Abstract
Model-based Analysis of ChIP-seq (MACS) is a computational algorithm that identifies genome-wide locations of transcription/chromatin factor binding or histone modification from ChIP-seq data. MACS consists of four steps: removing redundant reads, adjusting read position, calculating peak enrichment, and estimating the empirical false discovery rate. In this protocol, we provide a detailed demonstration of how to install MACS and how to use it to analyze three common types of ChIP-seq datasets with different characteristics: the sequence-specific transcription factor FoxA1, the histone modification mark H3K4me3 with sharp enrichment, and the H3K36me3 mark with broad enrichment. We also explain how to interpret and visualize the results of MACS analyses. The algorithm requires approximately 3 GB of RAM and 1.5 hours of computing time to analyze a ChIP-seq dataset containing 30 million reads, an estimate that increases with sequence coverage. MACS is open-source and is available from http://liulab.dfci.harvard.edu/MACS.
Keywords: MACS, ChIP-seq, peak calling, transcription factor, histone modification
INTRODUCTION
Researchers have widely used the process of chromatin immunoprecipitation (ChIP) coupled with high-throughput sequencing (ChIP-seq)1 to map transcription factor binding sites and histone modification status on a genome-wide scale2. ChIP comprises a few basic steps: crosslinking a protein to chromatin, shearing the chromatin, using a specific antibody to precipitate the protein of interest with its associated DNA, and purifying the associated DNA fragments3–6. ChIP usually yields several to a few hundred nanograms of DNA as 75- to 300-bp fragments surrounding transcription factor binding sites or histone mark locations. High-throughput sequencing often generates tens to hundreds of millions of 25- to 75-bp sequences (also called short reads) from the 5’ ends of ChIP-DNA fragments.
ChIP-seq data analysis typically begins by mapping the short reads back to a reference genome. Although many mapped reads are dispersed throughout the genome, others are found in clusters constituting read-enriched regions, which represent the locations of transcription factor binding or histone marks. For the majority of transcription factors and several histone modifications such as H3K4me3, ChIP-seq reads are often concentrated in narrow peaks of a few hundred base pairs. However, for some transcription factors such as RNA polymerase II and other histone modifications such as H3K36me3, read enrichment regions can be broad, spanning up to tens of thousands of base pairs. Factors such as GC content, read mappability, DNA repeats, copy number variations, and local chromatin structure can influence read distribution at different locations of the genome7–11. Therefore, sequencing control samples of sonicated chromatin is recommended as an additional measure to better eliminate background biases so as to reliably identify read-enriched regions obtained from ChIP-seq10,12.
Model-based Analysis of ChIP-seq (MACS) is a computational method that was designed to identify read-enriched regions from ChIP-seq data. According to Web of Science, since its first publication in 200813, MACS has been cited by more than 300 studies, including many influential works14–19. The MACS Google group contains approximately 3,000 active users. Over the years, MACS has continuously benefited from user feedback and contribution. We continue to add new functionality, fixing bugs and optimizing the algorithm based on user requests. In this protocol, we share the insights gained from collected user experience and demonstrate how to apply the latest stable version of MACS (1.4.2) to publicly available ChIP-seq data20 on a local computer. MACS is also available at the web-based ChIP-seq analysis portal Cistrome21, which provides a complete workflow for ChIP-seq and downstream analysis without the need for local installation and configuration.
Overview of the MACS algorithm
The MACS workflow is summarized in Figure 1. In the following sections, we detail the key steps of the MACS algorithm.
Figure 1.
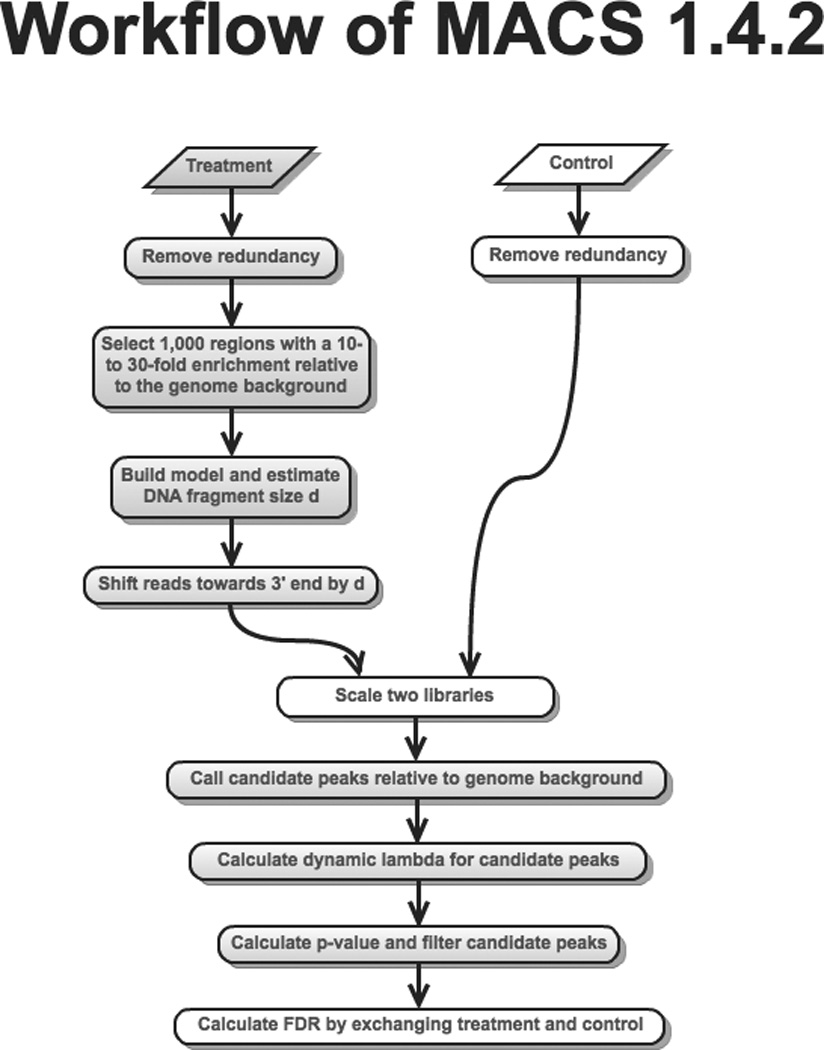
Workflow of MACS 1.4.2. If the control sample is missing, then the steps shown in white boxes will be skipped (Remove redundancy of the control sample, Scale two libraries, and Calculate FDR by exchanging treatment and control).
