Abstract
E2F transcription factors regulate the progression of the cell cycle by repression or transactivation of genes that encode cyclins, cyclin dependent kinases, checkpoint regulators, and replication proteins. Although some E2F functions are independent of the Retinoblastoma tumor suppressor (Rb) and related family members, p107 and p130, much of E2F-mediated repression of S phase entry is dependent upon Rb. We previously showed in cultured mouse embryonic fibroblasts that concomitant loss of three E2F activators with overlapping functions (E2F1, E2F2, and E2F3) triggered the p53-p21Cip1 response and caused cell cycle arrest. Here we report on a dramatic difference in the requirement for E2F during development and in cultured cells by showing that cell cycle entry occurs normally in E2f1-3 triply-deficient epithelial stem cells and progenitors of the developing lens. Sixteen days after birth, however, massive apoptosis in differentiating epithelium leads to a collapse of the entire eye. Prior to this collapse, we find that expression of cell cycle-regulated genes in E2F-deficient lenses is aberrantly high. In a second set of experiments, we demonstrate that E2F3 ablation alone does not cause abnormalities in lens development but rescues phenotypic defects caused by loss of Rb, a binding partner of E2F known to recruit histone deacetylases, SWI/SNF and CtBP-polycomb complexes, methyltransferases, and other co-repressors to gene promoters. Together, these data implicate E2F1-3 in mediating transcriptional repression by Rb during cell cycle exit and point to a critical role for their repressive functions in cell survival.
Keywords: Proliferation, Cell cycle, E2F, Rb, Lens, Repression, Cell survival, Transcription
Introduction
Cell cycle entry is guarded by cyclin dependent kinases (Cdk) which, upon activation by mitogenic signals, phosphorylate the Retinoblastoma pocket proteins, Rb, p107, and p130, and cause the release and accumulation of sequestered E2F transcription factors. E2Fs consist of a family of repressors and activators that together coordinately regulate cellular proliferation by controlling the transcriptional activity of over 130 known target genes that function to initiate the G1/S transition, DNA synthesis, DNA repair and mitosis (Bracken et al., 2004). Rb is thought to inhibit E2F activity and expression of cell cycle-regulated genes by association with co-repressors such as histone deacetylases, SWI/SNF and CtBP-polycomb complexes, histone methyltransferases, and DNA methyltransferases (Dahiya et al., 2001; Luo et al., 1998; Nielsen et al., 2001; Robertson et al., 2000; Vandel et al., 2001; Zhang et al., 2000). Classic paradigms of cell cycle regulation have consistently portrayed the three activators, E2F1, E2F2, and E2F3, as the final components of the Cdk-Rb signaling cascade that executes the transcriptional program necessary to commit cells to enter S phase. Although Rb-mediated cell cycle regulation has been studied extensively, the evidence that E2F1-3 are required for expression of genes critical for proliferation is based almost exclusively on analyses of lower eukaryotes and in vitro cell culture systems (Bracken et al., 2004; Hallstrom and Nevins, 2009; Morris et al., 2000; Rowland et al., 2002). Only recently has it been demonstrated that E2F1-3 are not essential for cell cycle entry, and that proliferation can proceed without E2F1-3 in the majority of cell types present during organ development and embryogenesis (Chen et al., 2009; Chong et al., 2009). In fact, there is now good evidence to suggest that these E2Fs are most critical for regulating survival in vivo (Chen et al., 2009; Chong et al., 2009).
The highly organized cellular architecture of the developing lens makes it an attractive system for the in vivo study of cell cycle and differentiation. Simply based upon spatial separation, epithelial cells that are proliferating can be distinguished easily from those that are exiting the cell cycle and terminally differentiating. At approximately 10 days of murine embryonic development (E10), morphological lens formation begins with the invagination of the surface ectoderm overlying the optic cup to form the lens pit, which subsequently closes to form the lens vesicle (Lovicu et al., 2004). Although initially all epithelial cells lining the lens vesicle maintain the capacity to proliferate, as development progresses, cycling cells are confined to a ring of epithelium slightly anterior to the lens equator. At the equatorial region (or bow) of the lens, rapidly proliferating cells move into a transition zone, wherein they begin to exit the cell cycle and differentiate. As these cells migrate toward the cortex of the lens, they terminally differentiate into fiber cells, losing their nuclei and organelles and gaining the translucent properties necessary for vision.
To test whether loss of E2F activators impact cell cycle entry in vivo, we examined the effects of E2F1, E2F2, and E2F3 triple deficiency during development in murine pre- and post-natal lenses. We find that morphological lens development occurs relatively normally through late gestation, illustrating that E2F activators are unnecessary for proliferation of lens epithelial cells. During migration of the epithelial cells to the equatorial zone, however, DNA double-strand breaks develop, p21Cip1 is upregulated, and cells exhibit signs of apoptotic cell death. Further, we find a dramatic increase in expression of E2F target genes in E2f-triply deficient lenses, pointing to the shared function that E2F1, 2, and 3 play in repressing expression of cell cycle genes during maturation of the lens epithelium. Aberrant expression of genes required for cell cycle exit and lens differentiation, along with upregulation of p19Arf and other E2F target genes, together culminates in collapse of the entire lens architecture between one to two weeks after birth. Importantly, we demonstrate a nearly complete rescue of Rb-deficiency phenotypes, including proliferation defects, with conditional ablation of E2F3, pointing to a context-dependent switch in the function of “activating” E2Fs to repress or activate transcription. These data illustrate that E2F-mediated activation of cell cycle regulated genes is not required for proliferation, and point to a critical role for E2F1-3 in cell survival and transcriptional repression in vivo.
Materials and Methods
Mouse Strains and Genotyping
E2f1, E2f2, E2f3 knockout mice and cry-cre (MLR10) transgenic (Zhao et al., 2004) and ROSA26 reporter (Gt(ROSA)26Sortm1Sor) (Soriano, 1999) mice were maintained on a mixed 129SvEv; C57BL/6; FVB background. PCR primers for genotyping are listed in Figure S1.
Proliferation Assays
Proliferation of lens epithelial or fiber cells was measured by incorporation of BrdU. Briefly, pregnant dams or pups were injected with BrdU 2 hours prior to harvest at 100 μg/g body weight. Tissue from embryos or neonates was fixed in 10% buffered formalin, dehydrated, and processed with paraffin. Sections were cut at 5-μm thickness and BrdU was detected by immunofluorescent staining.
