Abstract
Gene silencing agents such as small interfering RNA (siRNA) and microRNA offer the promise to modulate expression of almost every gene for the treatment of human diseases including cancer. However, lack of vehicles for effective systemic delivery to the disease organs has greatly limited their in vivo applications. In this study, we developed a high capacity polycation-functionalized nanoporous silicon (PCPS) platform comprised of nanoporous silicon microparticles functionalized with arginine-polyethyleneimine inside the nanopores for effective delivery of gene silencing agents. Incubation of MDA-MB-231 human breast cancer cells with PCPS loaded with STAT3 siRNA (PCPS/STAT3) or GRP78 siRNA (PCPS/GRP78) resulted in 91% and 83% reduction of STAT3 and GRP78 gene expression in vitro. Treatment of cells with a microRNA-18a mimic in PCPS (PCPS/miR-18) knocked down 90% expression of the microRNA-18a target gene ATM. Systemic delivery of PCPS/STAT3 siRNA in murine model of MDA-MB-231 breast cancer enriched particles in tumor tissues and reduced STAT3 expression in cancer cells, causing significant reduction of cancer stem cells in the residual tumor tissue. At the therapeutic dosage, PCPS/STAT3 siRNA did not trigger acute immune response in FVB mice, including changes in serum cytokines, chemokines and colony-stimulating factors. In addition, weekly dosing of PCPS/STAT3 siRNA for four weeks did not cause signs of sub-acute toxicity based on changes in body weight, hematology, blood chemistry, and major organ histology. Collectively, the results suggest that we have developed a safe vehicle for effective delivery of gene silencing agents.
Keywords: porous silicon, siRNA, microRNA, delivery, cancer therapy
Drug development has traditionally been focused on a limited number of targets such as enzymes, transporters, G-protein-coupled receptors, and secreted proteins.1 Most gene products are deemed as nondruggable since it is difficult to develop assays to measure their activities, and hence to carry out drug screening. As a result, many important proteins have been excluded from drug development efforts. This is especially true for cancer drug development. Many key cancer-causing genes encode transcription factors and these involved in protein-protein interaction.2-4 The discovery of RNA interference (RNAi) has opened the door for the development of a new class of therapeutic agents for the treatment of human diseases.5 RNAi is considered to have the capability of knocking down any gene in any cell type. Consequently, a large number of gene silencing agents including small inhibitory RNAs (siRNA) and microRNAs are being evaluated for their anti-cancer activities.
As unformulated nucleotide oligos are rapidly degraded in body fluids, they are delivered in nanocarriers to enable the gene silencing agents to reach the designated tissues and exert function in vivo. The most commonly used nanocarriers for delivery of cancer therapeutics include liposomes and polymer-based nanoparticles with a size of less than 200 nanometers.6 The leaky vasculature in the tumor tissue enables nanoparticles within a size range of 100-500 nm to permeate into the tumor interstitium, taking advantage of the enhanced permeability and retention (EPR) effect.7,8 Furthermore, oligonucleotides are negatively charged and cannot pass the cytoplasmic membrane. Nanoformulation effectively facilitates cellular entry of the gene silencing agent.
Multiple siRNA-based candidate cancer drugs are currently being tested in clinical trials for efficacy and safety evaluation.6 Liver cancer and liver metastases of other cancer types are the primary disease indications for many of the candidate drugs,9 since the drug carriers tend to accumulate in the liver. Although promising advancement has been made in recent years on siRNA delivery, 10,11 there is still an urgent demand for the design and development of new delivery systems to target other organs. We have previously shown that porous silicon can be tailored to produce particles with defined size and shape.12 The destiny of the particles inside the body is determined by the size, shape, and surface physical and chemical properties. For example, discoidal particles are more effective in tumor accumulation than spherical or cylindrical particles,13 and the 1,000 × 400 nm discoidal particles accumulate more in tumor tissues and less in the liver or spleen than the 600 × 200 nm particles in the first 30 minutes after i.v. administration.14 In a recent study with a murine model of melanoma, it was determined that up to 5% of total injected dose of the 1 μm discoidal particles with polyamine surface modification were enriched in the tumor tissue after systemic delivery.14 Tumor accumulation has also been confirmed in murine models of primary breast cancer and metastatic ovarian cancer.15,16 In addition, systemic administration of the discoidal porous silicon particles does not cause acute or sub-acute toxicity in wild-type mice.15 These results suggest the discoidal particles can serve as an efficient carrier for drug delivery to breast cancer, ovarian cancer, melanoma, and possible other types of solid tumors.
We hypothesize that the discoidal porous silicon (pSi) particles can serve as delivery vehicles for the RNA-based gene silencing agents if the surface of the nanopores is conjugated with polycation such as arginine (Arg), chitosan, dendrimer, and polyethyleneimine (PEI). The polycation-functionalized porous silicon (PCPS) should have a high binding capacity for oligonucleotides. Confinement inside the nanopores will prevent the polycation-bound oligonucleotide from interacting with the toll-like receptors to trigger innate immune responses. Once inside the body, the porous silicon will accumulate in tumor vasculature and gradually dissolve, releasing the oligonucleotide-bound polycation from confinement to form polyplex nanoparticles which carry siRNA/microRNA to tumor interstitium. In the current study, we first conjugated 3-aminopropyl-triethoxysilane (APTES) onto the surface of porous silicon, and covalently attached Arg and PEI onto APTES sequentially. The PCPS, pSi-Arg-PEI, was then tested for siRNA binding capacity, releasing kinetics, and knockdown efficacy in cancer cells. A STAT3 gene-specific siRNA was used to test delivery to the tumor and knockdown of gene expression in a murine model of breast cancer. In addition, we tested potential innate immunotoxicity and sub-acute toxicity of the PCPS loaded with STAT3 siRNA. These studies confirmed that we have developed an efficient system for in vivo delivery of gene silencing agents.
RESULTS AND DISSCUSSION
Fabrication of the PCPS Delivery System
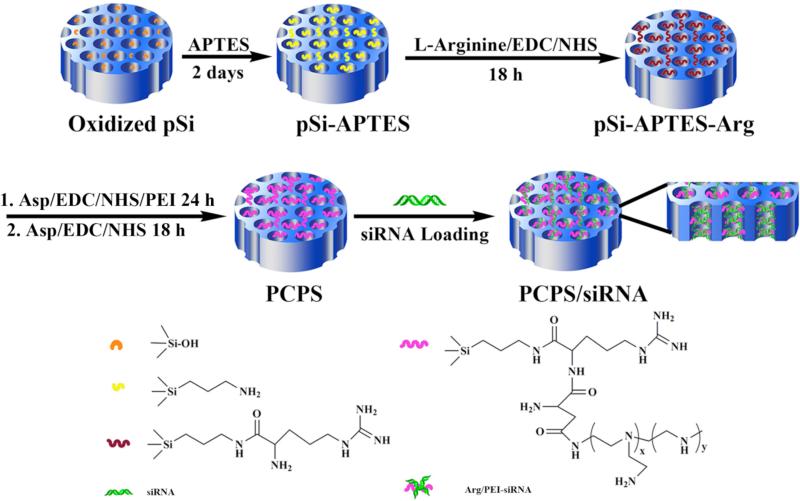
A four-step procedure for fabrication of the PCPS delivery vehicle is illustrated in Scheme 1. The surface of the porous silicon microparticle was first oxidized with H2O2/H2SO4 to expose a hydroxyl group that was used to conjugate APTES. Modification of porous silicon particle with APTES not only limits surface attack by water molecules and thus prevents the particle from rapid degradation, but also provides linkers for polycation conjugation. An Arg molecule was then conjugated to APTES, and the primary amino groups of PEI were subsequently attached to arginine. Loading of siRNA oligos into the nanopores was achieved through electrostatic interaction between the positively charged Arg-PEI and the negatively charged siRNA.