Removing redundant reads
Overamplification of ChIP-DNA by polymerase chain reaction (PCR) may cause the same original DNA fragment to be sequenced repeatedly, especially at a high sequencing depth. Our experience indicates that removing these redundant reads can yield more reliable peak calls for downstream analysis. Therefore, MACS removes redundant reads based on a user-specified parameter without changing the input files containing the mapped ChIP-seq reads and control reads. By default, MACS retains no more than one read per genomic location.
Adjusting read position based on fragment size distribution
ChIP-DNA fragments often encompass the minimal DNA sequence containing protein-DNA interactions, and the sequencer is equally likely to sequence the 5’ end of either strand. As a result, reads that map to the positive and negative strands often appear to the left and right of the protein-DNA interaction location, which leads to a bimodal enrichment pattern flanking the precise interaction location. It would be desirable to extend ChIP-seq reads to represent the original ChIP-DNA fragments, which necessitates the need to estimate the distribution of fragment size.
To estimate fragment size, denoted d, MACS first slides a window with a width of roughly twice the size of the sheared chromatin to identify regions of moderate enrichment. To avoid the influence of extremely enriched regions due to artifacts in PCR amplification or repetitive elements, MACS randomly samples 1,000 regions each having a 10- to 30-fold enrichment relative to the genome background as model peaks. For each peak, MACS separates reads mapped to the positive and negative strands and then calculates the reads’ respective mode positions. The midpoint between the positive and negative modes is then used to align all the reads belonging to model peaks. The alignment generates a bimodal pattern: most reads from the positive strand appear on the left, and most reads from the negative strand appear on the right. The distance between bimodal summits yields the estimated DNA fragment size d, and all reads are extended in the 3’ direction until d is obtained.
In the majority of cases, the above procedure, which is referred to as building the peak model, provides a reasonable estimate of fragment size. However, excessive chromatin shearing or transcription factors with broad enrichment may cause MACS to estimate a value of that is too small. When this happens (e.g., when d< 60 bp), we recommend that users rerun MACS with a specified distance based on the size selection in the sequencing library preparation. In addition, the value of d should be similar for multiple ChIP-seq samples corresponding to the same factor in the same study to ensure comparable downstream analyses among samples.
Calculate peak enrichment using local background normalization
Based on the position-adjusted reads, MACS slides a window of size 2d across the genome to identify regions that are significantly enriched relative to the genome background. Overlapping significant windows are then merged to form candidate regions for further analysis. Because many factors influence the local read enrichment distribution, MACS models the number of reads from a genomic region as a Poisson distribution with dynamic parameter λlocal. That is, instead of using a constant value of λ, λlocal values are allowed to vary along the genome. Specifically, the value of λlocal for a specific region is defined as max(λBG, [λregion, λlk], λ5k, λ10k), where λBG is a constant estimated from the genome background, λregion is estimated from the candidate region under consideration in the control sample, and the remaining λx are estimated from an x-bp window centered at the candidate region in the control sample. When ChIP-seq and control samples are sequenced at different depths, MACS either linearly scales down the larger sample (default behavior) or scales up the smaller sample. For example, after removing redundant reads, if the total number of reads in the control sample is greater than the number of reads obtained from ChIP-seq by a factor of r (r > 1), then when calculating the p-value, λlocal will be divided by r by default. When a control sample is not available, λlocal is calculated from the ChIP-seq sample, excluding λregion and λlk. Based on λlocal, MACS assigns every candidate region an enrichment p-value, and those passing a user-defined threshold (the default is 10−5) are reported as the final peaks.
Estimating the empirical false discovery rate by exchanging ChIP-seq and control samples
When a control sample is available, MACS can also estimate an empirical false discovery rate (FDR) for every peak by exchanging the ChIP-seq and control samples and identifying peaks in the control sample using the same set of parameters used for the ChIP-seq sample. Because the control sample should not exhibit read enrichment, any such peaks found by MACS can be regarded as false positives. For a particular p-value threshold, the empirical FDR is then calculated as the number of control peaks passing the threshold divided by the number of ChIP-seq peaks passing the same threshold.
Comparison with existing methods
Various methods that incorporate different strategies have been proposed for analyzing ChIP-seq data. For example, to find peak candidates, many methods including SISSRs22, USeq12, and MACS13 identify clusters consisting of reads that overlap or are located within a fixed distance. Alternatively, CisGenome23 and SICER24 use non-overlapping sliding windows to identify candidate regions. To identify binding sites more accurately, MACS extends reads in the 3’ direction until the estimated DNA fragment size is reached, a strategy also used by SICER24. The majority of methods utilize a background or null model to assign a significance score to each peak region identified by the method. PeakSeq8 models the number of reads mapped to a peak region using a binomial distribution. CisGenome23 applies a negative binomial distribution to model windows of low read count. MACS employs a Poisson distribution to accurately approximate a binomial distribution and calculates dynamic Poisson parameters for each region to obtain a distribution having more flexibility than the negative binomial distribution. FindPeaks25 works differently by implementing a Monte Carlo simulation to calculate the likelihood of observing a peak of a given height. To calculate an empirical FDR, methods such as USeq12 and QuEST26 identify false positive peaks by considering two inputs constructed from the control sample instead of exchanging the ChIP-seq and control samples as proposed by MACS. Previous reviews and systematic comparison studies provide further details regarding ChIP-seq experiments and comparisons of peak-calling algorithms2,27–32.
Applications and limitations
MACS can be applied to scenarios other than calling enriched regions from ChIP-seq data. MACS-1.4.2 and older versions can be used to identify differential read-enriched regions from two ChIP-seq samples by viewing one of the samples as a control, so that peak regions correspond to more enriched regions in the other sample. Alternatively, users can call peaks from two ChIP-seq data separately. A peak is considered exclusive to one ChIP-seq sample only when it is identified with a stringent p-value in the sample but not identified even with a loose p-value in the other sample. However, neither method is ideal for calling differential regions. To address this problem, we are developing a robust differential peak-calling method as a major functionality in the next significant version of MACS: MACS2. A test version of MACS2 is now available from https://github.com/taoliu/MACS/downloads. In addition to ChIP-seq data, MACS can be applied to other data types such as DNase-seq33, MeDIP-seq34, and MBD-seq35; however, we suggest using more specialized tools for these alternate purposes.
MATERIALS
EQUIPMENT
Hardware
Any computer running UNIX, Linux, or Mac OS can run MACS; the following examples were run on an Ubuntu 10.10 server with a 2.8 GHz CPU. A minimum of 4 GB of RAM is needed for 32 million reads. The RAM may need to be increased for more deeply sequenced ChIP-seq data. We do not recommend running MACS on Windows machines, although MACS does work on Cygwin, a Linux simulator for Windows. Users can also access MACS via the Cistrome web portal21.