Histology And Immunofluorescent Staining
Lens architecture was histologically examined by hematoxylin and eosin staining of lens sections. Proliferation was measured by immunostaining with BrdU-specific antibodies (DAKO; Bu20a), and apoptosis was detected by TUNEL assays using the Apoptag Plus Peroxidase In Situ Apoptosis Detection Kit (Chemicon International). DNA double-strand breaks were detected as a measure of DNA damage by immunofluorescent staining with phosphohistone H2AX (Ser139) antibodies (Upstate, NY; clone JBW301). Other antibodies used include PCNA (Santa Cruz Biotechnology, PC 10), Mcm3 (Santa Cruz Biotechnology, N-19), and cleaved caspase-3 (Cell Signaling Technology, Asp175). A minimum of three sections from each lens were quantified by comparison of the number of positive cells to the total number of nuclei in epithelial or fiber cells. Lens-specific markers of differentiation, including α-, β-, and γ-crystallin antibodies were a gift from Dr. S. Zigler.
Detection Of β-Galactosidase Activity
Cry-cre mice were crossed to the ROSA26 reporter (Gt(ROSA)26Sortm1Sor) line to test the activity of cre as described previously (Robinson et al., 1995). Briefly, eyes were surgically removed from embryos, fixed in 2% paraformaldehyde in 0.1M phosphate buffer pH 7.3, washed in PBS, and incubated overnight in the dark at room temperature in X-gal staining medium (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 1 mg/ml X-gal, 0.1% deoxycholate, 0.2% Nonidet P-40, and 2 mM MgCl2 in PBS). Eyes were post-fixed the next day in 10% buffered formalin for 2 hours and processed for paraffin embedding, sectioning, and counterstaining with nuclear fast red.
Global Gene Expression Analysis
Cry-Cre+/-; E2f1-/-; E2f2-/-; E2f3LoxP/LoxP and E2f1+/+; E2f2-/-; E2f3LoxP/LoxP lenses were microdissected from the eye at E17.5 and P0 with 18-gauge needles. RNA from both lenses of each embryo or neonate was isolated by TRIzol purification and processed for hybridization to Affymetrix Mouse Genome 430 2.0 Arrays. Expression values were adjusted by quantile normalization and log transformation with RMAExpress, and data was analyzed with BRB- Array Tools 3.7.0. (Fig. S2, (S3). Class comparison was used to select genes differentially expressed at a significance level of p < 0.0001 or p < 0.001, and gene set expression comparisons were used to identify gene ontologies and pathways impacted by loss of E2F (Fig. S4). DAVID (http://david.abcc.ncifcrf.gov) was used to further evaluate subsets of genes. CBRC TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html) and GATHER (http://gather.genome.duke.edu) aided in the identification of genes containing E2F consensus binding sites. Microarray data presented in this study has been deposited in the GEO public repository (GSE16533).
Real-Time RT-PCR
Both lenses from each embryo or neonate were placed in one microcentrifuge tube and homogenized in TRIzol reagent to obtain total RNA (Life Technologies). Five μg of total RNA was used to generate cDNA using Superscript III reverse transcriptase (Invitrogen). Quantitative RT-PCR was performed using the BioRad iCycler PCR machine. Each PCR reaction contained 0.5 μl of cDNA template, primers at a concentration of 100 nM, and 1× of SYBR Green Reaction Mix (BioRad). Reactions were performed in triplicate in a total volume of 25 μl and data were analyzed using the ΔCt method, where GAPDH served as the internal control. Each PCR reaction generated only the expected amplicon as shown by the melting-temperature profiles of the final products and gel electrophoresis. Primer sequences are listed in Fig. S1.
Results
Lens Epithelial Cells Proliferate in the Absence of E2F1-3
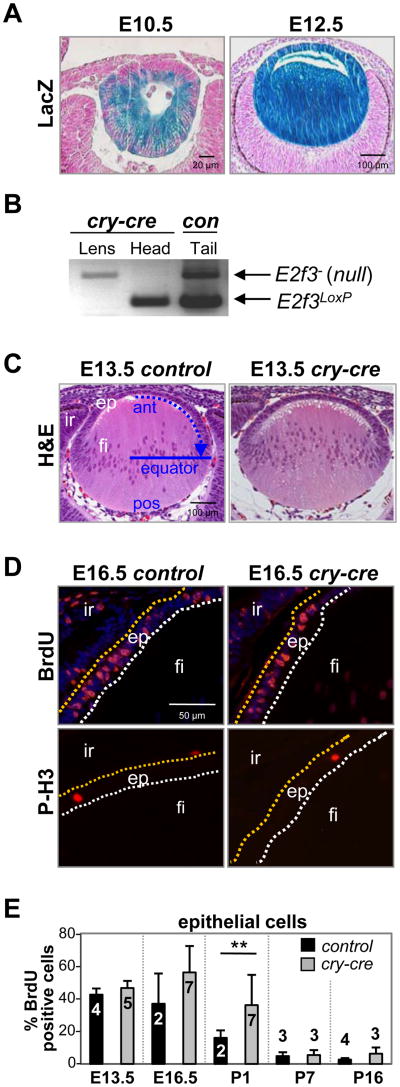
We utilized a well-characterized transgenic mouse that expresses cre in lens epithelial and fiber cells to delete a conditional allele of E2f3, either alone or in combination with E2f1 or E2f2, at the earliest stages of lens vesicle formation (cry-cre, also known as MLR10; Fig. 1A) (Zhao et al., 2004). This analysis showed that the combined deletion of any two of the three activator E2Fs does not adversely affect lens epithelial cell proliferation or lens development (Fig. S5A). To avoid compensation that could result from overlapping functions among E2F members (Tsai et al., 2008), the entire E2f1-3 subset was deleted. The efficient recombination of the E2f3LoxP allele was confirmed by PCR-genotyping of laser capture microdissected (LCM) lens tissue (Fig. 1B). Surprisingly, histological examination of E2f1-3 deficient (TKO) lenses revealed no conspicuous change in lens architecture prior to birth (Fig. 1C). Direct assessment of DNA replication and mitosis by BrdU incorporation and phosphorylated histone H3 (P-H3), respectively, did not identify significant differences in proliferation between control and TKO epithelial cells at most stages of lens development (E13.5-P16; Fig. 1D, 1E). Interestingly, we observed a doubling of BrdU incorporation in lens epithelial cells of newborn TKO pups (P1, p=0.002; Fig. 1E) and appearance of ectopic DNA synthesis in the associated lens fiber cells (0.5% to 2% BrdU positive), suggesting that cell cycle exit could have been impacted.
Fig. 1. Conditional deletion of E2f3 in cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses does not block epithelial cell proliferation.
