or disease status. The same allele associated with reduced alcohol intake also predicted faster longitudinal ventricular expansion at 1- and 2-year follow up.
The rs26907 polymorphism in RASGRF2 is associated with alcohol intake (Schumann et al., 2011; Stacey et al., 2012), and this study further links it to regional brain volumes, in several systems involved in alcohol reward – where cellular and molecular processes including dopamine release are also affected by this gene. It may seem surprising that the rs26907 allele associated with reduced alcohol intake – which is the less common one – predicts larger regional cortical volumes in the elderly at baseline but faster ventricular expansion rates over time, reflecting an accumulation of brain tissue loss globally, throughout multiple brain regions. In fact, baseline regional cortical volumes are likely to depend on factors unrelated to neurodegeneration, such as neurodevelopmental differences. Even so, if the lateral ventricles expand approximately linearly with age in healthy elderly individuals (Blatter et al., 1995), accelerated rates of ventricular expansion may be indicators or predictors of degenerative brain disorders (Apostolova et al., 2012).
The rs26907 polymorphism in RASGRF2 was associated with regional brain volumes and longitudinal ventricular expansion, but we are unable to provide mechanistic evidence for how this variant might affect brain structure. Rs26907 is an intronic SNP that is not linked with any known changes in gene function. SNPs in non-coding regions can affect gene splicing, and RASGRF2 may have one alternative splice variant (Feig, 2011). This polymorphism may affect transcription factor binding, mRNA degradation, and other molecular genetic processes that may relate to brain volumes. Data from the encyclopedia of DNA elements (“ENCODE”)3 as presented in HaploReg4, suggest that rs26907 alters the Rad21 regulatory motif. Since Rad21 is a transcription factor involved in apoptosis (Pati et al., 2002), programmed cell death represents a plausible mechanism by which this variant could affect brain volumes and rates of ventricular expansion, which indicate neuronal loss. Although we do not know the precise mechanisms that relate this polymorphism to brain structure, this particular variant may drive the observed associations, as there are no SNPs in high linkage disequilibrium (as defined by r2 > 0.8) with rs26907 in the 1000 Genomes Caucasian (CEU) panel5.
An important limitation of this study is that no alcohol consumption measures were available in these samples (although the cohorts excluded alcohol abusers). We cannot establish a relationship between the rs26907 variant and alcohol intake, or among brain volumes, longitudinal ventricular expansion, and alcohol intake in these particular cohorts. Some of these associations have been reported in other cohorts. Our experimental design does reveal whether the variant of interest directly affects the brain or just modifies drinking behaviors that affect brain structure and the rate of brain atrophy. This would be an interesting target of study. Nonetheless, the functional imaging literature may shed light on our results. A haplotype containing rs26907 as well as other SNPs in RASGRF2 was associated with ventral striatal activity during reward anticipation in healthy 14-year old boys, suggesting a hypersensitive reward system (Stacey et al., 2012). Follow-up at age 16 showed an association between this haplotype and number of drinking episodes (Stacey et al., 2012). The existence of differences, first in brain activation and later in drinking behaviors, between young carriers of risk alleles in RASGRF2 and carriers of protective alleles, suggests that this gene may directly affect the developing brain. This may result in different drinking patterns later in life. Along with other behaviors, this may further affect brain structure and function throughout the lifespan.