Software
MACS is a command-line program whose execution requires a terminal program, available on UNIX, Linux, or Mac OS. To run MACS on a remote server, an Internet connection and telnet or SSH are needed. MACS is coded in Python, an increasingly popular programming language in bioinformatics, which is pre-loaded with the majority of UNIX, Linux, or Mac OS installations. MACS works in Python version 2.6 or 2.7, and version 2.6.5 is recommended. To run MACS in a 64-bit environment, Python for the 64-bit CPU should be installed.
Optional software
Bowtie36, BWA37, or another aligner maps ChIP-seq and control sequencing reads to the reference genome. SAMtools38 merges sequencing data from replicates into one file. The R environment generates a PDF image of the DNA fragment size model. PeakSplitter39 can call sub-peaks and refine peak resolution from MACS output. Integrative Genomics Viewer (IGV)40 has a user-friendly interface for visualizing the original ChIP-seq data and MACS output on a local computer. Alternative visualization methods utilize the UCSC Genome Browser41 or Integrated Genome Browser (IGB)42.
Data
We selected several ChIP-seq and control datasets from ENCODE20 and made them accessible at http://cistrome.dfci.harvard.edu/MACSNatureProtocol/: the FoxA1 dataset for the T-47D cell line, with only one ChIP-seq replicate (from the HudsonAlpha Institute); the H3K4me3 dataset for the K562 cell line, with one ChIP-seq and one control replicate (from the University of Washington); the H3K4me3 dataset for the GM12878 cell line, with one ChIP-seq replicate and two controls (from the Broad Institute); and the H3K36me3 dataset for the GM12878 cell line, with two ChIP-seq and two control replicates (from the Broad Institute). The FoxA1 dataset uses the FASTQ format with raw sequencing reads, and the remaining datasets use the BAM format with reads previously mapped to the hg19 human genome.
PROCEDURE
Installing MACS ● TIMING 10 min
-
1|
Set up the necessary operating system and computing environment as listed under EQUIPMENT.
-
2|Download the MACS source code from https://github.com/downloads/taoliu/MACS/MACS-1.4.2-1.tar.gz. Locate the directory containing the downloaded source code package, and unpack the package using the following command:
> tar xvzf MACS-1.4.2-1.tar.gz
CRITICAL STEP: A precompiled MACS package for Debian or Ubuntu Linux is available for download from the above link.
-
3|Change the working directory to MACS-1.4.2 and use the standard installation command for Python packages as follows:
> cd MACS-1.4.2 > python setup.py install
The second command will install MACS globally, which requires root or administrator privileges. Alternatively, a user can install MACS to a specified directory in which the user has write privileges by using the following command:> python setup.py install --prefix /your_directory/
! CAUTION Do not install MACS in the source code directory.
? TROUBLESHOOTING
-
4|Configure the shell environment variable PATH (such as the Unix shell Bash) as shown below. If MACS is installed in a user-specified directory (in Step 3), then add the following lines to the user’s configuration file .bashrc in the home directory:
> export PATH=/your_directory/bin:$PATH > export PYTHONPATH=/your_directory/lib/python2.X/site-packages/:$PYTHONPATH
Here, python2.X represents the version of Python used for the setup script in Step 3. To determine the current version of Python, type the following:> python --version
For example, if the output is Python 2.7.1, then 2.X must be replaced by 2.7. Load the configuration file by typing either source ~/.bashrc or bash on the command line to reload Bash. To temporarily change the environment variables, type the above two export commands in Bash.
? TROUBLESHOOTING
Installing optional software ● TIMING 30 min
-
5|
Download Bowtie from http://bowtie-bio.sourceforge.net/manual.shtml, a pre-built index of hg19 for Bowtie from ftp://ftp.cbcb.umd.edu/pub/data/bowtie_indexes/hg19.ebwt.zip, SAMtools from http://samtools.sourceforge.net/, R from http://cran.r-project.org/, PeakSplitter_Cpp_1.0.tar.gz from http://www.ebi.ac.uk/bertone/software.html, and IGV from http://www.broadinstitute.org/igv/. Install each software package according to the corresponding instructions.
Running MACS to call peaks
-
6|
We use 4 different ChIP-Seq datasets to illustrate how to run MACS using varying parameters: use option A to call FoxA1 peaks; option B to call H3K4me3 peaks with fragment size estimation turned on; option C to call H3K4me3 peaks with a specified DNA fragment size; or option D to call H3K36me3 peaks. Finally, we show how to load the results generated by MACS into IGV.
A. Calling FoxA1 peaks ● TIMING 90 min
-
Locate the downloaded pre-built index for Bowtie, and unpack the package using the following command:
> unzip hg19.ebwt.zip
This command will generate several files with names prefixed by ‘hg19’ in the current directory.
-
Download the HudsonAlpha Institute FoxA1 raw reads from http://cistrome.dfci.harvard.edu/MACSNatureProtocol/HAIB_T47D_FoxA1.tar.gz, locate the download directory, unpack the compressed file, and map the raw reads to the reference genome using Bowtie by entering the following two commands:
> tar xvzf HAIB_T47D_FoxA1.tar.gz > bowtie –m 1 -S -q /path_to/hg19 HAIB_T47D_FoxA1.fastq HAIB_T47D_FoxA1.sam
In these commands,- -m 1 specifies that reads with only one hit on the genome are retained;
- -S specifies the output format as SAM;
- -q specifies the input format as FASTQ;
- /path_to/ is the directory containing the unzipped bowtie pre-built indexes; and
- HAIB_T47D_FoxA1.fastq contains the downloaded raw reads for FoxA1.
Please refer to the Bowtie manual for more information.
-
Run MACS in the same directory by entering the following command:
> macs14 -t HAIB_T47D_FoxA1.sam -n HAIB_T47D_FoxA1 –g hs -B -S - -call-subpeaks
The meanings of the parameters in this command are as follows:- -t specifies the file name for the ChIP-seq sample read alignment. MACS supports and can automatically detect any of the following file formats: SAM, BAM, BED, ELAND, ELANDMULTI, ELANDMULTIPET, ELANDEXPORT, and BOWTIE. The user-specified parameter --format can override the automatic format detection.
- -g specifies the genome size. The hs parameter is a shortcut for the approximate effective genome size of humans, which equals 2.7e9.
- -n applies the prefix ‘HAIB_T47D_FoxA1’ to the output file names.