(A) Cre recombinase activity in cry-cre;ROSA26Sortm1Sor mice is detectable as a blue stain (LacZ expression) in embryonic lens epithelial and fiber cells. (B) PCR genotyping of E2f3 on lens or head tissue collected by laser capture microdissection. Control tail DNA was collected from a weanling E2f3LoxP/- mouse. (C) E13.5 lenses of E2f1-/-;E2f2-/-;E2f3LoxP/LoxP(control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) mice were stained by H&E to examine nuclear distribution and lens architecture. Blue arrow indicates direction of epithelial migration toward equatorial line; ant, anterior; pos, posterior; ep, epithelium; fi, fiber cells; ir, iris. (D) Immunodetection of BrdU incorporation and phosphorylated histone H3 show normal patterns of DNA synthesis and proliferation in E16.5 control and cry-cre lenses. Epithelial cells positive for BrdU or phosphohistone H3 stain red. Nuclei are stained by DAPI in blue (top quadrants). (E) The percentage of BrdU-positive epithelial cells in lenses at the indicated ages. A minimum of three sections near the central plane of cry-cre lenses were analyzed at E13.5 (n=5), E16.5 (n=7), P1 (n=7), P7 (n=3), and P16 (n=3) and of control lenses at E13.5 (n=4), E16.5 (n=2), P1 (n=2), P7 (n=3), and P16 (n=4). Error bars represent standard deviation and significance of unpaired T-test is indicated by ** p < 0.01.
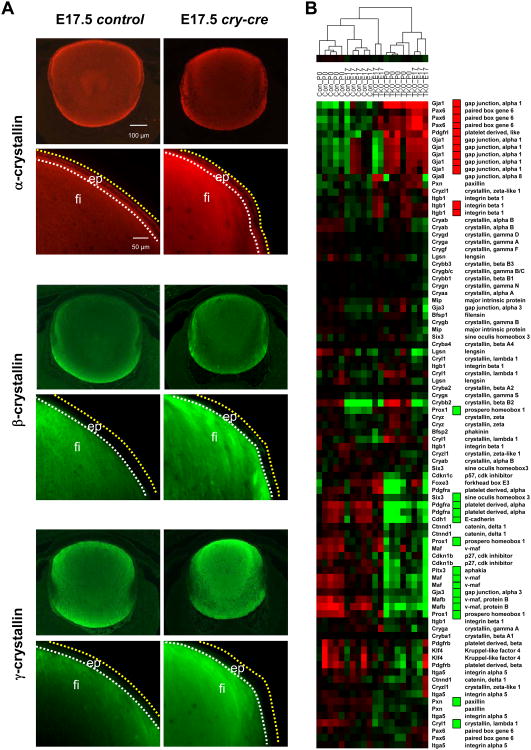
Perinatal differentiation appeared to be relatively normal, as measured by the expression and distribution of α-, β-, and γ-crystallin proteins in TKO lenses (Fig. 2A); however, careful inspection of a panel of classical markers of lens differentiation and associated factors by microarray revealed significant changes in gene expression of a number of genes required for proper lens development (Fig. 2B). At E17.5, a master regulator of eye development, Pax6, was increased by approximately 1.5- to 2-fold in TKO lenses (p < 0.001, Fig. 2B). Targeted overexpression of Pax6 in mice causes anomalies in fiber cell morphology and packing, increases in proteins involved in cell signaling and adhesion, and eventual breakdown of cellular structure (Duncan et al., 2000). Mutation of Pax6 in human patients has also been found to be responsible for ocular disorders such as aniridia and cataract (Cai et al., 2010; Hanson et al., 1999). Additionally, we found significant downregulation of a tumor suppressor essential for proper cell cycle exit and fiber cell differentiation, Prox1. Prox1 activates Cdk inhibitors, p27Kip1 and p57Kip2, in lens epithelium at the transition zone and its inactivation causes ectopic DNA synthesis and cycling in lens fiber cells (Wigle et al., 1999). Interestingly, both Pax6 and Prox1 have several E2F consensus binding sites upstream of their transcriptional start sites. By P0, many more genes critical for differentiation and cell cycle control were altered in the TKO lens, including Gja1, Pdgfrl, Cryl1, Pitx3, Pdgfra, Gja3, and Six3, (p < 0.001). In fact, a host of other genes required for proper lens development were consistently altered, most notably genes downstream of Pax6 (Itgb1 and Pxn), Prox1 (Cdkn1b (p27Kip1)), and others required for proper differentiation, fiber cell elongation, and cell survival, such as Foxe3, Cdh1, Maf, and Mafb (p < 0.01, Fig. 2B). Together, these observations illustrate that E2F1-3 are not essential for the proliferation of lens epithelial stem cells or progenitors but they may be required for terminal differentiation.
Fig 2. Loss ofE2f1-3 causes transcriptional deregulation of a subset of lens-specific genes.
(A) Immunostaining of α-, β-, and γ-crystallin in control and cry-cre lenses. The distribution of positive staining ranges from the anterior face of the lens to the transition zone. β- and γ-crystallins are only detectable in the fiber cell compartment. Low magnification images of immunostaining with α-, β-, and γ-crystallin antibodies shows specificity for lens. Surrounding retina provides reference for background levels of fluorescence. (B) Lens differentiation markers were analyzed by hierarchical clustering of gene expression and visualized by heatmap. Statistically significant changes (p < 0.001) were determined by class comparison analysis and are indicated by red boxes (upregulated) and green boxes (downregulated).