RASGRF2 regulates alcohol-induced reinforcement and alcohol preference is associated with whole-brain RASGRF2 mRNA expression (Stacey et al., 2012). During neurodevelopment, carriers of the rs26907 allele associated with reduced drinking may have developed neurological circuits conferring less sensitivity to alcohol reward. This could be one factor associated with their volume differences in the medial prefrontal cortex, which plays a role in the mesolimbic reward system (Baltazar et al., 2013). As a result, these individuals may have been more likely to abstain from drinking throughout their lives. In our study, this may be linked to the faster lateral ventricle expansion rates (typically associated with neurodegeneration across the brain) in elderly carriers of the allele. Elderly individuals who abstain from drinking have higher rates of brain abnormalities and infarcts than moderate drinkers (Mukamal et al., 2001; den Heijer et al., 2004; Mukamal, 2004). This framework remains speculative, and future studies relating alcohol consumption data to brain imaging and genetic data, should clarify the direction of these relationships. Even so, our findings, combined with earlier functional imaging investigations of the same gene (Stacey et al., 2012) point to interdependent and reciprocal interactions between genes, brain, and drinking behaviors.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
AUTHOR CONTRIBUTIONS
Florence F. Roussotte formulated the hypothesis, conducted the analyses, and wrote the manuscript. Boris A. Gutman developed the novel Linear Discriminant Analysis (LDA)-based approach for segmenting the lateral ventricles, described in Section Longitudinal Ventricular Expansion Study. Derrek P. Hibar helped with data analysis and interpretation, and helped revise the manuscript. Neda Jahanshad helped with data analysis and interpretation, and helped revise the manuscript. Sarah K. Madsen helped with data analysis and interpretation, and helped revise the manuscript. Clifford R. Jack helped revise the manuscript. Michael W. Weiner is the principal investigator of the ADNI initiative and helped revise the manuscript. Paul M. Thompson is the principal investigator of this study, and overviewed all phases of the investigation and manuscript preparation.
Acknowledgments
Florence F. Roussotte was supported, in part, by a postdoctoral fellowship from the A. P. Giannini Foundation. Additional support was provided by NSF grant DGE-0707424 to Derrek P. Hibar. This work was additionally supported by National Institute of Health grants (R01 MH097268, R01 AG040060) to Paul M. Thompson. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Footnotes
Data used preparating this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but most of them did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
REFERENCES
- Apostolova L. G., Green A. E., Babakchanian S., Hwang K. S., Chou Y. Y., Toga A. W., et al. (2012). Hippocampal atrophy and ventricular enlargement in normal aging, mild cognitive impairment (MCI), and Alzheimer Disease. Alzheimer Dis. Assoc. Disord. 26 17–27 10.1097/WAD.0b013e3182163b62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltazar R. M., Coolen L. M., Webb I. C. (2013). Diurnal rhythms in neural activation in the mesolimbic reward system: critical role of the medial prefrontal cortex. Eur. J. Neurosci. 38 2319–2327 10.1111/ejn.12224 [DOI] [PubMed] [Google Scholar]
- Blaine S., Claus E., Harlaar N., Hutchison K. (2013). TABR1 genotypes predict fMRI response to alcohol cues and level of alcohol dependence. Alcohol. Clin. Exp. Res. 37 E125–E130 10.1111/j.1530-0277.2012.01923.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter D. D., Bigler E. D., Gale S. D., Johnson S. C., Anderson C. V., Burnett B. M., et al. (1995). Quantitative volumetric analysis of brain MR: normative database spanning 5 decades of life. AJNR Am. J. Neuroradiol. 16 241–251 [PMC free article] [PubMed] [Google Scholar]