- -B generates signal files in the bedGraph format containing the extended read pileup at every base pair. This step is very time consuming and memory intensive; therefore, only specify -B if bedGraph output files are needed.
- -S generates a single bedGraph file for the whole genome; otherwise, signal files will be generated for each chromosome separately.
- --call-subpeaks asks MACS to call PeakSplitter automatically after peak calling, if the latter has been installed properly.
! CAUTION Make sure that the character ‘/’ does not appear in the specified file prefix after the -n option, as MACS will interpret the string before ‘/’ as a directory (causing an error if this directory does not exist).
? TROUBLESHOOTING
-
Check the screen output for the running status of MACS in the terminal. MACS generates warnings and progress reports similar to the following:
INFO @ Sun, 03 Jun 2012 23:36:03: # ARGUMENTS LIST: # name = HAIB_T47D_FoxA1 # format = AUTO # ChIP-seq file = HAIB_T47D_FoxA1.sam # control file = None # effective genome size = 2.70e+09 # band width = 300 # model fold = 10,30 # pvalue cutoff = 1.00e-05 # Large dataset will be scaled towards smaller dataset. # Range for calculating regional lambda is: 10000 bps INFO #1 read tag files... INFO #1 read treatment tags... INFO Detected format is: SAM <Several lines are skipped> INFO #2 Build Peak Model... INFO #2 number of paired peaks: 16586 INFO #2 finished! INFO #2 predicted fragment length is 114 bps INFO #2.2 Generate R script for model : HAIB_T47D_FoxA1_model.r INFO #3 Call peaks... INFO #3 shift treatment data INFO #3 merge +/− strand of treatment data INFO #3 save the shifted and merged tag counts into bedGraph file... INFO write to a bedGraph file <Several lines are skipped> INFO #3 call peak candidates INFO #3 use self to calculate local lambda and filter peak candidates... INFO #3 Finally, 74761 peaks are called! INFO #4 Write output xls file... HAIB_T47D_FoxA1_peaks.xls INFO #4 Write peak bed file... HAIB_T47D_FoxA1_peaks.bed INFO #4 Write summits bed file... HAIB_T47D_FoxA1_summits.bed INFO #5 Done! Check the output files! INFO #6 Try to invoke PeakSplitter... INFO #6 Please check HAIB_T47D_FoxA1_peaks.subpeaks.bed file for PeakSplitter output!
The messages provide information such as the date (the first line), key parameters (lines starting with ‘#’), and the run progress (lines starting with ‘INFO’). For lines indicating run progress, we have removed the date information and several lines to make the screen output more concise. If MACS encounters exceptions (e.g., if MACS estimates a fragment size that is too small), then warning messages appear in the list.
! CAUTION Although warning messages do not affect the success of a MACS run, the majority should still be carefully evaluated. For example, the warning message ‘unbalanced reads between treatment and control’ means that the FDR of the resulting peaks will be overestimated when the control sample has more reads and will be underestimated when the ChIP-seq sample is sequenced more deeply. The message ‘Fewer paired peaks X than 1000’ means that MACS only identified X model peaks and may indicate potential data quality issues because 1,000 model peaks are needed to robustly estimate ChIP-DNA fragment size. The message ‘missing chromosome X data’ might suggest that the raw input file for that chromosome is incomplete.
-
Generate a PDF figure for the peak model using the following command (assuming that R has been installed properly):
> Rscript HAIB_T47D_FoxA1_model.r
This command will produce a PDF image named HAIB_T47D_FoxA1_model.pdf in the current working directory. This image illustrates the distribution of reads on positive and negative strands in the model peaks and the estimated fragment size.
Verify the existence of the files listed in Table 1 in the current directory. Details of the output files are described in the ANTICIPATED RESULTS section.
Table 1.
Files generated by MACS for the HudsonAlpha Institute FoxA1 dataset
| File Name | Description |
|---|---|
| HAIB_T47D_FoxA1_model.r | An R script for producing a PDF illustrating the peak model |
| HAIB_T47D_FoxA1_model.pdf | The PDF image of the read distribution in model peaks and fragment size estimation, which is available only after executing Step 6Av |
| HAIB_T47D_FoxA1_peaks.xls | Key parameters used by MACS and detailed information of every peak identified by MACS |
| HAIB_T47D_FoxA1_peaks.bed | Peak locations in BED format |
| HAIB_T47D_FoxA1_peaks.subpeaks.bed | Subpeak locations in BED-like format. This file is generated by PeakSplitter, which is called by MACS |
| HAIB_T47D_FoxA1_summits.bed | Summit locations of the peaks in BED format |
| HAIB_T47D_FoxA1_MACS_bedGraph | Directory where the BedGraph files are generated. For each control or ChIP-seq sample, a BedGraph file describes the read distribution along the whole genome |
B. Calling H3K4me3 peaks with fragment size estimation turned on ● TIMING 90 min
- Download the University of Washington H3K4me3 dataset from http://cistrome.dfci.harvard.edu/MACSNatureProtocol/UW_K562_H3K4me3.tar.gz. This dataset contains one control replicate and one ChIP-seq replicate. Locate the directory where the downloaded file has been stored. Extract the bundle using the following command:
> tar xvzf UW_K562_H3K4me3.tar.gz
-
In the same directory, run MACS as follows:
> macs14 -t UW_K562_H3K4me3.bam -c UW_K562_H3K4me3_Control.bam - g hs -n UW_K562_H3K4me3 -B -S --call-subpeaks
The parameter -c specifies the file name for the control sample read alignment. The other parameters follow the same convention as described in Step 6Aiii, and see Box 1.
? TROUBLESHOOTING
- Check the screen output generated by MACS as described in Step 6Aiv. MACS will report a successful model build by displaying the following messages:
<Several lines are skipped> INFO : #2 Build Peak Model... INFO : #2 number of paired peaks: 12267 INFO : #2 finished! INFO : #2 predicted fragment length is 156 bps <Several lines are skipped> INFO : #3 use control data to filter peak candidates... INFO : #3 Finally, 20632 peaks are called! INFO : #3 find negative peaks by swapping treat and control INFO : #3 Finally, 4006 peaks are called! <Several lines are skipped>
- Generate a PDF figure named UW_K562_H3K4me3_model.pdf for the read distribution in model peaks and the estimation of fragment size by applying the following command:
> Rscript UW_K562_H3K4me3_model.r
Verify that all output files are present as described in Step 6Avi except that they should have the file name prefix ‘UW_K562_H3K4me3’ instead of ‘HAIB_T47D_FoxA1’. Because a control sample is available in this case, another file (‘UW_K562_H3K4me3_negative_peaks.xls’) is generated that contains the peaks called by comparing the control sample to the ChIP-seq sample using the same parameters. These peaks are used by MACS for estimating the FDR of each reported ChIP-seq peak.