Scheme 1.
Schematic illustration of fabrication of PCPS as a delivery carrier for gene silencing agents.
Characterization of the PCPS Delivery System
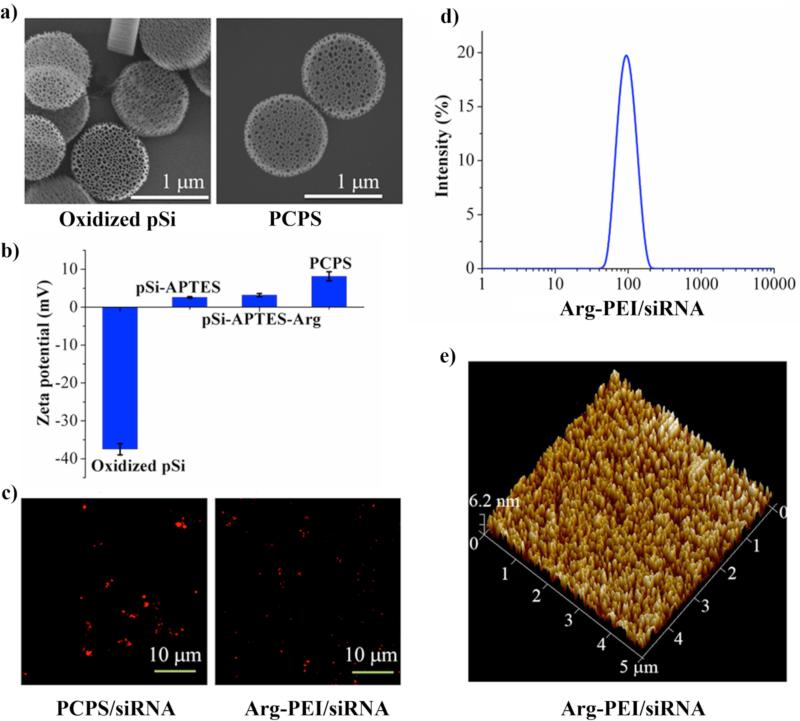
Scanning electron microscopy (SEM) was applied to analyze morphological changes of the original porous silicon particles and the PCPS particles (Figure. 1a). The 1 μm discoidal particles contain 45 to 80 nm nanopores with a porosity of about 80%.17 The nanopores with clearly defined structures and edges distributed evenly across the particle (Figure. 1a, left panel). They were partially filled in the final product PCPS (Figure. 1a, right panel), indicating a substantial amount of Arg-PEI was conjugated inside the pores. Surface chemical modification of the particles was confirmed by changes in surface charge (Figure. 1b). APTES modification brought the zeta potential from -37.5 mV to the positive territory, and Arg-PEI contributed significantly to the positive value of zeta potential as a result of the cooperative effect of PEI and the guanidine residue in Arg (+2.38 mV for pSi-APTES, +3.72 mV for pSi-APTES-Arg, and +8.18 mV for pSi-APTES-Arg-PEI). SEM analysis revealed that the PCPS particles were more stable than the unmodified pSi particles in phosphate buffer saline (PBS, Figure S1). PCPS degradation was minimal in the initial 4 days of incubation, comparing to massive degradation for pSi during the same time. To characterize siRNA and nanoparticle formation, siRNA oligos labeled with the Alexa555 fluorescent dye were mixed with PCPS for complete siRNA-polycation binding. Confocal microscopic analysis confirmed the presence of Alexa555-siRNA in the microparticles (Figure 1c, left panel). These particles were then incubated in PBS. Arg-PEI/siRNA was released and formed polyplex nanoparticles when the silicon dissolved (Figure 1c, right panel). Fluorescence from the siRNA ployplex nanoparticles was not as bright as in PCPS/siRNA where the Alexa555-siRNA was concentrated. The size of released polyplex nanoparticles was within 40 to 200 nm range with a median size of 102 nm as determined by dynamic light scattering (Figure 1d). This size range was confirmed by atomic force microscopy (Figure 1e). It is interesting to note that the median size of released polyplex was larger than that of the nanopores in porous silicon, indicating that the final polyplex structure formed during or after porous silicon degradation, but not inside the confinement of the porous silicon particle.
Figure 1.
Characterization of PCPS as a delivery carrier for gene silencing agents. (a) SEM images of oxidized porous silicon (pSi) as the starting material and PCPS as the final product. (b) Changes in zeta potential of particles at various stages of fabrication. Results are presented as the mean of five measurements ± standard deviation. (c) Confocal microscopic images of PCPS/siRNA (left panel) and the released Arg-PEI-siRNA polyplex nanoparticles. Red fluorescence was from the Alexa555-conjugated siRNA. (d) Size distribution of released siRNA polyplex nanoparticles measured by dynamic light scattering. (e) Atomic force microscopic image for size analysis of siRNA polyplex nanoparticle.
