Abstract
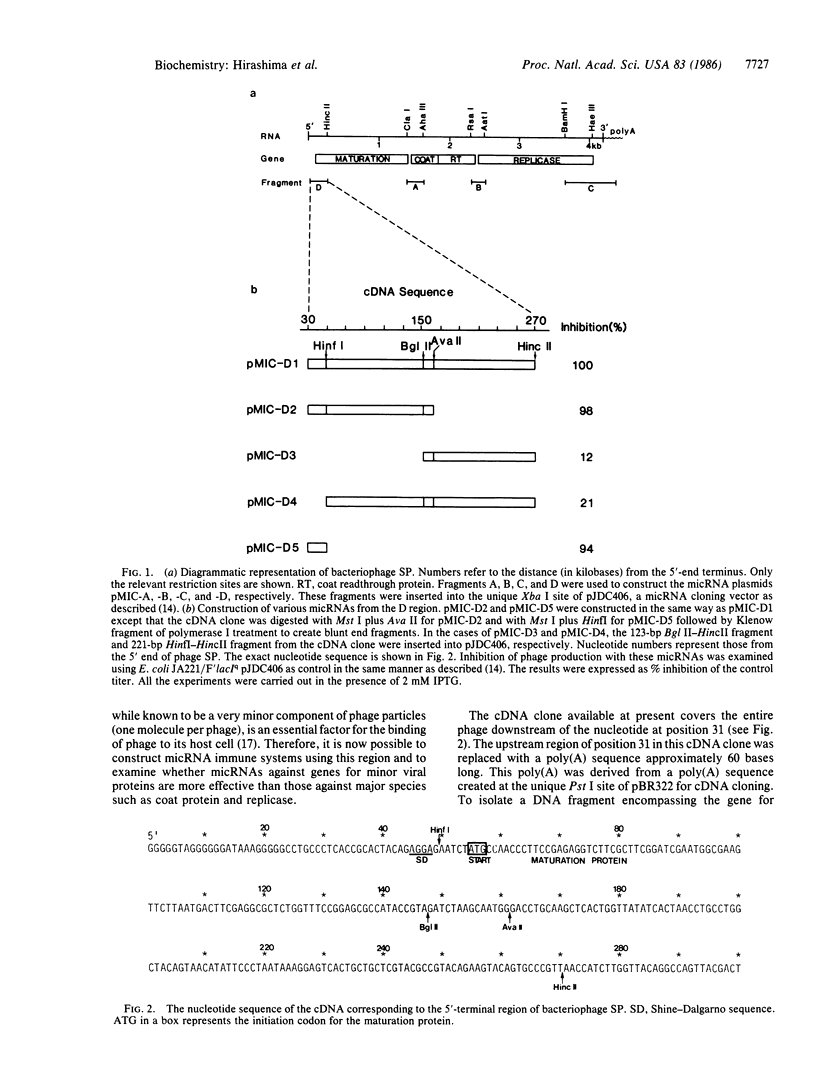
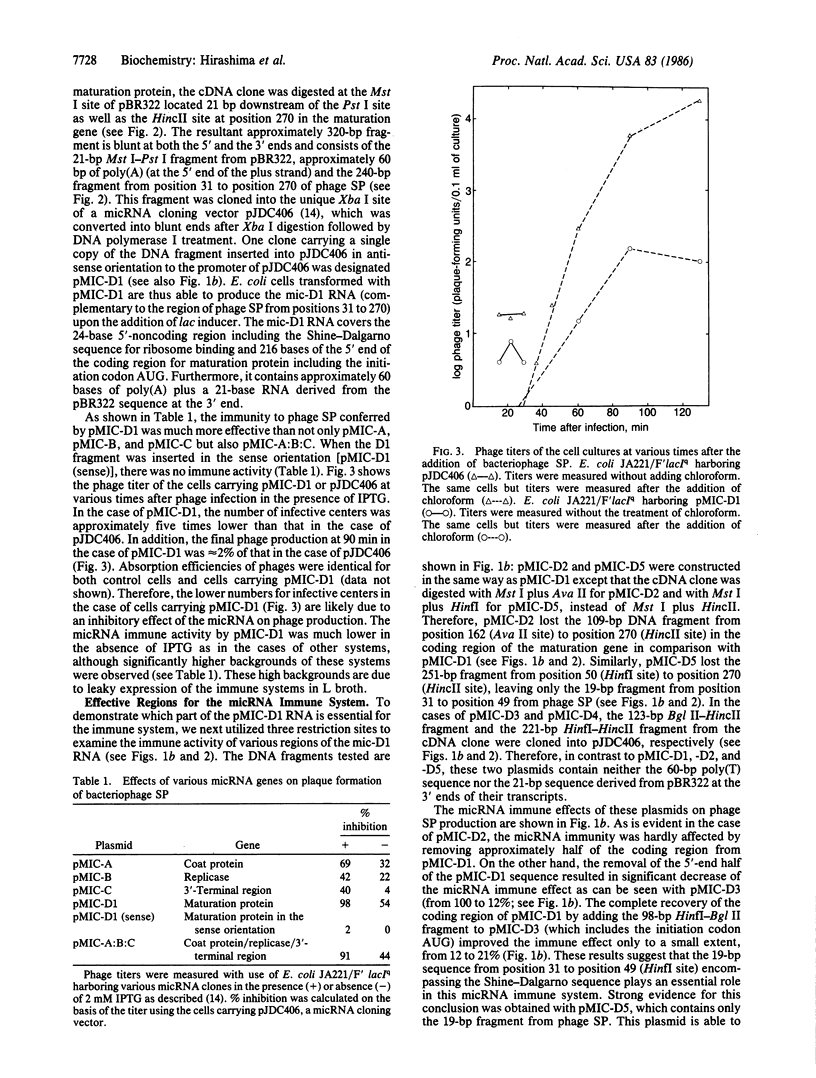
Creation of an artificial mRNA-interfering complementary RNA (micRNA) immune system, utilizing anti-sense RNAs to inhibit viral gene expression, has been shown to be an effective way to prevent viral infection. In the RNA coliphage SP, the gene for the maturation protein was found to be the best target for this type of immune system; mRNA-interfering complementary RNAs specific to the genes for coat protein and replicase were less effective in preventing infection. The greatest inhibitory effect was observed with a 240-base sequence encompassing the 24-base noncoding region of the maturation gene plus the 216-base coding sequence. Significantly, even a 19-base sequence covering only the Shine-Dalgarno sequence (ribosome-binding region) without the coding region exerted a strong inhibitory effect on phage proliferation. In contrast to the highly specific action against phage SP exhibited by the longer mRNA-interfering complementary RNA, the specificity with the shorter mRNA-interfering complementary RNA was broadened to phages Q beta and GA as well as SP, all of which are classified in the different groups of RNA coliphages. Therefore, this type of anti-viral reagent may be designed to have a particular breadth of specificity, thus increasing its value in various research and possibly clinical applications.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Coleman J., Green P. J., Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984 Jun;37(2):429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- Coleman J., Hirashima A., Inokuchi Y., Green P. J., Inouye M. A novel immune system against bacteriophage infection using complementary RNA (micRNA). Nature. 1985 Jun 13;315(6020):601–603. doi: 10.1038/315601a0. [DOI] [PubMed] [Google Scholar]
- Furuse K., Hirashima A., Harigai H., Ando A., Watanabe K., Kurosawa K., Inokuchi Y., Watanabe I. Grouping of RNA coliphages based on analysis of the sizes of their RNAs and proteins. Virology. 1979 Sep;97(2):328–341. doi: 10.1016/0042-6822(79)90344-1. [DOI] [PubMed] [Google Scholar]
- Green P. J., Mitchell C., Inouye M. Effect of mic gene structure on repressor activity in the OmpA system. Biochimie. 1985 Jul-Aug;67(7-8):763–767. doi: 10.1016/s0300-9084(85)80165-6. [DOI] [PubMed] [Google Scholar]
- Green P. J., Pines O., Inouye M. The role of antisense RNA in gene regulation. Annu Rev Biochem. 1986;55:569–597. doi: 10.1146/annurev.bi.55.070186.003033. [DOI] [PubMed] [Google Scholar]