Box 1. Additional important MACS parameters.
Several additional important parameters that could be used to run MACS in Step 6Aiii are as follows:
--bw sets the ‘bandwidth’, which is half of the sliding window size used in the model-building step.
--mfold specifies an interval of high-confidence enrichment ratio against the background on which to build the model. The default value ‘10, 30’ means that a model will be built based on regions having read counts that are 10- to 30-fold of the background.
--pvalue establishes a threshold p-value: only peaks surpassing the threshold will be reported. The default threshold is 10−5. Users can first set a loose p-value cutoff so that a sufficient number of peaks will be reported and then select peaks having the smallest p-values for downstream analyses.
C. Calling H3K4me3 peaks with a specified DNA fragment size ● TIMING 90 min
- Download the Broad Institute H3K4me3 dataset from http://cistrome.dfci.harvard.edu/MACSNatureProtocol/BROAD_GM12878_H3K4me3.tar.gz. This dataset contains one ChIP-seq replicate and two control replicates. MACS runs either on a single ChIP-seq sample or on a single ChIP-seq sample having a single control; in this case, the two control replicates must be concatenated. Extract the bundle, and merge the two control replicates using the following two commands:
> tar xvzf BROAD_GM12878_H3K4me3.tar.gz > samtools merge BROAD_GM12878_H3K4me3_Control.bam BROAD_GM12878_H3K4me3_Control_1.bam BROAD_GM12878_H3K4me3_Control_2.bam
- Run MACS as follows:
> macs14 -t BROAD_GM12878_H3K4me3.bam -c BROAD_GM12878_H3K4me3_Control.bam -g hs -n BROAD_GM12878_H3K4me3 -B -S --call-subpeaks
-
Check the screen output of MACS; it should contain the following lines:
<Several lines are skipped> INFO : #2 number of paired peaks: 31077 INFO : #2 finished! INFO : #2 predicted fragment length is 53 bps <Several lines are skipped>
The model built by MACS has a fragment length of 53, which is unusually short in a typical ChIP-seq experiment. Therefore, it is preferable to rerun MACS with modified parameters, as described in Steps 6Cv.
- If MACS is still running, terminate it by typing Ctrl+C (hold the Control key and press C). Then, remove the directory BROAD_GM12878_H3K4me3_MACS_bedGraph from the previous MACS run using the following command:
> rm -rf BROAD_GM12878_H3K4me3_MACS_bedGraph
-
Rerun MACS using modified parameters, as follows:
> macs14 -t BROAD_GM12878_H3K4me3.bam -c BROAD_GM12878_H3K4me3_Control.bam -g hs -n BROAD_GM12878_H3K4me3 --nomodel --shiftsize 73 -B -S --call-subpeaks
This command uses --nomodel to instruct MACS not to estimate the fragment size. --shiftsize 73 tells MACS to use a fixed DNA fragment size of 146 = 73 × 2.
CRITICAL STEP: The fragment size is set to 146 because a nucleosome is wrapped in a DNA sequence that is approximately 146 bp in length, extending reads mapped to either DNA strand in the 3’ direction by 146 bp. Users can also specify the fragment size according to their sequencing library preparation, often in the range of 150–200 bp.
- Check the MACS screen output. The following messages relay that MACS did not build the model but instead used shiftsize 73:
<Several lines are skipped> INFO : #2 Build Peak Model... INFO : #2 Skipped... INFO : #2 Use 73 as shiftsize, 146 as fragment length <Some lines are skipped>
Verify all the generated files except the R script as in Step 6Avi, where file names must contain the prefix ‘BROAD_GM12878_H3K4me3’ instead of ‘HAIB_T47D_FoxA1’. The file BROAD_GM12878_H3K4me3_negative_peaks.xls contains the peaks identified in the control sample over the ChIP-seq sample. In this case, because MACS did not build the peak model, no R script is generated.
D. Calling H3K36me3 peaks ● TIMING 90 min
- Download the Broad Institute H3K36me3 dataset from http://cistrome.dfci.harvard.edu/MACSNatureProtocol/BROAD_GM12878_H3K36me3.tar.gz. This dataset contains two control replicates and two ChIP-seq replicates. Extract the bundle and merge the replicates using the following commands:
> tar xvzf BROAD_GM12878_H3K36me3.tar.gz > samtools merge BROAD_GM12878_H3K36me3.bam BROAD_GM12878_H3K36me3_1.bam BROAD_GM12878_H3K36me3_2.bam > samtools merge BROAD_GM12878_H3K36me3_Control.bam BROAD_GM12878_H3K36me3_Control_1.bam BROAD_GM12878_H3K36me3_Control_2.bam
-
Run MACS to call the peaks using the following command:
> macs14 -t BROAD_GM12878_H3K36me3.bam -c BROAD_GM12878_H3K36me3_Control.bam -g hs -n BROAD_GM12878_H3K36me3 --nomodel --shiftsize 73 -B -S --pvalue 1e-3 --call-subpeaks
Compared with Step 6Cv, this command sets a less stringent p-value cutoff (--pvalue 1e-3) than the default (1e-5). Because H3K36me3 ChIP-seq data often form broader but less enriched regions, the parameters --nomodel and --shiftsize 73 are preferred.
? TROUBLESHOOTING
Verify all the generated files as in Step 6Cvii using the file name prefix ‘BROAD_GM12878_H3K36me3’ rather than ‘BROAD_GM12878_H3K4me3’.
E. Loading results generated by MACS into IGV ●TIMING 10 min
To load a bedGraph generated by MACS into IGV, the user must first decompress and rename the bedGraph file. As an example, consider the results of MACS on the FoxA1 dataset. First, locate the bedGraph in the directory HAIB_T47D_FoxA1_MACS_bedGraph/treat.
- Unzip the file as follows:
> gzip -d HAIB_T47D_FoxA1_treat_afterfiting_all.bdg.gz
- Change the extension of the file to “bedGraph”, which can be recognized by IGV, via the following command:
> mv HAIB_T47D_FoxA1_treat_afterfiting_all.bdg HAIB_T47D_FoxA1_treat_afterfiting_all.bedGraph
Load HAIB_T47D_FoxA1_treat_afterfiting_all.bedGraph into IGV following IGV manual.