Loading and Release Kinetics
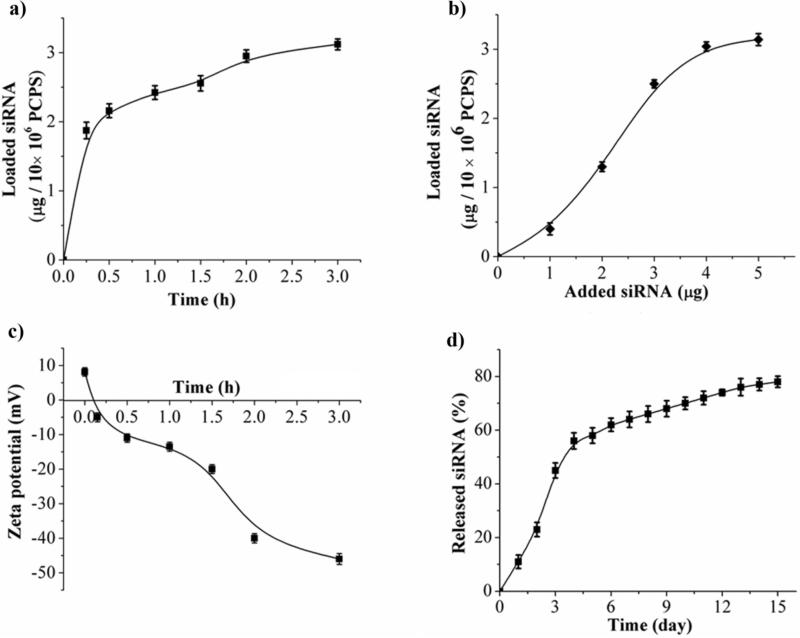
Binding of siRNA to polycation in PCPS can be monitored by decrease in UV absorbance in supernatant or decrease in zeta potential value. Within the first 30 minutes of incubation, 1.8 μg of siRNA oligos was loaded into 10×106 PCPS particles (Figure 2a). Another 1.2 μg siRNA was loaded when the incubation time was extended to 3 hours (Figure 2a). The maximum loading capacity was 3.14 μg siRNA/10×106 PCPS (Figure 2b). This was a dramatic improvement over the existing siRNA carriers. We have previously used the porous silicon-based multistage vector system to deliver siRNA packaged in nanoliposomes. 15,16 In order to deliver 15 μg siRNA, 6×108 1 μm size particles were needed to load liposomal siRNA. The number of particles would be reduced by 90% with the current platform. Increase in binding of the negatively charged siRNA oligos to PCPS correlated with decrease in surface charge of the complex (Figure 2c). Release of siRNA from the carrier was biphasic in 10% fetal bovine serum (Figure 2d). A quick release profile was observed in the first 4 days. Approximately 60% of the siRNA was released within this period of time, and another 20% was released over the next 10 days (Figure 2d). Since siRNA is released as polyplex nanoparticles upon porous silicon degradation (Figure 1), the release curve indicates that porous silicon degraded gradually, resulting in sustained release of siRNA nanoparticle polyplex. In a parallel experiment, we loaded PEI/siRNA polyplex nanoparticles into pSi-APTES-Arg, and monitored particle release (Figure S2). Most particles left the pSi-APTES-Arg carrier within the first 3 days. The results indicate that the Arg-PEI bond is essential for sustained release of PEI/siRNA polyplex nanoparticles. Thus, we have developed a delivery system with high nucleotide binding capacity for sustained release of gene silencing agents.
Figure 2.
Loading of siRNA into PCPS and release of siRNA polyplex nanoparticle. (a) Time-dependent loading of siRNA into PCPS. Scramble siRNA was used to test time-dependent loading, and UV absorbance of the supernatant at 260 nm was measured to assess loading capacity. (b) Dose-dependent loading of siRNA into PCPS. (c) Zeta potential changes during siRNA loading into PCPS. (d) Release of siRNA nanoparticle from the carrier. PCPS/Alexa555-siRNA was incubated in PBS with 10% fetal bovine serum. Supernatant was separated from the particles, and was used to measure fluorescent intensity at Ex543/Em590. Results are presented as the mean of five measurements ± standard deviation.
Cellular Uptake of PCPS/siRNA and Intra-cellular Trafficking of siRNA Polyplex Nanoparticle
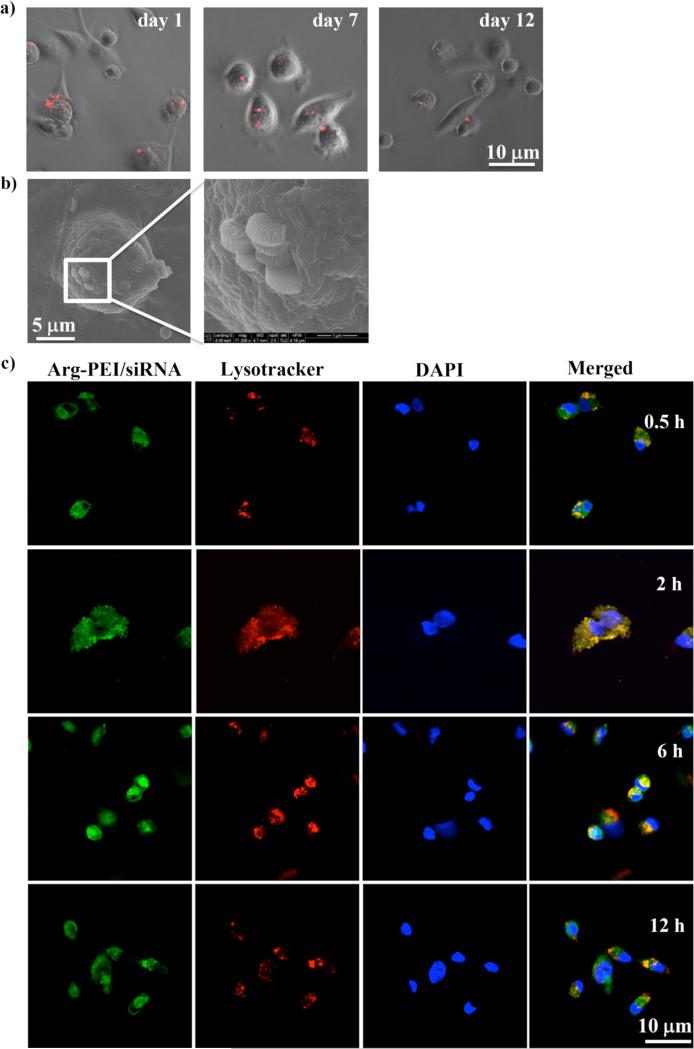
PSPC particles loaded with Alexa555 siRNA were incubated with MDA-MB-231 human breast cancer cells in culture, and the fluorescent particles were monitored under a confocal microscope. Uptake of particles by the breast cancer cells was apparent one day after inoculation (Figure 3a). SEM analysis captured an image of particles at various stages of cellular entry (Figure 3b). Multiple fluorescent particles could still be visualized on day 7 and 12 (Figure 3a).
Figure 3.
Cellular uptake of PCPS/siRNA and intra-cellular trafficking of Arg-PEI/siRNA polyplex nanoparticles. (a) Time-dependent release of Alexa555-siRNA inside tumor cells. PCPS/Alexa555 siRNA particles (in red) were added into MDA-MB-231 cell culture, and fluorescence from particles was monitored with a confocal microscope on days 1, 7, and 12 after incubation. (b) SEM images of cellular internalization of PCPS/siRNA. Multiple particles were at various stages of cellular entry. (c) Intra-cellular trafficking of Arg-PEI/siRNA polyplex nanoparticles. Arg-PEI/FAM-siRNA polyplex nanoparticles (in green) were added into MDA-MB-231 cell culture. Cells were harvested at the indicated time points, and stained with Lysotracker for late endosomes/lysosomes (in red) and DAPI for nuclei (in blue). Fluorescent images was captured with a confocal microscope.