- Chou Y. Y., Lepore N., Avedissian C., Madsen S. K., Parikshak N., Hua X., et al. (2009). Mapping correlations between ventricular expansion and CSF amyloid and tau biomarkers in 240 subjects with Alzheimer’s disease, mild cognitive impairment and elderly controls. Neuroimage 46 394–410 10.1016/j.neuroimage.2009.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y. Y., Lepore N., de Zubicaray G. I., Carmichael O. T., Becker J. T., Toga A. W., et al. (2008). Automated ventricular mapping with multi-atlas fluid image alignment reveals genetic effects in Alzheimer’s disease. Neuroimage 40 615–630 10.1016/j.neuroimage.2007.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M. K., Worsley K. J., Paus T., Cherif C., Collins D. L., Giedd J. N., et al. (2001). A unified statistical approach to deformation-based morphometry. Neuroimage 14 595–606 10.1006/nimg.2001.0862 [DOI] [PubMed] [Google Scholar]
- Coffey C. E., Ratcliff G., Saxton J. A., Bryan R. N., Fried L. P., Lucke J. F. (2001). Cognitive correlates of human brain aging: a quantitative magnetic resonance imaging investigation. J. Neuropsychiatry Clin. Neurosci. 13 471–485 10.1176/appi.neuropsych.13.4.471 [DOI] [PubMed] [Google Scholar]
- Collins D. L., Neelin P., Peters T. M., Evans A. C. (1994). Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J. Comput. Assist. Tomogr. 18 192–205 10.1097/00004728-199403000-00005 [DOI] [PubMed] [Google Scholar]
- den Heijer T., Vermeer S. E., van Dijk E. J., Prins N. D., Koudstaal P. J., van Duijn C. M., et al. (2004). Alcohol intake in relation to brain magnetic resonance imaging findings in older persons without dementia. Am. J. Clin. Nutr. 80 992–997 [DOI] [PubMed] [Google Scholar]
- ENIGMA2 (2012). “ENIGMA2 Genetics support team, ENIGMA2 1KGP cookbook (v3) [Online],” in Enhancing Neuroimaging Genetics Through Meta-Analysis (ENIGMA) Consortium. Available at: http://enigma.ini.usc.edu/wp-content/uploads/2012/07/ENIGMA2_1KGP_cookbook_v3.pdf [Google Scholar]
- Fasano S., Brambilla R. (2011). Ras-ERK signaling in behavior: old questions and new perspectives. Front. Behav. Neurosci. 5:79 10.3389/fnbeh.2011.00079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L. A. (2011). Regulation of neuronal function by Ras-GRF exchange factors. Genes Cancer 2 306–319 10.1177/1947601911408077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Medarde A., Santos E. (2011). The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim. Biophys. Acta 1815 170–188 10.1016/j.bbcan.2010.11.001 [DOI] [PubMed] [Google Scholar]
- Ferrarini L., Palm W. M., Olofsen H., van der Landen R., van Buchem M. A., Reiber J. H., et al. (2008). Ventricular shape biomarkers for Alzheimer’s disease in clinical MR images. Magn. Reson. Med. 59 260–267 10.1002/mrm.21471 [DOI] [PubMed] [Google Scholar]
- Freeborough P. A., Fox N. C. (1998). Modeling brain deformations in Alzheimer disease by fluid registration of serial 3D MR images. J. Comput. Assist. Tomogr. 22 838–843 10.1097/00004728-199809000-00031 [DOI] [PubMed] [Google Scholar]
- Girault J. A., Valjent E., Caboche J., Herve D. (2007). ERK2: a logical AND gate critical for drug-induced plasticity? Curr. Opin. Pharmacol. 7 77–85 10.1016/j.coph.2006.08.012 [DOI] [PubMed] [Google Scholar]
- Gutman B. A., Hua X., Rajagopalan P., Chou Y. Y., Wang Y., Yanovsky I., et al. (2013). Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage 70 386–401 10.1016/j.neuroimage.2012.12.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman B. A., Wang Y., Thompson P. M., Rajagopalan P., Toga A. W. (2012). “Shape matching with medial curves and 1-D group-wise registration,” in Biomedical Imaging (ISBI), 9th IEEE International Symposium Barcelona: 10.1109/ISBI.2012.6235648 [DOI] [Google Scholar]