TIMING
The time required to download the program and data depends on the user’s network bandwidth. Each step that requires running MACS (i.e., Steps 6Aiii, 6Bii, 6Cv, and 6Dii) requires approximately 70 minutes.
Steps 1–4, installing MACS: 10 min
Step 5, installing optional software: 30 min
Step 6A, calling FoxA1 peaks: 90 min
Step 6B, calling H3K4me3 peaks with fragment size estimation turned on: 90 min
Step 6C, calling H3K4me3 peaks with a specified DNA fragment size: 90 min
Step 6D, calling H3K36me3 peaks: 90 min
Step 6E, loading results generated by MACS into IGV: 10 min
? TROUBLESHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 3 | Installation error | The user has no write permission to the installation directory | Pay attention to the installation messages to understand the reason for the failure. The installation messages will indicate where the setup script copies the Python files. Change the installation path by using --prefix as illustrated in the text |
| 4 | Import Error | Python cannot locate certain library files from MACS. This error is usually caused by the existence of multiple versions of Python on the system. It can also occur when the user installed MACS to a specified path but forgot to set the environment variable PYTHONPATH | Check the last message in the installation message. It should read as follows:
Writing /PATH_TO_MACS_LIB/MACS-1.4.2-py2.X.egg-info
In this message, PATH_TO_MACS_LIB is the library path where MACS is installed, which depends on the operating system. X is the Python version, e.g., X is 6 if Python 2.6 is installed. Change the environment variable PYTHONPATH by executing the following command: > export PYTHONPATH=PATH_TO_MACS_LIB Change the privileges of PATH_TO_MACS_LIB using the following command: > chmod –R 755 PATH_TO_MACS_LIB |
| 6Aiii 6Bii | MACS returns an error when reading the input sequence alignment files | The input files are corrupted | Check the file integrality. For a BAM or SAM format file, the user can simply run samtools flagstat to determine whether the sequence statistics appear reasonable. For a BED format file, the user can count the number of lines using the command wc -l or check the end of file using the command tail to ensure that the file is complete |
| 6Dii | Other errors | The format automatically detected by MACS is not correct | Pay attention to the running messages pertaining to the file format and read length detected by MACS. When necessary, the user can explicitly set the file format and read length using the commands --format and --tsize |
ANTICIPATED RESULTS
Common results for all datasets
Although we set different parameters for different ChIP-seq datasets, the types and formats of the output files generated by MACS are similar. As an example, we describe in detail the MACS results for the H3K4me3 dataset from the University of Washington.
The most important output file is UW_K562_H3K4me3_peaks.xls, which contains all of the information about the peaks identified by MACS. Both UW_K562_H3K4me3_summits.bed and UW_K562_H3K4me3_peaks.bed contain partial information for all of the peaks in BED format to expedite downstream analyses, such as visualization in IGV or in the UCSC genome browser. The top lines of UW_K562_H3K4me3_peaks.xls are as follows:
# This file is generated by MACS version 1.4.2 20120305 # ARGUMENTS LIST: # name = UW_K562_H3K4me3 # format = AUTO # ChIP-seq file = UW_K562_H3K4me3.bam # control file = UW_K562_H3K4me3_Control.bam # effective genome size = 2.70e+09 # band width = 300 # model fold = 10,30 # pvalue cutoff = 1.00e-05 # Large dataset will be scaled towards smaller dataset. # Range for calculating regional lambda is: 1000 bps and 10000 bps # tag size is determined as 36 bps # total tags in treatment: 15465586 # tags after filtering in treatment: 13913615 # maximum duplicate tags at the same position in treatment = 1 # Redundant rate in treatment: 0.10 # total tags in control: 14653281 # tags after filtering in control: 14444786 # maximum duplicate tags at the same position in control = 1 # Redundant rate in control: 0.01 # d = 156 chr start end length summit tags -10*log10(pvalue) fold_enrichment FDR(%) chr1 137660 138139 480 153 62 635.75 41.05 0.00 chr1 138380 139613 1234 680 118 1038.81 41.05 0.00 chr1 712724 715549 2826 1877 433 3100.00 98.27 0.00 chr1 752449 752902 454 151 15 83.67 12.44 6.45 chr1 760913 763271 2359 1901 296 2878.46 83.33 0.00 chr1 805080 805697 618 385 18 54.29 8.13 15.71 chr1 839086 841012 1927 465 175 1154.41 24.76 0.00
The lines starting with ‘#’ contain the key parameters used by MACS and the basic data statistics that the program computes, information critical for reproducing results. ‘Redundant rate’ indicates the proportion of reads that are removed due to redundancy. This section is followed by detailed peak information. The first four columns contain each peak’s length and genome coordinates. Note that the coordinates in this file are 1-based, which differs from files in the BED format, in which coordinates are 0-based. The fifth column (‘summit’) gives the position having the highest fragment pileup in each peak region (e.g., the summit coordinate of the first peak is chr1, position 137812, which is 137660-1+153), which may not necessarily represent the center of a peak. The ‘tags’ column shows the number of reads aligned to each peak region. The ‘-10*log10(pvalue)’ column lists the transformed p-value of each peak, which makes peak sorting easier. For example, a p-value of 1e-5 would be transformed to 50. The ‘fold_enrichment’ column shows the ratio of the ChIP-seq read count to the local value of lambda within each peak. The ‘FDR(%)’ column contains the empirical FDR percentage for each peak. For example, the fourth peak in the list has an ‘FDR(%)’ value of ‘6.45’, and ‘-10*log10(pvalue)’ value of ‘83.67’; using the same p-value cutoff of 4.4e-09= 1083.67 / −10, the ratio of the number of peaks identified by MACS after and before exchanging control and ChIP-seq samples is 6.45:100. The FDR column is only available when the control sample is available. For example, using the HudsonAlpha Institute FoxA1 dataset, this column would not appear in the corresponding xls file.