Loss of E2F1-3 causes DNA double-strand breaks and apoptosis
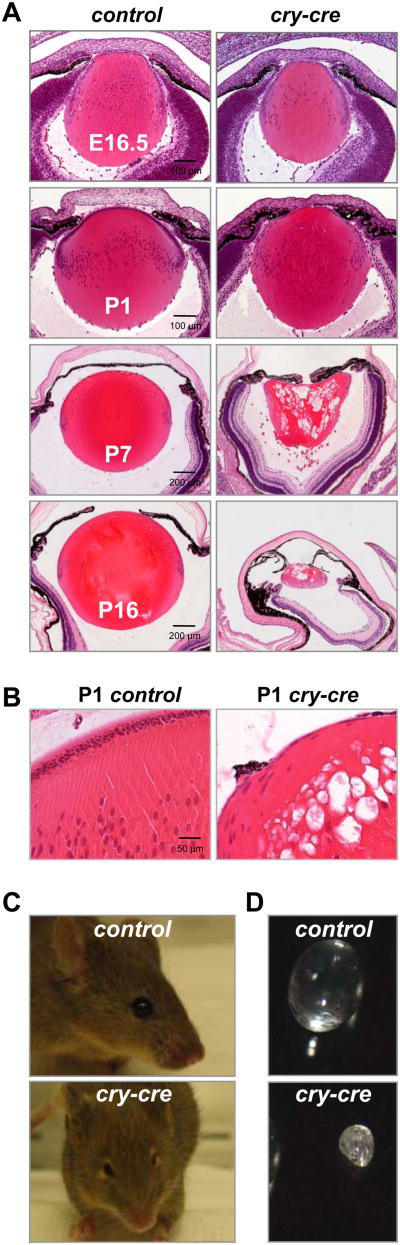
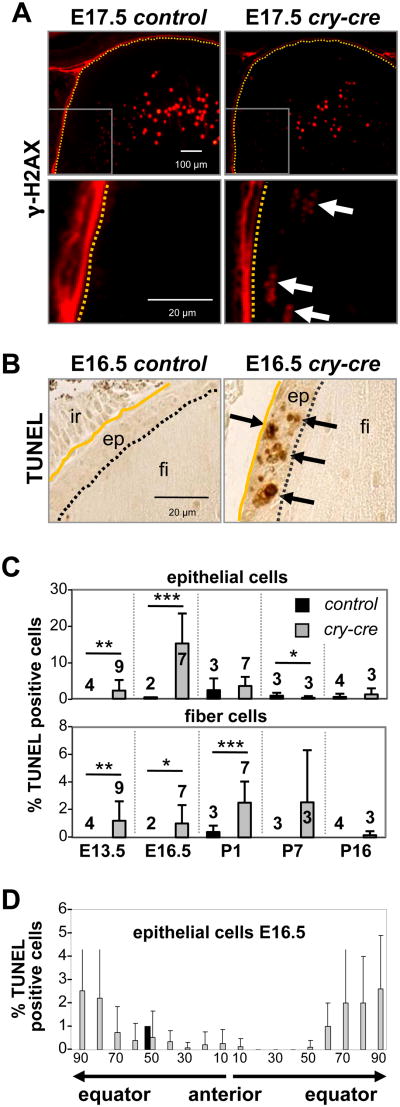
Soon after birth (P0), TKO lenses exhibited signs of cellular breakdown and hypotrophy within the fiber compartment (Fig. 3A, 3B, S5B). After 7 days of development (P7), TKO lenses had undergone extensive vacuolation, and at just over 2 weeks old (P16), lenses were clouded, shriveled and approximately one-eighth the size of control lenses (Fig. 3A-D, S5B). Not surprisingly, many of these features resembled murine models of Pax6 overexpression (Duncan et al., 2000). Given the protracted nature of TKO lens breakdown, we examined the integrity of genomic DNA and apoptosis in TKO lenses prior to birth, before the manifestation of these phenotypes. Immunohistochemical staining of E13.5 lenses showed a marked increase in cells that stained positive for phosphorylated histone H2AX (γ-H2AX). Positivity for γ-H2AX was particularly acute in newly differentiating fiber cells at the equatorial plane of the TKO lenses, whereas γ-H2AX positivity was high in both TKO and control lenses during late stages of fiber cell differentiation (Fig. 4A). DNA double-strand breaks in terminally differentiating fiber cells within the lens cortex are presumably a result of normal nuclear degeneration that must occur for maturation of the translucent fibers. Apoptotic cell death, as measured by TUNEL and cleaved caspase-3 immunofluorescence, was apparent in the epithelium and fiber compartments of E13.5 TKO lenses but not in control lenses (Fig. 4B, 4C, and Fig. S5C). At E16.5, apoptosis was most pronounced near the equatorial transition zone of E16.5 TKO lenses, consistent with the distribution of γ-H2AX-positive cells (Fig. 3D). As a result, the anterior perimeter of E17.5 TKO lens sections contained 36% fewer epithelial cells than control lenses (p = 0.0002; Fig. S5D). From these observations, we conclude that postnatal architectural collapse of TKO lenses is accompanied by massive breakdown of genetic material and initiation of an apoptotic program.
Fig. 3. Loss of E2F1-3 causes degradation and opacification of the lens.
(A) Hematoxylin and eosin staining of lenses from of E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) embryos (E16.5) and neonates (P1, P7 and P16). (B) High magnification of P1 control and cry-cre lenses. Note the prominent vacuolation in the cortex of the TKO fiber compartment. (C) control and cry-cre mice and (D) dissected lenses from control and cry-cre mice at 3 weeks old. Normal adult lenses are spherical and transparent, whereas TKO lenses are distinguished by decreased size, irregular surface and opacity (cataract).
Fig. 4. Loss of E2F1-3 causes increased cell death without affecting differentiation.
(A) Epithelial cells of E17.5 E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) lenses stained with antibodies for γ-phosphorylated H2AX, a form of H2AX protein recruited to DNA double-strand breaks. Note that H2AX- positivity is present during normal degradation of nuclear contents, required for organelle degradation and maturation of lens fiber cells. Aberrant positivity was observed near the bow region of the mutant lenses. (B) TUNEL staining at E16.5 shows apoptosis in the lens. ep, epithelium; fi, fiber cells; ir, iris. (C) Quantification of the percentage of TUNEL-positive cells shows elevated levels of cell death in the epithelium and fiber compartment. A minimum of three sections near the central plane were analyzed for cry-cre lenses (grey bars) at E13.5 (n=9), E16.5 (n=7), P1 (n=7), P7 (n=3), and P16 (n=3) and for control lenses (black bars) at E13.5 (n=4), E16.5 (n=2), P1 (n=3), P7 (n=3), and P16 (n=4). ep, epithelium; fi, fiber cells; ir, iris. Error bars represent standard deviation. Significance of unpaired T-test indicated by * p < 0.05, ** p < 0.01, and *** p < 0.001. (D) Spatial distribution of TUNEL-positive cells in control lenses (black bars) and cry-cre lenses (grey bars); note the increased apoptosis near the transition zones of cry-cre lens equators.