- Hibar D. P., Stein J. L., Ryles A. B., Kohannim O., Jahanshad N., Medland S. E., et al. (2013). Genome-wide association identifies genetic variants associated with lentiform nucleus volume in N = 1345 young and elderly subjects. Brain Imaging Behav. 7 102–115 10.1007/s11682-012-9199-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Hibar D. P., Ching C. R., Boyle C. P., Rajagopalan P., Gutman B. A., et al. (2013). Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage 66 648–661 10.1016/j.neuroimage.2012.10.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Leow A. D., Parikshak N., Lee S., Chiang M. C., Toga A. W., et al. (2008a). Tensor-based morphometry as a neuroimaging biomarker for Alzheimer’s disease: an MRI study of 676 AD, MCI, and normal subjects. Neuroimage 43 458–469 10.1016/j.neuroimage.2008.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X., Leow A. D., Lee S., Klunder A. D., Toga A. W., Lepore N., et al. (2008b). 3D characterization of brain atrophy in Alzheimer’s disease and mild cognitive impairment using tensor-based morphometry. Neuroimage 41 19–34 10.1016/j.neuroimage.2008.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hvidtfeldt U. A., Tolstrup J. S., Jakobsen M. U., Heitmann B. L., Gronbaek M., O’Reilly E., et al. (2010). Alcohol intake and risk of coronary heart disease in younger, middle-aged, and older adults. Circulation 121 1589–1597 10.1161/CIRCULATIONAHA.109.887513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack C. R., Jr., Bernstein M. A., Fox N. C., Thompson P., Alexander G., Harvey D., et al. (2008). The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27 685–691 10.1002/jmri.21049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S. X., Feig L. A. (2010). Long-term potentiation in the CA1 hippocampus induced by NR2A subunit-containing NMDA glutamate receptors is mediated by Ras-GRF2/Erk map kinase signaling. PLoS ONE 5:e11732 10.1371/journal.pone.0011732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J., Czanner S., Greve D., Haley E., van der Kouwe A., Gollub R., et al. (2006). Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. Neuroimage 30 436–443 10.1016/j.neuroimage.2005.09.046 [DOI] [PubMed] [Google Scholar]
- Lander E. S., Schork N. J. (1994). Genetic dissection of complex traits. Science 265 2037–2048 10.1126/science.8091226 [DOI] [PubMed] [Google Scholar]
- Langers D. R., Jansen J. F., Backes W. H. (2007). Enhanced signal detection in neuroimaging by means of regional control of the global false discovery rate. Neuroimage 38 43–56 10.1016/j.neuroimage.2007.07.031 [DOI] [PubMed] [Google Scholar]
- Leow A., Huang S. C., Geng A., Becker J., Davis S., Toga A., et al. (2005). Inverse consistent mapping in 3D deformable image registration: its construction and statistical properties. Inf. Process Med. Imaging 19 493–503 10.1007/11505730_41 [DOI] [PubMed] [Google Scholar]
- Leow A. D., Klunder A. D., Jack C. R., Jr., Toga A. W., Dale A. M., Bernstein M. A., et al. (2006). Longitudinal stability of MRI for mapping brain change using tensor-based morphometry. Neuroimage 31 627–640 10.1016/j.neuroimage.2005.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow A. D., Yanovsky I., Chiang M. C., Lee A. D., Klunder A. D., Lu A., et al. (2007). Statistical properties of Jacobian maps and the realization of unbiased large-deformation nonlinear image registration. IEEE Trans. Med. Imaging 26 822–832 10.1109/TMI.2007.892646 [DOI] [PubMed] [Google Scholar]
- Madsen S. K., Gutman B. A., Joshi S. H., Toga A. W., Jack C. R. J., Weiner M. W., et al. (2013). “Mapping dynamic changes in ventricular volume onto baseline cortical surface maps in normal aging, MCI, and Alzheimer’s disease,” in MICCAI MBIA Workshop Nagoya: 10.1007/978-3-319-02126-3_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazziotta J., Toga A., Evans A., Fox P., Lancaster J., Zilles K., et al. (2001). A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. B Biol. Sci. 356 1293–1322 10.1098/rstb.2001.0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J. L., Nyman E., Loukola A., Rose R. J., Kaprio J., Dick D. M. (2013). The association between DRD2/ANKK1 and genetically informed measures of alcohol use and problems. Addict. Biol. 18 523–536 10.1111/j.1369-1600.2012.00490.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukamal K. J. (2004). Alcohol consumption and abnormalities of brain structure and vasculature. Am. J. Geriatr. Cardiol. 13 22–28 10.1111/j.1076-7460.2004.01334.x [DOI] [PubMed] [Google Scholar]