- Harland R., Weintraub H. Translation of mRNA injected into Xenopus oocytes is specifically inhibited by antisense RNA. J Cell Biol. 1985 Sep;101(3):1094–1099. doi: 10.1083/jcb.101.3.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inokuchi Y., Takahashi R., Hirose T., Inayama S., Jacobson A. B., Hirashima A. The complete nucleotide sequence of the group II RNA coliphage GA. J Biochem. 1986 Apr;99(4):1169–1180. doi: 10.1093/oxfordjournals.jbchem.a135580. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Constitutive and conditional suppression of exogenous and endogenous genes by anti-sense RNA. Science. 1985 Jul 26;229(4711):345–352. doi: 10.1126/science.2990048. [DOI] [PubMed] [Google Scholar]
- Izant J. G., Weintraub H. Inhibition of thymidine kinase gene expression by anti-sense RNA: a molecular approach to genetic analysis. Cell. 1984 Apr;36(4):1007–1015. doi: 10.1016/0092-8674(84)90050-3. [DOI] [PubMed] [Google Scholar]
- Kim S. K., Wold B. J. Stable reduction of thymidine kinase activity in cells expressing high levels of anti-sense RNA. Cell. 1985 Aug;42(1):129–138. doi: 10.1016/s0092-8674(85)80108-2. [DOI] [PubMed] [Google Scholar]
- Melton D. A. Injected anti-sense RNAs specifically block messenger RNA translation in vivo. Proc Natl Acad Sci U S A. 1985 Jan;82(1):144–148. doi: 10.1073/pnas.82.1.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A. 1984 Apr;81(7):1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Masui Y., Inouye M. Use of a lac promoter-operator fragment as a transcriptional control switch for expression of the constitutive lpp gene in Escherichia coli. J Mol Appl Genet. 1982;1(4):289–299. [PubMed] [Google Scholar]
- Pestka S., Daugherty B. L., Jung V., Hotta K., Pestka R. K. Anti-mRNA: specific inhibition of translation of single mRNA molecules. Proc Natl Acad Sci U S A. 1984 Dec;81(23):7525–7528. doi: 10.1073/pnas.81.23.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg U. B., Preiss A., Seifert E., Jäckle H., Knipple D. C. Production of phenocopies by Krüppel antisense RNA injection into Drosophila embryos. Nature. 1985 Feb 21;313(6004):703–706. doi: 10.1038/313703a0. [DOI] [PubMed] [Google Scholar]
- Rubenstein J. L., Nicolas J. F., Jacob F. L'ARN non sens (nsARN): un outil pour inactiver spécifiquement l'expression d'un gène donné in vivo. C R Acad Sci III. 1984;299(8):271–274. [PubMed] [Google Scholar]
- Weissmann C., Billeter M. A., Goodman H. M., Hindley J., Weber H. Structure and function of phage RNA. Annu Rev Biochem. 1973;42:303–328. doi: 10.1146/annurev.bi.42.070173.001511. [DOI] [PubMed] [Google Scholar]