E2F target gene expression is upregulated with ablation of E2F “activators”
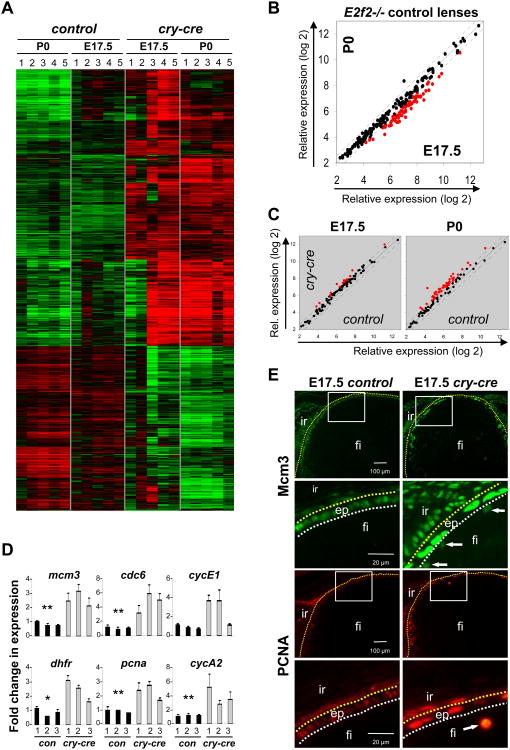
In order to identify the molecular pathways impacted by loss of E2F1-3 in vivo, we compared global gene expression profiles in control (E2f2-/-) and TKO lenses at two developmental stages, E17.5 and P0, which precede the physical breakdown of the mutant lens. We implemented unsupervised class comparison analyses of all probesets on the array to identify genes differentially expressed between lenses. We also used an unbiased approach similar to Gene Set Enrichment Analysis (Mootha et al., 2003) to identify a priori defined groups of genes that were significantly differentially expressed. Expression changes observed among samples were attributable to two variables: developmental time (E17.5 vs. P0) and genotype (control vs. TKO). Comparison of control lenses at the two developmental stages identified 408 genes that were downregulated and 174 that were upregulated in P0 lenses (> 1.5 fold and p < 0.001) (Fig. 5A, Fig. S3). The expression of known E2F target genes, as defined by expression and chromatin immunoprecipitation assays, decreased with age, consistent with a decrease in proliferation as lenses develop from E17.5 to birth (Black et al., 2005; Bracken et al., 2004; Kong et al., 2007; Ren et al., 2002; Vernell et al., 2003; Weinmann et al., 2002; Wells et al., 2002; Xu et al., 2007) (Fig. 5B, Fig. S2A). Comparison between control and TKO lenses revealed a significant number of gene expression changes at both developmental time points analyzed (Fig. 5A, Fig. S2, S4). Significantly changed gene ontology categories illustrated a bias for differential expression of genes involved in metabolism and DNA processing (Eisen et al., 1998) (Least square and Kolmogorov-Smirnov permutation values of p < 0.001) (Fig. S2B, S4). Genes that were downregulated in TKO lenses, depicted in the lower third of the heatmap in Figure 5A,were enriched for functions in the mitochondrion and oxidation reduction at E17.5 and, atP0, for roles in the extracellular region, including proteins critical for cell adhesion and structural matrix, such as thrombospondin, procollagen, albumin, nidogen, perlecan, and laminin (Fig. S6). In contrast, genes contributing to DNA replication were uniformly upregulated in TKO tissues, including E2F targets such as chromatin licensing and DNA replication factor 1(Cdt1) and the minichromosome maintenance deficient genes (Mcm2,Mcm3, Mcm4, Mcm5, Mcm7, Mcm8). In fact, expression of nearly all E2F target genes was significantly higher in TKO than in control lenses at both E17.5 and P0 (Fig. 5C, 5D). Quantitative analysis of gene expression by real-time RT PCR was used to validate a subset of changes identified by Affymetrix oligo arrays (Fig. 5D). To characterize the spatial nature of E2F target misexpression, lenses were sectioned and probed with antibodies specific for several known downstream effectors of E2F. As shown in Figure 5E, Mcm3 and PCNA protein levels were undetectable in control E17.5 lens fibers, whereas in TKO tissues these proteins were apparent in fiber cells and conspicuously elevated in the epithelium. Together, the gene expression data and protein immunostaining of E2F targets points to a central role for E2F1-3 in transcriptional repression.
Fig. 5. E2F target genes are upregulated in lenses triply deleted for E2F1-3.
(A) Heatmap of hierarchical cluster analysis of key genes upregulated (red) or downregulated (green) in the lens at the indicated ages and genotypes (p < 0.0001 and greater than 1.5-fold change, n = 5 for each group). (B) Scatter plot of known E2F target genes shows downregulation of some genes (highlighted in red) in E2f2-/- (control) lenses with increasing age from E17.5 to P0. (C) Scatter plots comparing the levels of known E2F target genes in cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) and E2f2-/- (control) lenses. Red points represent those genes that are upregulated more than 1.5 fold. (D) Quantitative RT-PCR at E17.5 was used to confirm upregulation of differentially regulated genes in control and cry-cre lenses. Statistical significance of unpaired T-test is indicated * p < 0.05 and ** p < 0.01. (E) Photomicrographs of lens sections stained with Mcm3- and PCNA-specific antibodies; immunofluorescence shows increased intensity of staining in lens epithelium and ectopic expression within the fiber compartment (arrows). ep, epithelium; fi, fiber cells; ir, iris.
Activation of pro-apoptotic gene expression in the p53 pathway
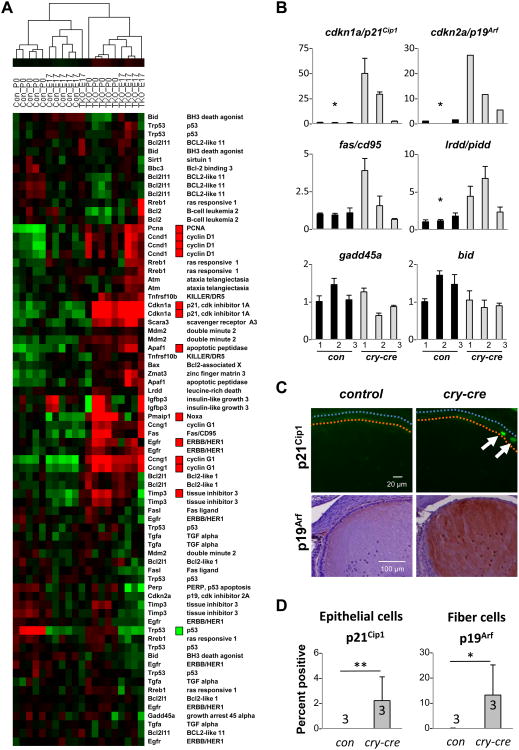
Previously, we identified a role for E2F in regulation of the p53-p21Cip1 axis (Sharma et al., 2006; Timmers et al., 2007). This relationship between E2F ablation and upregulation of p53 activity was further supported by recent evidence implicating the deacetylase Sirtuin 1 as a critical modulator of p53 in the murine retina (Chen et al., 2009). In the current study, we observe increased expression of a number of regulators and downstream effectors of p53 (Fig.6A). Most notably, p21Cip1 and p19Arf were significantly upregulated in TKO lenses (Fig. 6B-D). Ectopic expression of p21Cip1 protein was observed in the epithelium, loosely corresponding to an area approximately 60 degrees above the lens equator (just anterior to TUNEL-positive enrichment depicted in Fig. 4D) (Fig. 6C). In contrast, p19Arf was localized to the fiber compartment, and appeared throughout the fiber cytoplasm and in the nuclei of nucleated lens fibers (Fig. 6C). The fiber-specific distribution of p19Arf is intriguing, not only for its recognized role in stabilizing p53 through inhibition of Mdm2, but also as a modulator of proliferation. It has been demonstrated in the murine eye that p19Arf regulates Pdgf signaling, a pathway essential for cycling of the lens epithelium. Pdgfra and Pdgfrl are both misexpressed in TKO lenses, suggesting either that E2F directly regulates these genes or that p19Arf is playing a role in suppression of cell cycling via Pdgfr modulation (Fig. 2B). This cascade could be stimulated by signals initiated by p21Cip1 in the epithelium or by ectopic proliferation and DNA synthesis in the fiber compartment (Fig. 4C, 6C).