We loaded FAM-labeled siRNA into PCPS, and collected Arg-PEI/siRNA following PCPS degradation in PBS. The green siRNA polyplex nanoparticles were incubated with MDA-MB-231 cells, and time-dependent intracellular localization was monitored (Figure 3c). Confocal microscopic images showed that the FAM-siRNA had reached endosomes/lysosomes (in red) after 30 minutes of incubation, as the green fluorescent FAM-siRNA co-localized with the red fluorescent endosomes/lysosomes. By the 2-hour time point, almost all green FAM-siRNA co-stained with the red endosomes/lysosomes, indicating maximum accumulation of siRNA in these organelles. Some siRNA molecules had exited these organelles by 6 hour, and FAM-siRNA could be visualized in multiple areas inside the cells by 12 hour, suggesting successful endosomal escape of siRNA. Effective endosomal escape of the Arg-PEI/siRNA enabled siRNA molecules to exert their biological activities inside the cell. It has been speculated that polyamine molecules can serve as proton sponges that break endosomes.18 It is very likely that breakage of endosomes by Arg-PEI facilitated siRNA exit from late endosomes.
Knockdown of Gene Expression in Human Cancer Cells by PCPS/siRNA
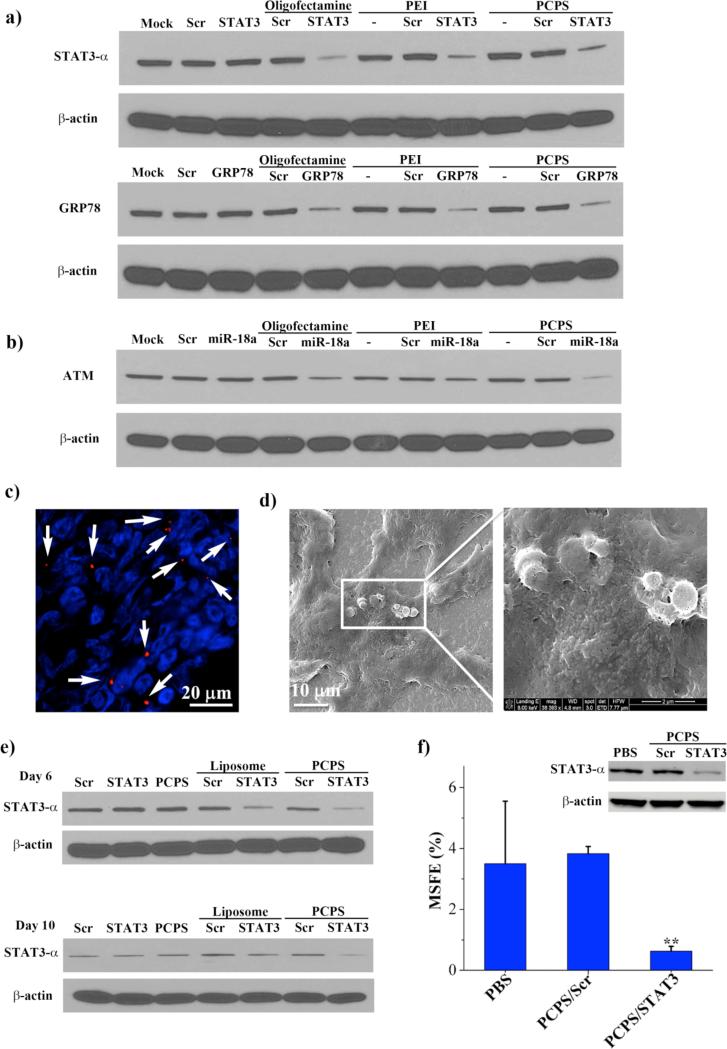
In order to test knockdown efficiency in human cancer cells and to identify the optimal ratio between siRNA oligo and polycation, we incubated MDA-MB-231 cells with an increasing amount of PCPS particles (5×106 - 15×106) loaded with the same amount (1.4 μg) of siRNA oligos specific to the human STAT3-α gene. This amount of siRNA oligo in a 2 mL cell culture was equivalent to 50 nM final concentration, a level commonly used in cell-based RNAi studies.18 STAT3 encodes a transcription factor that plays a significant role in mediating the JAK/STAT signal transduction pathway.20 A recent study indicated that STAT3 is an essential gene for breast cancer stem cells that are responsible for therapy resistance, tumor recurrence and metastasis.21 Thus knockdown of this gene might facilitate elimination of breast cancer stem cells. As controls of this study, MDA-MB-231 cells were also transfected with 1.4 μg STAT3 siRNA or scramble siRNA (Scr) using oligofectamine, a commonly used transfection reagent for siRNA.19 Cells were harvested 72 hours post transfection, and STAT3 expression was measured by Western blot analysis (Figure S3-a). Excellent knockdown of STAT3 expression was achieved in cells treated with 1.4 μg siRNA loaded into 8×106 - 15×106 PCPS particles (lanes 5 to 7: 86%, 74%, and 91% knockdown), but not in the cells treated with 5×106 particles (lane 4: 55% knockdown), suggesting that a certain level of nitrogen (from Arg-PEI) to phosphate (from siRNA) ratio (N/P ratio) has to be reached in the final product in order to achieve optimal biological activity. In a follow-up study, we treated MDA-MB-231 cells with PCPS particles loaded with STAT3 or scramble siRNA at the optimized ratio (1.4 μg siRNA in 15×106 PCPS particles). The controls were the same amount of free siRNA alone, siRNA transfected with oligofectamine, or PEI/siRNA polyplex. As anticipated, unprotected STAT3 siRNA alone did not have any effect on gene expression. Similar knockdown efficiency was observed in cells transfected with oligofectamine, or treated with PEI/STAT3 or PCPS/STAT3 (Figure 4a).
Figure 4.
Knockdown of gene expression in vitro and in murine model of MDA-MB-231 primary tumor. (a) Western blot analyses on knockdown of STAT3 and GRP78 expression in MDA-MB-231 cells following siRNA treatment. Cells were treated with the indicated agents, and harvested 72 hours later for protein analysis. The β-actin level indicates equal protein loading. (b) Knockdown of ATM expression in MCF-7 cells. (c) Accumulation of PCPS/siRNA in primary MDA-MB-231 tumor. Mice bearing MDA-MB-231 primary tumor were administrated with 150 × 106 PCPS/Alexa555 siRNA by tail vein injection. They were sacrificed 6 hours later, and tumor samples were processed for confocal analysis. PCPS/siRNA particles were in red (Alexa555) and highlighted by the white arrows, and nuclei of tumor cells were stained in blue by DAPI. (d) SEM image of PCPS/siRNA in tumor tissue. (e) Knockdown of STAT3-α expression in vivo. Each mouse bearing MDA-MB-231 primary tumor received 150×106 PCPS loaded with 15 μg siRNA by i.v. on day 1. Mice were sacrificed 6 or 10 days later for expression analysis by Western blot. (f) Mammosphere formation efficiency (MSFE) in MDA-MB-231 primary tumor cells treated with PCPS/siRNA. The inserted Western blot result shows gene expression levels in the cells used for the MSFE assay. MSFE results were average from 3 mice/group.