- Mukamal K. J., Chung H., Jenny N. S., Kuller L. H., Longstreth W. T., Jr., Mittleman M. A., et al. (2006). Alcohol consumption and risk of coronary heart disease in older adults: the Cardiovascular Health Study. J. Am. Geriatr. Soc. 54 30–37 10.1111/j.1532-5415.2005.00561.x [DOI] [PubMed] [Google Scholar]
- Mukamal K. J., Longstreth W. T., Jr., Mittleman M. A., Crum R. M., Siscovick D. S. (2001). Alcohol consumption and subclinical findings on magnetic resonance imaging of the brain in older adults: the Cardiovascular Health Study. Stroke 32 1939–1946 10.1161/hs0901.095723 [DOI] [PubMed] [Google Scholar]
- Olfson E., Bierut L. J. (2012). Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol. Clin. Exp. Res. 36 2086–2094 10.1111/j.1530-0277.2012.01843.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati D., Zhang N., Plon S. E. (2002). Linking sister chromatid cohesion and apoptosis: role of Rad21. Mol. Cell. Biol. 22 8267–8277 10.1128/MCB.22.23.8267-8277.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddle W. R., Li R., Fitzpatrick J. M., DonLevy S. C., Dawant B. M., Price R. R. (2004). Characterizing changes in MR images with color-coded Jacobians. Magn. Reson. Imaging 22 769–777 10.1016/j.mri.2004.01.078 [DOI] [PubMed] [Google Scholar]
- Rimm E. B., Williams P., Fosher K., Criqui M., Stampfer M. J. (1999). Moderate alcohol intake and lower risk of coronary heart disease: meta-analysis of effects on lipids and haemostatic factors. BMJ 319 1523–1528 10.1136/bmj.319.7224.1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussotte F. F., Jahanshad N., Hibar D. P., Sowell E. R., Kohannim O., Barysheva M., et al. (2013). A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: replication in elderly and young populations. Hum. Brain Mapp. 10.1002/hbm.22247 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann G., Coin L. J., Lourdusamy A., Charoen P., Berger K. H., Stacey D., et al. (2011). Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. U.S.A. 108 7119–7124 10.1073/pnas.1017288108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sled J. G., Zijdenbos A. P., Evans A. C. (1998). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging 17 87–97 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- Stacey D., Bilbao A., Maroteaux M., Jia T., Easton A. C., Longueville S., et al. (2012). RASGRF2 regulates alcohol-induced reinforcement by influencing mesolimbic dopamine neuron activity and dopamine release. Proc. Natl. Acad. Sci. U.S.A. 109 21128–21133 10.1073/pnas.1211844110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. L., Hibar D. P., Madsen S. K., Khamis M., McMahon K. L., de Zubicaray G. I., et al. (2011). Discovery and replication of dopamine-related gene effects on caudate volume in young and elderly populations (N=1198) using genome-wide search. Mol. Psychiatry 16 927–937 881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. L., Hua X., Morra J. H., Lee S., Hibar D. P., Ho A. J., et al. (2010). Genome-wide analysis reveals novel genes influencing temporal lobe structure with relevance to neurodegeneration in Alzheimer’s disease. Neuroimage 51 542–554 10.1016/j.neuroimage.2010.02.068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein J. L., Medland S. E., Vasquez A. A., Hibar D. P., Senstad R. E., Winkler A. M., et al. (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nat. Genet. 44 552–561 10.1038/ng.2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P. M., Giedd J. N., Woods R. P., MacDonald D., Evans A. C., Toga A. W. (2000). Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature 404 190–193 10.1038/35004593 [DOI] [PubMed] [Google Scholar]
- Tian X., Gotoh T., Tsuji K., Lo E. H., Huang S., Feig L. A. (2004). Developmentally regulated role for Ras-GRFs in coupling NMDA glutamate receptors to Ras, Erk and CREB. EMBO J. 23 1567–1575 10.1038/sj.emboj.7600151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich P. A., Wang H., Pluta J., Das S. R., Craige C., Avants B. B., et al. (2010). Nearly automatic segmentation of hippocampal subfields in in vivo focal T2-weighted MRI. Neuroimage 53 1208–1224 10.1016/j.neuroimage.2010.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]