Fig. 6. Cell cycle inhibitors are activated by loss of E2F.
(A) Heatmap rendering of changes in p53 signaling as determined by global geneexpression profiling. Statistically significant changes (p < 0.001) that were 1.5-fold orgreater were determined by class comparison analysis and are indicated by red boxes (upregulated) and green boxes (downregulated). (B) Gene expression changes in key p53targets were validated by realtime RT PCR. Statistical significance of unpaired T-test isindicated * p < 0.05. (C) Immunodetection of p19Arf and p21Cip1 on lens sectionsconfirms activation of these downstream targets of p53. (D) Quantification ofimmunostaining of p21Cip1 in epithelial cells and of p19Arf in fiber cells. Statistical significance of unpaired T-test is indicated * p < 0.05 and ** p < 0.01.
Rb deficiency provides context for E2F's role in proliferation
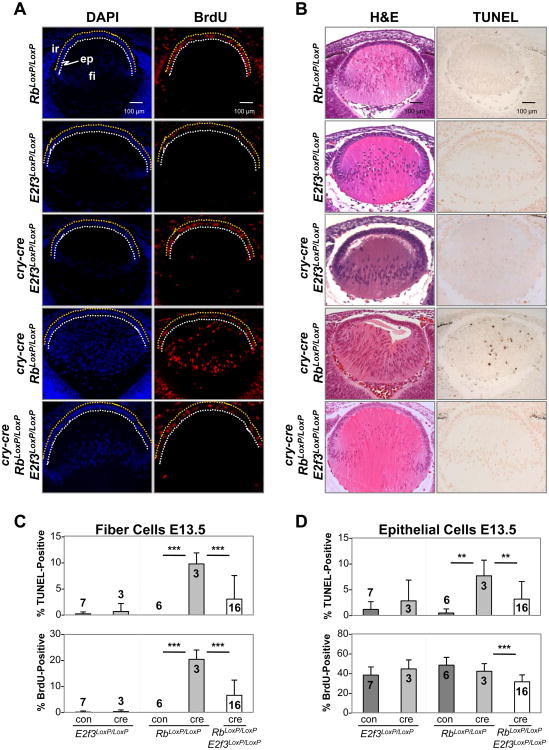
The Rb tumor suppressor is a known regulator of E2Fs that is critical for the balanced proliferation of epithelial cells of the lens (Jacks et al., 1992). Rb-null embryos exhibit ectopic cell proliferation in multiple tissues, including lens cells destined to exit the cell cycle at the equatorial plane. The loss of E2f3 suppresses the ectopic proliferation in Rb-deficient lenses (Saavedra et al., 2002; Ziebold et al., 2001) without impacting overall development of the lens, presumably because other E2F family members could compensate for loss of E2F3 by providing redundant functions needed for basal levels of cell proliferation in Rb/E2f3 doubly-deficient lenses. While these findings have been interpreted as evidence that E2f3 is critical for cell proliferation, this inference disregards the possibility that the requirements for normal and ectopic cell proliferation caused by Rb deficiency could differ. Thus, our current observation that E2F1-3 repress E2F targets and are dispensable for cell proliferation appears to contradict the general conclusions drawn from previous analyses of lenses that were singly or doubly deficient for Rb and E2f3 (Saavedra et al., 2002; Ziebold et al., 2001). We therefore sought to explore the mechanistic relationship between Rb and E2F3 using the cry-cre in vivo system, such that results could be uniformly compared across different genetic configurations. We found that deletion of Rb in cry-cre;RbLoxP/LoxP embryos resulted in ectopic proliferation and apoptosis of fiber cells, culminating in a profound disruption of lens architecture (Fig. 7A-D). As shown in Figure 5, loss of E2f3 suppressed the ectopic proliferation and apoptosis of Rb mutant fiber cells, but did not decrease proliferation or cell numbers below that observed in wild type lenses (Fig. 7C, 5D). These results parallel previous observations in Rb-/- embryos (Saavedra et al., 2002; Ziebold et al., 2001) but refine and extend the analysis by tissue-specific ablation to demonstrate that Rb functions in the lens are cell autonomous. Together with the analysis of E2f triply-deficient lenses, these latter results support the idea that E2F3 promotes ectopic cell proliferation specifically in the context of Rb deficiency.
Fig. 7. Cry-cre mediated deletion of RbLoxP recapitulates hallmark phenotypes of Rb-null lenses and confirms cell autonomous function of Rb in the E13.5 lens.
(A) Immunodetection of BrdU incorporation illustrates the dramatic defect in the pattern of DNA synthesis in cry-cre; RbLoxP/LoxP lenses. Cellular organization within the fiber compartment is restored by concurrent loss of E2f3. ep, epithelium; fi, fiber cells; ir, iris. TUNEL detection of apoptotic cells and staining with hematoxylin and eosin show an almost complete rescue of the Rb phenotype by loss of E2f3. (C) The percentage of TUNEL- and BrdU-positive cells in lenses of indicated genotypes shows a profound increase in cell death and proliferation in fiber cells of cry-cre; RbLoxP/LoxP but not cry-cre; RbLoxP/LoxP; E2f3LoxP/LoxP lenses. (D) Quantification of the percentage of TUNEL- and BrdU-positive cells in the lens epithelium. Error bars represent standard deviation. Significance of unpaired T-test indicated by ** p < 0.01 and *** p < 0.001.
Discussion
Contrary to many previous studies performed in cell culture, the data presented here suggest that E2F1-3 are not required for proliferation of lens epithelial cells (Sharma et al., 2006; Timmers et al., 2007; Wu et al., 2001). Instead, we show that E2F1-3 play a critical role in cell survival. It remains to be determined whether E2Fs are required in other tissues of the adult mouse, but it would appear from parallel analyses of neuroectodermal derivatives of the retina and endodermal derivatives of the small intestine (Chen et al., 2009; Chong et al., 2009) that epithelial lineages in general do not require E2F1-3 to proliferate but do require them for cell survival. Indeed, it is also very likely that the majority of embryonic cell types do not require E2F1-3 for proliferation since triple mutant embryos can survive to E9.5 (Chong et al., 2009).