We also treated MDA-MB-231 cells with siRNA specific for the GRP78 gene in order to confirm that the N/P ratio-dependent knockdown was not restricted to the STAT3 siRNA. GPR78 encodes a 78-kDa glucose-regulated protein, also known as heat shock 70-kDa protein 5, which modulates the activity of multiple endoplasmic reticulum stress proteins.22 As with STAT3, efficient knockdown was achieved at the right N/P ratio (Figure 3S-a). At the optimal ratio, PCPS/GRP78 was as efficient on knockdown of gene expression as in cells transfected with oligofectamine or treated with PEI/GRP78 polyplex (Figure 4a).
MCF-7 human breast cancer cells were treated with a microRNA-18a (miR-18a) mimic oligo. It has been previously reported that miR-18a regulates expression of the ATM gene.23 So we assessed ATM expression following miR-18a treatment. As expected, ATM expression in the positive control cells transfected with miR-18a was dramatically suppressed (71% knockdown, Figure S3-b). Treatment with PCPS/miR-18a resulted in a more significant knockdown (85% to 90%) of ATM expression when the optimal number of PCPS particles were used (Figure 4b and Figure S3-b).
In addition, we measured changes in mRNA levels following transfection with oligofectamine or coincubation with PEI or PCPS. The mRNA levels of STAT3, GRP78, and ATM in the post-treatment MDA-MB-231 and MCF-7 cells (Fig. S4) correlated with changes in protein expression by Western blot analysis (Fig. 4). These results confirmed effective knockdown of gene expression by the PCPS-delivered siRNA and microRNA.
Delivery of siRNA In Vivo and Knockdown of Gene Expression in Tumor Tissues
We generated a murine model of MDA-MB-231 human breast cancer to examine whether PCPS was effective in delivering gene silencing agents to solid tumors. Since tumor cells were inoculated into the mammary gland fat pad of female athymic nude mice, this model mimicked the pathological condition of human primary breast cancer. To test tumor distribution of particles, PCPS loaded with Alexa-555 siRNA were systemically administrated into the tumor mice. Mice were sacrificed 6 hour later, and tumor tissues were collected for histological analysis. The presence of red fluorescent particles in tumor could be visualized under a fluorescent microscope (Figure 4c). SEM analysis of tissue blocks also revealed clusters of particles inside the tumor (Figure 4d). In a separate study, mice bearing MDA-MD-231 primary tumors were treated once by tail vein injection with PCPS carrying siRNA specific for the human STAT3 gene. We used 100×106 PCPS particles to deliver 15 μg siRNA, a ratio within the optimal N/P range from the above in vitro studies (Figure 3S). In the control groups, mice were dosed i.v. with free siRNA or liposomal siRNA every 3 days. The animals were sacrificed on 6 or 10 days after the first dosing, and tumor tissues were collected for STAT3 expression analysis. While both PCPS/STAT3 and liposomal STAT3 caused reduction of STAT3 expression, Western blot analysis showed that the knockdown efficiency was more profound with PCPS/STAT3 treatment in both sets of samples (Figure 4e). As expected, unprotected siRNA did not have any effect on STAT3 expression. Tumor samples from day 10 were also digested for single cell isolation, and mammosphere formation efficiency (MSFE), one of the key features of breast cancer stem cells,24 was measured. Knockdown of STAT3 expression by PCPS/STAT3 resulted in a significant reduction in MSFE (Figure 4f), demonstraing the power of this new delivery system on delivery of siRNA to target key cancer genes.
Evaluation of Acute Immunotoxicity In Vitro and In Vivo
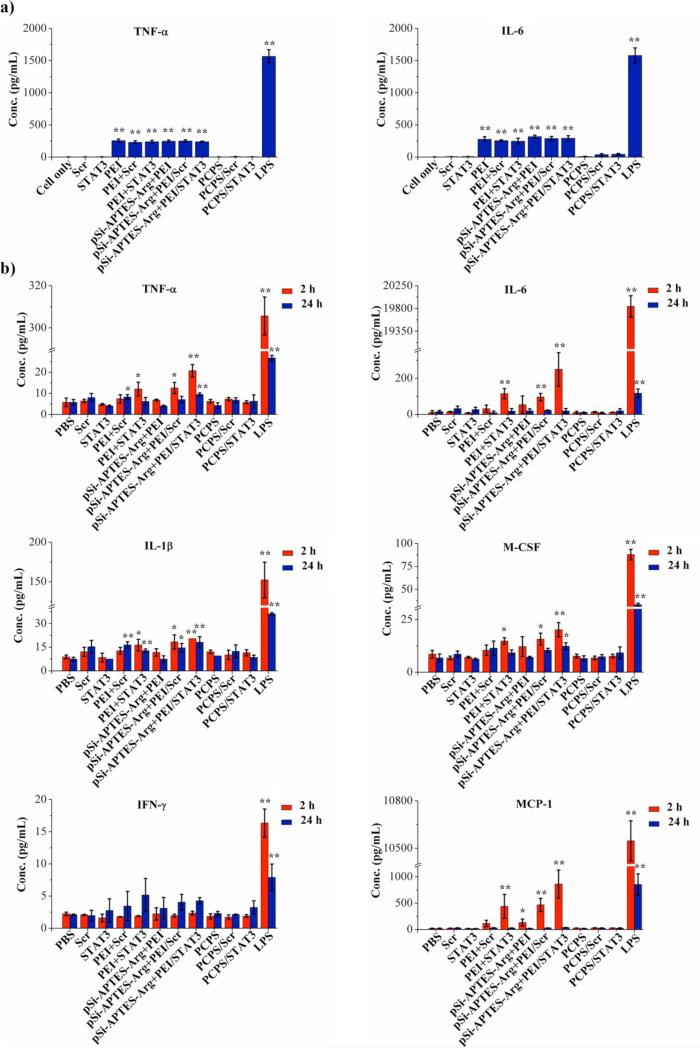
Potential immunotoxicity associated with this new delivery system was evaluated in cell culture and with wild-type mice. In the in vitro setting, both the empty vehicle and PCPS/siRNA were co-cultured with Raw264.7 murine macrophage cells, and levels of two key inflammatory mediators, tumor necrosis factor-alpha (TNF-α) and interleukin-6 (IL-6), were measured. Lipopolysaccharide (LPS) was used as a positive control for this assay, as its potential to induce innate immune response has been well documented.25 Treatment with LPS triggered a surge in both TNF-α and IL-6 levels (Figure 5a). PEI or PEI/siRNA polyplex treatment also caused significant increases of both cytokines in cell culture media. Simple mix of pSi-APTES-Arg with PEI/siRNA polyplex did not prevent stimulation of cytokine production. Addition of these reagents into culture of MDA-MB-231 cells also caused cell death (Figure S5). In contrast, treatment with empty PCPS or PCPS/siRNA did not elevate levels of TNF-α or IL-6 in Raw264.7 cells (Figure 5a). Moreover, they did not cause cytotoxicity to MDA-MB-231 cells either (Figure S5).