Specific results for each dataset
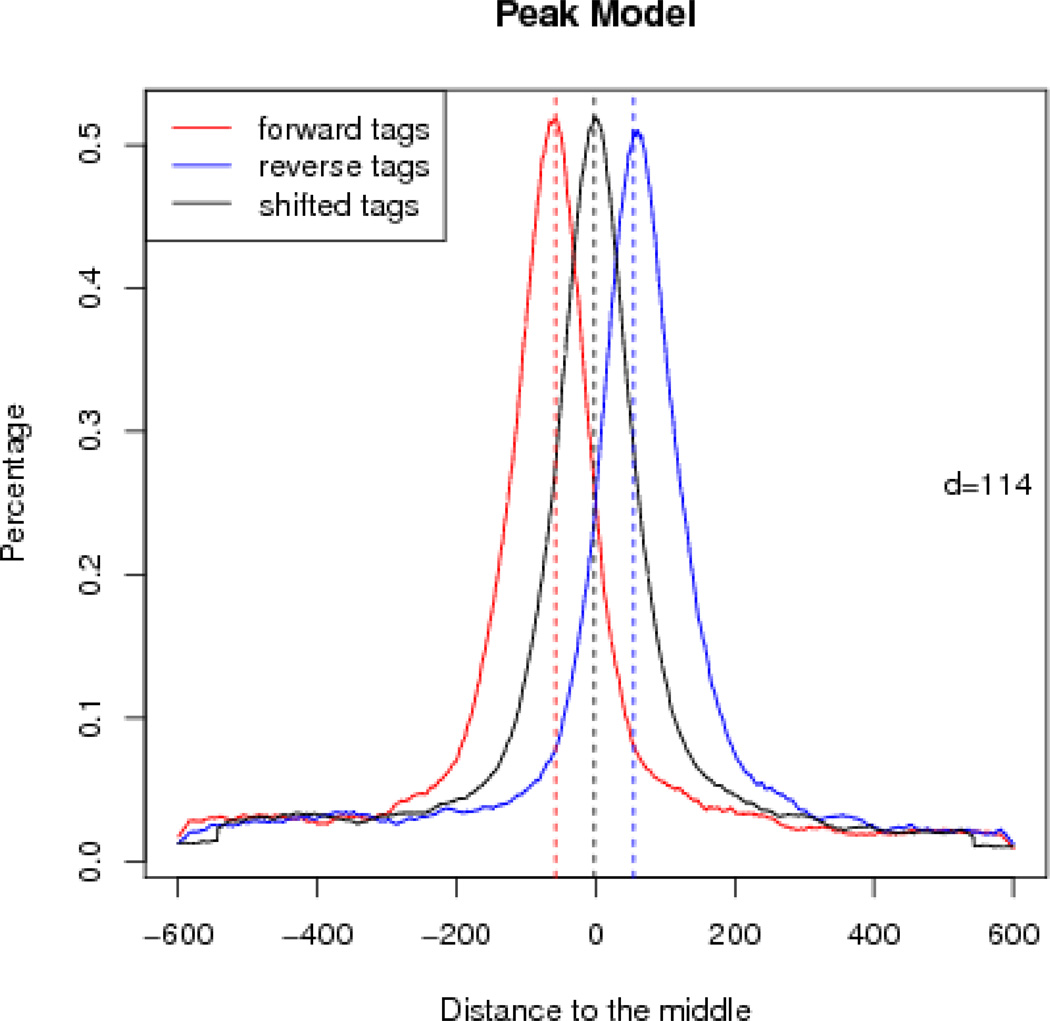
For a typical transcription factor such as FoxA1, the peak model can often be built successfully by MACS, i.e., when the detected DNA fragment size is not too small (e.g., less than 60 bp). We can check the model by inspecting the file HAIB_T47D_FoxA1_model.pdf, which is generated in Step 6Av. Figure 2 illustrates that reads on the positive or negative strand are enriched at the left or right of the paired peak center, respectively. The detected DNA fragment length is 114 bp, which may vary among different ChIP-seq libraries.
Figure 2.
Peak model built by MACS using the FoxA1 dataset. d=114 represents the estimated DNA fragment size. The red curve represents the percentage of positive strand reads at each base pair, and the blue curve models reads on the negative strand. The black curve illustrates the distribution of reads after shifting them towards the 3’ end by 57=114/2 bp.
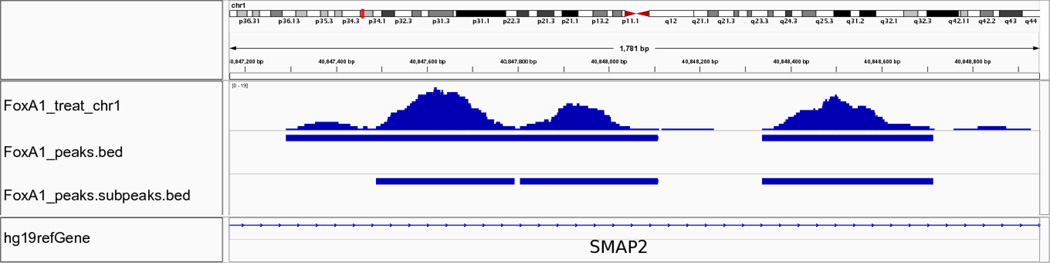
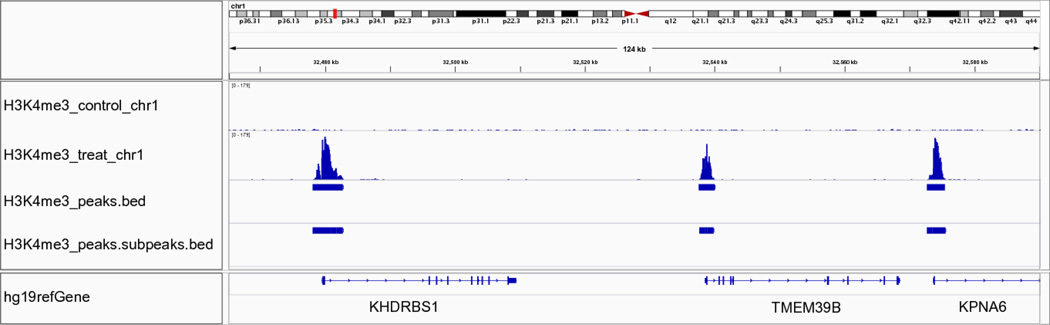
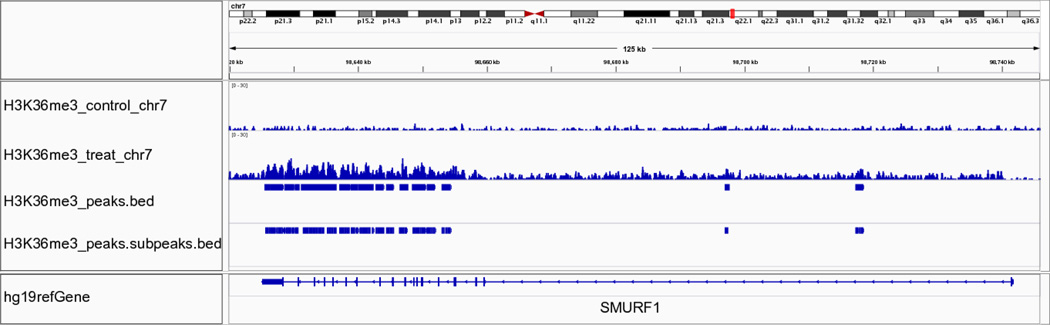
We can visualize the peak regions in detail using IGV, as exemplified in Figures 3, 4, and 5. Figure 3 displays the results of running MACS on the FoxA1 dataset. The figure has three tracks: one bedGraph track for the fragment pileup of the ChIP-seq sample, peak regions called by MACS, and sub-peaks refined by PeakSplitter. The peak track illustrates three peaks identified by MACS that correspond to three enriched regions shown in the bedGraph track. Figure 4 presents an example region of the University of Washington H3K4me3 dataset, on which reads are enriched in gene promoter regions, as indicated by the gene annotation track. Figure 5 illustrates the Broad Institute H3K36me3 dataset, where H3K36me3 is enriched at the 3’ end of the gene body, especially at exons.