Since lens morphogenesis proceeds normally in the absence of E2F1-3 until late gestation, it is unlikely that the transcriptional control of fiber cell differentiation is disrupted prior to this stage. Primary fiber cell differentiation appears to have taken place normally and major crystallin proteins are present with no noticeable disruption of distribution within the lens. In late gestation, however, there are some significant changes in the abundance of key transcripts in lens development (Fig 2B). Most notably, lens epithelial cell associated transcripts (Pax6 and connexin 43 (Gja1)) were elevated and lens fiber cell associated transcripts (Prox1 and connexin 46 (Gja3)) were reduced. These changes in gene expression may represent a fundamental shift in the requirement of E2Fs in secondary versus primary fiber cell differentiation, or may result as a secondary effect of decreased lens cell survival. Whether any of these transcripts represent direct E2F target genes will require further investigation.
Our results support the view that E2F1-3 family members normally function as transcriptional repressors at a time in development when cells are exiting the cell cycle (Aslanian et al., 2004; Leone et al., 2000). We show that loss of E2F1-3 in the lens results in ectopic expression of E2F target genes, many of which are required for nucleotide metabolism and DNA synthesis. It is interesting to note that upregulation of Mcm3 and PCNA was particularly acute in cells transitioning to G0 and that the Cdk inhibitors p27 and p57 were downregulated (Fig. 2B, 5E). Further, this inappropriate expression of E2F targets was accompanied by a doubling of the number of epithelial cells incorporating BrdU and the appearance of DNA replication in fiber cells that are quiescent in normal lenses (Fig. 1D). Given recent work establishing redundant roles for E2Fs in embryonic development (Hurst et al., 2008; Tsai et al., 2008), we would suggest that overlapping functions of E2F1-3 lie primarily in gene repression.
The analysis of Rb and Rb-E2f3 mutant lenses suggests that the previously described requirement for E2Fs in proliferation and transcriptional activation may be restricted to specific cellular contexts where Rb protein is inactivated. Such situations could include abnormal proliferation in response to genetic alterations in Rb or normal proliferation induced in response to liver damage, immune cell activation, and acute growth factor activation. Earlier work has characterized the lenses of Rb-E2F3-deficient embryos (Saavedra et al., 2002; Ziebold et al., 2001), yet the conditional deletion strategy here demonstrates for the first time that the apoptotic defect found in Rb-null lenses is not a secondary defect caused by placental deficiencies (de Bruin et al., 2003; Wenzel et al., 2007; Wu et al., 2003) but rather is due to a cell autonomous requirement for Rb function to balance transcriptional activation by E2F3. In summary, the analysis of TKO lenses provides compelling in vivo evidence for a role of E2F1, E2F2 and E2F3 in transcriptional repression and cell survival during normal developmental programs, and for a role in transcriptional activation and cell proliferation during specialized circumstances restricted to inactivation of Rb and its associated repressive cofactors, including histone deacetylases, SWI/SNF, lysine methyltransferases, arginine methyltransferases, and DNA methyltransferases.
Supplementary Material
Fig. S1. Primers used for real-time RT-PCR and genotyping.
(A) List of validated E2F target genes used for analysis of microarray data. Genes were identified as bona fide E2F targets only if confirmed by reports of direct regulation and binding as demonstrated by chromatin immunoprecipitation assays. (B) Gene ontology (GO) categories representing genes differentially expressed between control and cry-cre lenses include those involved in metabolism and DNA processing. GO categories are classified as Biological Process (BP), Molecular Function (MF), or Cellular Component (CC).
Fig. S3. Tables of differentially expressed genes in control and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses as determined by class comparison analyses.
Fig. S4. Tables of gene ontology categories representing genes differentially expressed between control and cry-cre lenses.
(A) BrdU incorporation as a measure of DNA synthesis in E13.5 lenses deficient for various combinations of E2f1, E2f2, and E2f3. Numbers of embryos analyzed are indicated within value bars. (B) Number of embryos recovered for analysis of phenotype penetrance. (C) Immunostaining for cleaved caspase-3 in E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) lenses show that deletion of E2F1-3 induces an apoptosis in epithelial cells approaching the transition zone. ep, epithelium; fi, fiber cells; ir, iris. (D) The total number of epithelial cells per section in cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses is reduced from the normal number found in E2f2-/- control lenses. A minimum of 4 sections were counted for each embryo to acquire total epithelial cell numbers (n=3 embryos for cre-cry and n=3 embryos for control). Statistical significance of unpaired T-test is indicated * p < 0.05.
Groups of genes in the mitochondrion, oxidation reduction, and extracellular region gene ontology categories are depicted with probe set identification and gene names.
Acknowledgments
We thank J. Moffitt and L. Rawahneh for histological support. This work was funded by NIH grants to G.L. (R01CA85619, R01CA82259, R01HD047470, P01CA097189) and M.L.R. (R01EY012995) and an NIH training grant to P.L.W. (5 T32 CA106196-04). G.L. is the recipient of The Pew Charitable Trust Scholar Award and the Leukemia & Lymphoma Society Scholar Award.