Figure 5.
In vitro and in vivo acute toxicity. (a) Raw-264.7 mouse macrophage cells were incubated with the indicated agents for 24 hours, and TNF-α and IL-6 levels in supernatant were measured by ELISA. Results are presented as the mean of three measurements ± standard deviation. b) Changes in levels of selected serum cytokine/chemokine/colony-stimulating factors in post-treatment mice. Blood samples were collected 2 or 24 hours after intravenous dosing of treatment agents. A multiplexed bead-based immunoassay was used to measure levels of the cytokines/chemokines/colony-stimulating factors. Results were the average from 3 mice/group. *p <0.05. **p <0.01.
In the in vivo seting, FVB mice were treated once with 1x therapeutic dosage (15 μg siRNA) of PCPS/siRNA by i.v. administration, and serum levels of chemokines and cytokines were measured. As in cell culture, LPS treatment caused dramatic increases in TNF-α within 2 hours and IFN-γ at the 24-hour time point (Figure 5b). It also triggered secretion of most of the proinflammatory cytokines and chemokines such as IL-1β, interferon-γ, and MCP-1. Free PEI polyplxes and mixtures of PEI polyplex with pSi-APTES-Arg also significantly elevated serum levels of most of the proinflammatory cytokines (Figure 5b and Figure S6). However, neither PCPS/scramble nor PCPS/STAT3 siRNA caused significant changes in serum level of any of these factors (Figure 5b and Figure S6). These results demonstrated lack of immunotoxicity from PCPS or PCPS/siRNA.
Evaluation of Sub-acute Toxicity
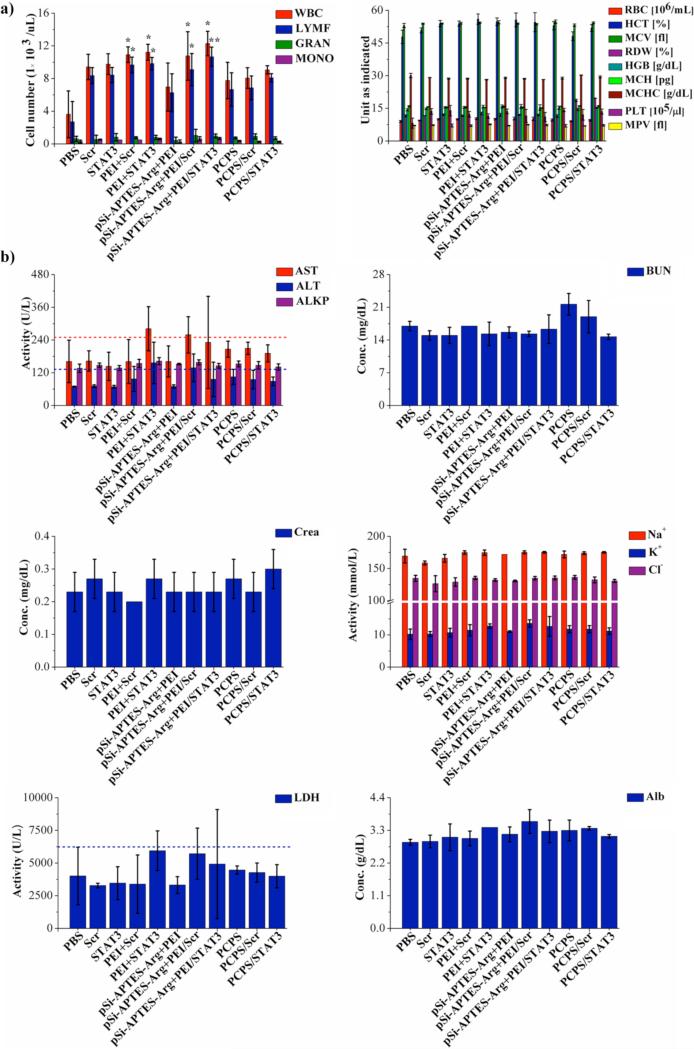
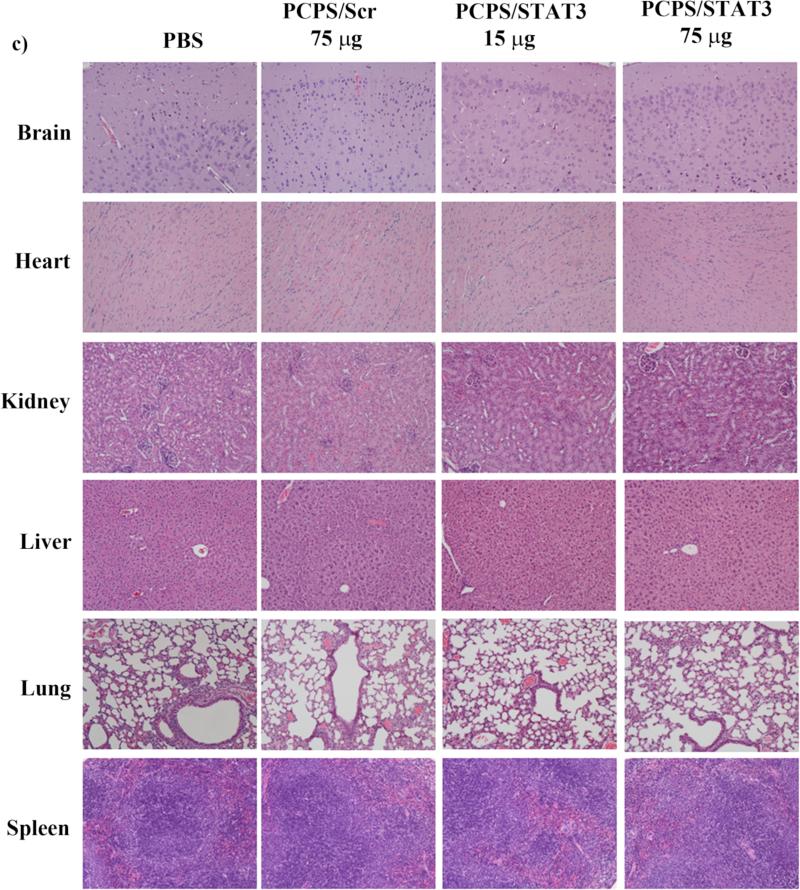
FVB mice were treated weekly by i.v. administration for four weeks with unprotected siRNA, PEI/siRNA polyplex, mixture of PEI/siRNA polyplex and pSi-APTES-Arg, or PCPS/siRNA. At the end of the treatment, whole blood samples were collected and cell counts were performed. Serum samples were collected and used to measure biomarkers associated with functions of the liver, kidney, and heart. Biomarkers for liver function included aspartate aminotransferase (AST), alanine aminotransferase (ALT), albumin (ALB), and alkaline phosphatase (ALKP). Parameters for renal function were blood urea nitrogen (BUN), creatine, Na+, K+ and Cl−. Treatment with PEI/siRNA polyplex alone or in mixture with pSi-APTES-Arg resulted in significant elevation of red blood cells (Figure 6a). These treatments also raised plasma AST and lactate dehydrogenase (LDH) levels to or above the normal ranges (Figure 6b). The results indicate that PEI/siRNA polyplex has the potential to cause damages to major organs such as the liver. On the other hand, no hematological and biochemical values were altered in the empty PCPS or PCPS/siRNA treatment groups. To further confirm lack of toxicity from PCPS/siRNA, we treated FVB mice repeatly with 5-fold the normal dosage of PCPS/siRNA (75 μg siRNA in 0.5 billion PCPS particles per week for four weeks). Histological analysis of the major organs including brain, heart, kidney, liver, lung, and spleen supported the notion that particle treatment did cause damages to these organs (Figure 6c). Taken together, repeated administrations of PCPS carrying scramble or STAT3 siRNA did not cause any detectable sub-acute toxicity.