Figure 3.
IGV visualization of MACS results using the FoxA1 dataset. This region is selected from chromosome 1, as shown at the top of the figure. The middle section of the figure illustrates the pileup signal after extending all reads to the estimated fragment size in the top track (labeled FoxA1_treat_chr1). Below this, the middle track, labeled FoxA1_peaks.bed, shows two peaks identified by MACS. The bottom track, labeled FoxA1_peaks.subpeaks.bed, shows three sub-peaks generated by PeakSplitter. The bottom track, labeled hg19refGene, shows the gene annotation of the human genome assembly of version hg19.
Figure 4.
IGV visualization of MACS results using the University of Washington H3K4me3 dataset. The region on chromosome 1 (shown in the top section of the figure) shows three peaks are identified by MACS in the middle section. These three peaks are located in the promoter regions of three genes, shown in the bottom part of the figure.
Figure 5.
IGV visualization of MACS results using the Broad Institute H3K36me3 dataset. The selected region spans the whole gene body of SMURF1 shown in the bottom part of the figure. The middle section of the figures shows that H3K36me3 signal is more enriched in exon regions, as demonstrated in the second track (H3K36me3_treat_chr7). MACS identifies such enriched regions as multiple peaks, shown in the third track (H3K36me3_peaks.bed).
ACKNOWLEDGMENTS
This project was supported by Chinese NSF grants 31028011 and 31071114, US NIH grant HG4069, the Shanghai Key Laboratory of Intelligent Information Processing, China (Grant No. 20102662), and the Excellent Young Teachers Program of Tongji University (Grant No. 2010KJ041).
Footnotes
AUTHOR CONTRIBUTIONS
Y.Z., T.L., and X.S.L. developed the original MACS algorithm. T.L. developed the current version of the MACS program. J.F. and B.Q. performed the data analysis. J.F, T.L., and X.S.L. wrote the initial manuscript. All authors contributed to the discussion and writing of the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Mardis ER. ChIP-seq: welcome to the new frontier. Nat Meth. 2007;4:613–614. doi: 10.1038/nmeth0807-613. [DOI] [PubMed] [Google Scholar]
- 2.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 4.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 5.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 7.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Research. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rozowsky J, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotech. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vega VB, Cheung E, Palanisamy N, Sung W-K. Inherent Signals in Sequencing-Based Chromatin-ImmunoPrecipitation Control Libraries. PLoS ONE. 2009;4:e5241. doi: 10.1371/journal.pone.0005241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ET, Pott S, Huss M. Q&A: ChIP-seq technologies and the study of gene regulation. BMC Biol. 2010;8:56. doi: 10.1186/1741-7007-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teytelman L, et al. Impact of chromatin structures on DNA processing for genomic analyses. PLoS ONE. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nix DA, Courdy SJ, Boucher KM. Empirical methods for controlling false positives and estimating confidence in ChIP-Seq peaks. BMC Bioinformatics. 2008;9:523. doi: 10.1186/1471-2105-9-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Model-based Analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137–R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavares L, et al. RYBP-PRC1 Complexes Mediate H2A Ubiquitylation at Polycomb Target Sites Independently of PRC2 and H3K27me3. Cell. 2012;148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–1550. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nature Genetics. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Genetic analysis of variation in transcription factor binding in yeast. Nature. 2010;464:1187–1191. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noordermeer D, et al. The Dynamic Architecture of Hox Gene Clusters. Science. 2011;334:222–225. doi: 10.1126/science.1207194. [DOI] [PubMed] [Google Scholar]
- 19.Welboren W-J, et al. ChIP-Seq of ER|[alpha]| and RNA polymerase II defines genes differentially responding to ligands. The EMBO Journal. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu T, et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biology. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jothi R, Cuddapah S, Barski A, Cui K, Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotech. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fejes AP, et al. FindPeaks 3.1: a tool for identifying areas of enrichment from massively parallel short-read sequencing technology. Bioinformatics. 2008;24:1729–1730. doi: 10.1093/bioinformatics/btn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valouev A, et al. Genome-Wide Analysis of Transcription Factor Binding Sites Based on ChIP-Seq Data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laajala TD, et al. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics. 2009;10:618. doi: 10.1186/1471-2164-10-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilbanks EG, Facciotti MT. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS ONE. 2010;5:e11471. doi: 10.1371/journal.pone.0011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barski A, Zhao K. Genomic location analysis by ChIP-Seq. J. Cell. Biochem. 2009;107:11–18. doi: 10.1002/jcb.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone BM, Tan F, Bridges SM, Peng Z. Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS ONE. 2011;6:e25260. doi: 10.1371/journal.pone.0025260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nature methods. 2012 doi: 10.1038/nmeth.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stitzel ML, et al. Global epigenomic analysis of primary human pancreatic islets provides insights into type 2 diabetes susceptibility loci. Cell Metab. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sati S, et al. High Resolution Methylome Map of Rat Indicates Role of Intragenic DNA Methylation in Identification of Coding Region. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N, et al. Whole genome DNA methylation analysis based on high throughput sequencing technology. Methods. 2010;52:203–212. doi: 10.1016/j.ymeth.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmon-Divon M, Dvinge H, Tammoja K, Bertone P. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics. 11:415. doi: 10.1186/1471-2105-11-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, et al. Integrative genomics viewer. Nat Biotech. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicol JW, Helt GA, Blanchard SG, Jr, Raja A, Loraine AE. The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics. 2009;25:2730–2731. doi: 10.1093/bioinformatics/btp472. [DOI] [PMC free article] [PubMed] [Google Scholar]