Footnotes
The authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aslanian A, Iaquinta PJ, Verona R, Lees JA. Repression of the Arf tumor suppressor by E2F3 is required for normal cell cycle kinetics. Genes Dev. 2004;18:1413–1422. doi: 10.1101/gad.1196704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black EP, Hallstrom T, Dressman HK, West M, Nevins JR. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc Natl Acad Sci U S A. 2005;102:15948–15953. doi: 10.1073/pnas.0504300102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29:409–417. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Cai F, Zhu J, Chen W, Ke T, Wang F, Tu X, Zhang Y, Jin R, Wu X. A novel PAX6 mutation in a large Chinese family with aniridia and congenital cataract. Mol Vis. 2010;16:1141–1145. [PMC free article] [PubMed] [Google Scholar]
- Chen D, Pacal M, Wenzel P, Knoepfler PS, Leone G, Bremner R. Division and apoptosis of E2f-deficient retinal progenitors. Nature. 2009;462:925–929. doi: 10.1038/nature08544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JL, Wenzel PL, Saenz-Robles MT, Nair V, Ferrey A, Hagan JP, Gomez YM, Sharma N, Chen HZ, Ouseph M, Wang SH, Trikha P, Culp B, Mezache L, Winton DJ, Sansom OJ, Chen D, Bremner R, Cantalupo PG, Robinson ML, Pipas JM, Leone G. E2f1-3 switch from activators in progenitor cells to repressors in differentiating cells. Nature. 2009;462:930–934. doi: 10.1038/nature08677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the Rb and polycomb pathways. Mol Cell. 2001;8:557–569. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- de Bruin A, Wu L, Saavedra HI, Wilson P, Yang Y, Rosol TJ, Weinstein M, Robinson ML, Leone G. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci. 2003;100:6546–6551. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan MK, Kozmik Z, Cveklova K, Piatigorsky J, Cvekl A. Overexpression of PAX6(5a) in lens fiber cells results in cataract and upregulation of (alpha)5(beta)1 integrin expression. J Cell Sci. 2000;113(Pt 18):3173–3185. doi: 10.1242/jcs.113.18.3173. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8:532–535. doi: 10.4161/cc.8.4.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- Hurst CD, Tomlinson DC, Williams SV, Platt FM, Knowles MA. Inactivation of the Rb pathway and overexpression of both isoforms of E2F3 are obligate events in bladder tumours with 6p22 amplification. Oncogene. 2008;27:2716–2727. doi: 10.1038/sj.onc.1210934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Kong LJ, Chang JT, Bild AH, Nevins JR. Compensation and specificity of function within the E2F family. Oncogene. 2007;26:321–327. doi: 10.1038/sj.onc.1209817. [DOI] [PubMed] [Google Scholar]
- Leone G, Nuckolls F, Ishida S, Adams M, Sears R, Jakoi L, Miron A, Nevins JR. Identification of a novel E2F3 product suggests a mechanism for determining specificity of repression by Rb proteins. Mol Cell Biol. 2000;20:3626–3632. doi: 10.1128/mcb.20.10.3626-3632.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovicu FJ, Steven P, Saika S, McAvoy JW. Aberrant lens fiber differentiation in anterior subcapsular cataract formation: a process dependent on reduced levels of Pax6. Invest Ophthalmol Vis Sci. 2004;45:1946–1953. doi: 10.1167/iovs.03-1206. [DOI] [PubMed] [Google Scholar]
- Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Morris L, Allen KE, La Thangue NB. Regulation of E2F transcription by cyclin E-Cdk2 kinase mediated through p300/CBP co-activators. Nat Cell Biol. 2000;2:232–239. doi: 10.1038/35008660. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Robinson ML, Overbeek PA, Verran DJ, Grizzle WE, Stockard CR, Friesel R, Maciag T, Thompson JA. Extracellular FGF-1 acts as a lens differentiation factor in transgenic mice. Development. 1995;121:505–514. doi: 10.1242/dev.121.2.505. [DOI] [PubMed] [Google Scholar]
- Rowland BD, Denissov SG, Douma S, Stunnenberg HG, Bernards R, Peeper DS. E2F transcriptional repressor complexes are critical downstream targets of p19(ARF)/p53-induced proliferative arrest. Cancer Cell. 2002;2:55–65. doi: 10.1016/s1535-6108(02)00085-5. [DOI] [PubMed] [Google Scholar]
- Saavedra HI, Wu L, de Bruin A, Timmers C, Rosol TJ, Weinstein M, Robinson ML, Leone G. Specificity of E2F1, E2F2 and E2F3 in mediating Rb function. Cell Growth Differ. 2002;13:215–225. [PubMed] [Google Scholar]
- Sharma N, Timmers C, Trikha P, Saavedra HI, Obery A, Leone G. Control of the p53-p21CIP1 Axis by E2f1, E2f2, and E2f3 is essential for G1/S progression and cellular transformation. J Biol Chem. 2006;281:36124–36131. doi: 10.1074/jbc.M604152200. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Timmers C, Sharma N, Opavsky R, Maiti B, Wu L, Wu J, Orringer D, Trikha P, Saavedra HI, Leone G. E2f1, E2f2, and E2f3 control E2F target expression and cellular proliferation via a p53-dependent negative feedback loop. Mol Cell Biol. 2007;27:65–78. doi: 10.1128/MCB.02147-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SY, Opavsky R, Sharma N, Wu L, Naidu S, Nolan E, Feria-Arias E, Timmers C, Opavska J, de Bruin A, Chong JL, Trikha P, Fernandez SA, Stromberg P, Rosol TJ, Leone G. Mouse development with a single E2F activator. Nature. 2008;454:1137–1141. doi: 10.1038/nature07066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernell R, Helin K, Muller H. Identification of target genes of the p16INK4A-pRB-E2F pathway. J Biol Chem. 2003;278:46124–46137. doi: 10.1074/jbc.M304930200. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells J, Graveel CR, Bartley SM, Madore SJ, Farnham PJ. The identification of E2F1-specific target genes. Proc Natl Acad Sci U S A. 2002;99:3890–3895. doi: 10.1073/pnas.062047499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel PL, Wu L, de Bruin A, Chong JL, Chen WY, Dureska G, Sites E, Pan T, Sharma A, Huang K, Ridgway R, Mosaliganti K, Sharp R, Machiraju R, Saltz J, Yamamoto H, Cross JC, Robinson ML, Leone G. Rb is critical in a mammalian tissue stem cell population. Genes Dev. 2007;21:85–97. doi: 10.1101/gad.1485307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Wu L, de Bruin A, Saavedra HI, Starovic M, Trimboli A, Yang Y, Opavska J, Wilson P, Thompson JC, Ostrowski MC, Rosol TJ, Woollett L, Weinstein M, Cross JC, Robinson ML, Leone G. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414:457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]
- Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, Luo RX, Harbour JW, Dean DC. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45:1930–1939. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
- Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001;15:386–391. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Primers used for real-time RT-PCR and genotyping.
(A) List of validated E2F target genes used for analysis of microarray data. Genes were identified as bona fide E2F targets only if confirmed by reports of direct regulation and binding as demonstrated by chromatin immunoprecipitation assays. (B) Gene ontology (GO) categories representing genes differentially expressed between control and cry-cre lenses include those involved in metabolism and DNA processing. GO categories are classified as Biological Process (BP), Molecular Function (MF), or Cellular Component (CC).
Fig. S3. Tables of differentially expressed genes in control and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses as determined by class comparison analyses.
Fig. S4. Tables of gene ontology categories representing genes differentially expressed between control and cry-cre lenses.
(A) BrdU incorporation as a measure of DNA synthesis in E13.5 lenses deficient for various combinations of E2f1, E2f2, and E2f3. Numbers of embryos analyzed are indicated within value bars. (B) Number of embryos recovered for analysis of phenotype penetrance. (C) Immunostaining for cleaved caspase-3 in E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (control) and cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP (cry-cre) lenses show that deletion of E2F1-3 induces an apoptosis in epithelial cells approaching the transition zone. ep, epithelium; fi, fiber cells; ir, iris. (D) The total number of epithelial cells per section in cry-cre;E2f1-/-;E2f2-/-;E2f3LoxP/LoxP lenses is reduced from the normal number found in E2f2-/- control lenses. A minimum of 4 sections were counted for each embryo to acquire total epithelial cell numbers (n=3 embryos for cre-cry and n=3 embryos for control). Statistical significance of unpaired T-test is indicated * p < 0.05.
Groups of genes in the mitochondrion, oxidation reduction, and extracellular region gene ontology categories are depicted with probe set identification and gene names.