Figure 6.
Analysis on subacute toxicity from PCPS/STAT3 siRNA. Mice (n = 3/group) were treated once a week for four weeks with the indicated agents. They were sacrificed 24 hours after the final treatment, and (a) hematological analysis, (b) blood chemistry analysis, (c) histological analysis were performed to evaluate potential toxicity. ALT, alanine aminotransferase; ALB, albumin; ALKP, alkaline phosphatase; Arg, arginine; AST, aspartate aminotransferase; BUN, blood urea nitrogen; GRAN, granulocytes; HCT, hematocrit; HGB, hemoglobin; LDH, lactate dehydrogenase; LYMPH, lymphocytes; MCH, Mean Corpuscular Hemoglobin; MCHC, mean corpuscular hemoglobin concentration; MCV, mean corpuscular volume; MONO, monocytes; MPV, mean platelet volume; PBS, phosphate buffer saline; PLT, platelet count; RBC, red blood cells; RDW, red blood cell distribution width; WBC, white blood cells.
Two key considerations in design of delivery systems are efficiency and toxicity. Here, we have shown that PCPS is very effective in knocking down gene expression in vitro and in vivo (Figure 4). We have further demonstrated that knockdowm of STAT3 expression in MDA-MB-231 primary tumors results in significant reduction on cancer stem cell population. Since these cells are responsible for tumor recurrence and metastasis, 26,27 reduction of the cancer stem cell population by PCPS/STAT3 siRNA may have synergy with other cancer drugs that kill the bulk of non-stem cell cancer cells. Further validation awaits functional studies on combination treatment with clinically available cancer drugs.
Previous reports have indicated that high molecular weight PEI can cause severe cytotoxicity.28 Certain siRNA oligos, when packaged in cationic or liposomal carriers, trigger toll-like receptors and induce interferons and inflammatory cytokines.30,31 In the current study, we have confirmed cytotoxicity and immunotoxicity from free PEI and PEI/siRNA polyplex (Figure 5). Subacute toxicities have also been observed in mice treated with these agents (Figure 6). In comparison, there was no sign of such toxicities from either the empty PCPS delivery carrier or siRNA-loaded PCPS. No acute toxicity from PCPS was detected in both in vitro and in vivo settings (Figure 5). Furthermore, repeated treatments with increased dosages did not cause damages to major organs in wild-type mice (Figure 6c). Several factors in the design of this delivery system might have contributed to the lack of toxicity. It is well known that PEI causes toxicity by interaction with glycocalyx on the cell surface, resulting in the formation of large clusters. In addition, the primary amine moieties of PEI are more cytotoxic than the secondary or tertiary amines. 28,31 Conjugation of PEI into the nanopores inside porous silicon shields the molecule from interacting with cells. In addition, covalent linkage with arginine eliminates the primary amine group in the molecule. Furthermore, an Arg-PEI/siRNA nanoparticle enters the cell via vesicular transport, reducing the chance of interaction with molecules on cell surface. Slow degradation of the silicon particles also limited the amount of PEI exposure to cells at a given time. These results point to a desirable safety profile for the new delivery system.
CONCLUSIONS
In conclusion, we have developed a new carrier for the delivery of gene silencing agents. It has a high oligonucleotide-binding capacity, and lacks any detectable toxicity. This system is easy to operate, and can be used to deliver siRNA or microRNA to achieve knockdown of key cancer genes in vivo. Such gene silencing agents will play a dominant role in the treatment of human cancers in the upcoming years. It has been estimated there are up to 100 mutations in a breast cancer, many of them driving mutations.32 Large number of mutations has also been identified in other cancer types. 32,33 On top of that, each patient carries a unique mutation spectrum, making the total number of combination of cancer-causing genes incredibly large. On the other hand, the current cancer drugs only target a handful of gene products. This gap can only be bridged by the development of gene silencing agents as cancer therapeutics. This new platform offers an excellent enabling platform in the era of personalized medicine.
EXPERIMENTAL SECTION
Materials
Branched polyethyleneimine (MW 25 kDa), arginine, 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), isopropyl alcohol (IPA), (3-aminopropyl)triethoxysilane (APTES) and N-(tert-butoxycarbonyl)-L-aspartic acid (Boc-Asp-OH) were purchased from Sigma-Aldrich. Dulbecco's Modified Eagle Medium high glucose (DMEM) and fetal bovine serum (FBS) were from Fisher Scientific. Small interfering RNA (siRNA)-Alexa Fluor 555 was purchased from Qiagen. Scrambled siRNA, GRP78 siRNA, miRNA-18a mimic, and STAT3 siRNA were ordered from Sigma. Female athymic nu/nu nude mice and FVB mice were acquired from Charles River.
Cell Lines
The human breast carcinoma cell lines MDA-MB-231 and MCF-7, and murine RAW 264.7 cells were obtained from ATCC (Rockville, MD). Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin under the condition of 5% CO2 and 95% humidity at 37°C.
Fabrication of Polycation-functionalized Nanoporous Silicon Microparticles
Fabrication of discoidal nanoporous silicon microparticles based on a combination of photolithography and electrochemical etch was described previously, 17,34 and surface APTES modification was as reported before.35
Arginine was covalently conjugated to APTES using EDC/NHS chemistry. Briefly, the acid group of arginine (0.1 nmol) was activated using EDC/NHS (0.1 nmol/0.1 nmol) in 20 mL ethanol for 4 hours. pSi-APTES (10 billion particles) was then added, and the reaction was kept for 18 hours at 20°C.
PEI was conjugated to the arginine group of pSi-APTES-Arg through an aspartic acid linker. Briefly, the N-tert-butoxycarbonyl (N-BOC) protected aspartic acid (0.1 nmol) was activated with EDC/NHS (0.1 nmol/0.1 nmol) in 20 mL ethanol for 4 hours at 4°C. After acid activation, PEI (50 mg) was dissolved in 10 mL ethanol and added into the reaction mixture. The reaction was allowed to take place for 24 hours at 20°C. In the second step of the reaction, the other acid group of aspartic acid was activated using EDC/NHS (0.1 nmol/0.1 nmol) at 4°C for 6 hours. After acid activation, pSi-APTES-Arg (10 billion particles) was then added into the reaction mixture, and the reaction was kept for 18 hours at 20°C. The mixture was centrifuged, washed three times with ethanol, and polycation-functionalized particles were dried in vacuum. The size and zeta potential of the polycation-functionalized particles were measured with a Malvern multi-purpose titrator.
Loading of siRNA into PCPS Particles and Release of Arg-PEI/siRNA Polyplex Nanoparticles
PCPS particles were mixed with siRNA in nuclease-free water, and incubated for 3 hours at 4°C for complete binding of siRNA to polycation. The suspension was then centrifuged to remove free siRNA oligos in the supernatant. To measure release profile of siRNA, PCPS particles loaded with Alexa555 siRNA were incubated in PBS with 10% FBS at 37°C in a shaker with a speed of 100 rpm. Supernatant was collected at different time points, and fluorescence intensity was measured with a BioTek H4 synergy hybrid plate reader.
Cellular Uptake of PCPS/Alexa555 siRNA Microparticles
Cellular uptake of PCPS particles loaded with Alexa555 siRNA was examined by confocal microscopy. MDA-MB-231 cells were plated in 4-well slide chambers at a seeding density of 1×104 cells/well and incubated for 24 hours. PCPS/siRNA particles (1 million/well) were added into cell culture. Cells were harvested and fixed at different time points, and confocal microscopic images were obtained using a Fluo View TM 1000 Confocal Microscope (Olympus Inc., USA).
Intra-cellular Trafficking of Arg-PEI/siRNA Polyplex Nanoparticle
To monitor intra-cellular trafficking of siRNA polyplex, PCPS/FAM-labeled siRNA particles were incubated in PBS for 48 hours. The released Arg-PEI/FAM siRNA polyplex nanoparticles were then collected by centrifugation, and added into culture medium of MDA-MB-231 cells. Cells were harvested at different time points (0.5 h, 2 h, 6h, and 12 h), and fixed with 4% paraformaldehyde. The samples were then blocked with 1% bovine serum albumin (BSA) in PBS with 0.1% Tween-20 (PBST) solution, and stained with Lysotracker (Invitrogen) for late endosomes/lysosomes and DAPI for nuclei.
Gene Silencing In Vitro
MDA-MB-231 or MCF-7 cells were seeded in a 6-well tissue culture dish at 2 × 105 cells/well in 2 mL DMEM containing 10% FBS. Cells were incubated with PCPS/ siRNA for 72 hours before they were harvested for Western blot analysis. Proteins were extracted from cells using a M-PER protein extraction reagent (Pierce Inc., USA), separated on a 12% gel by SDS-PAGE, and transferred to a nitrocellulose membrane (Bio-Rad Inc., USA). Antibodies for Western blot analysis were rabbit anti-human STAT3 antibody (Cell Signaling, 1:1,000 dilution), anti-human GRP78 antibody (Cell Signaling, 1:1,000 dilution), and anti-human ATM antibody (Cell Signaling, 1:1,000 dilution). The amount of protein was quantitated by densitometry, and ratio to β-actin level was calculated.
Quantitative RT-PCR analysis in vitro
Real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis was performed to examine STAT3, GRP78, and ATM mRNA expression with the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, CA, USA). β-Actin served as the housekeeping gene for qPCR analysis, and the relative mRNA levels of target genes were normalized to the mRNA level of β-actin. Cells were seeded in 6-well plates (2 × 105 cells/well), treated with siRNA or microRNA, and harvested 48 hours later. Total RNA was isolated using the TRIzol reagent following the manufacturer's suggested protocol (Qiagen, Vlencia, CA). Two micrograms of total RNA was transcribed into cDNA using the PrimeScript first strand cDNA synthesis Kit (Takare, Dalian, China), and 2 μL of cDNA was subjected to qPCR analysis using the SYBR Premix Ex Taq. Primer sequences for mRNA amplification are listed below. STAT3 forward: CAGCAGCTTGACACGGTA, STAT3 reverse: AAACACCAAAGTGGCATGTGA; GRP78 forward: CACAGTGGTGCCTACCAAGA, GPR78 reverse: TGTCTTTTGTCAGGGGTCTTT; ATM forward: TTCAAAGGATTCATGGTCCAG, ATM reverse: GCTGTGAGAAAACCATGGAA; β-actin forward: AAATCGTGCGTGACATTAA, β-actin reverse: CTCGTCATACTCCTGCTTG. All reactions were performed in triplicates.
Knockdown of STAT3 Expression and Mammosphere Formation Efficiency Assay In Vivo
All experimental procedures for animal studies were performed in accordance with regulation in the Houston Methodist Research Institute for the care and use of laboratory animals. Nude mice bearing orthotopic MDA-MB-231 breast tumors were divided into seven treatment groups, and each mouse received treatment with free Scr siRNA (15 μg siRNA/injection, every 3 days), free STAT3 siRNA (15 μg siRNA/injection, every 3 days), liposome/Scr siRNA (15 μg siRNA/injection, every 3 days), liposome/STAT3 siRNA (15 μg siRNA/injection, every 3 days), empty PCPS (100×106 PCPS microparticles on day 1), PCPS/Scr siRNA (15 μg siRNA in 100×106 PCPS microparticles on day 1), or PCPS/STAT3 siRNA (15 μg siRNA in 100×106 PCPS microparticles on day 1). Treatment was initiated when the average tumor size reached 150-200 mm3. Mice were sacrificed on day 6 or day 10 after the first treatment, and tumor samples were collected for expression analysis by Western blot and for mammosphere formation.
For mammosphere formation assay, tumor samples were minced and digested with 200 u/mL collagenase in DMEM-F12 for 1.5 h at 37°C. Single cells were collected, and seeded in ultra-low attachment plates at a density of 1,000 cells/well in the mammocult TM basal medium for 1-2 weeks at 37°C. The number of mammospheres (cells ≥ 50) were counted under the microscope. Percentage of MSFE was calculated as number of mammospheres/number of total seeded cells.
In Vitro Immunotoxicity Assay
RAW 264.7 cells were seeded in a 96-well tissue culture plate at 2 × 104 cells/well in 100 μL DMEM containing 10% FBS. LPS served as the positive control (0.5 μg/mL), and the untreated cell as the negative control. Cells were treated with free siRNA, PEI/siRNA, mixture of pSi-Arg with PEI/siRNA, or PCPS/siRNA for 24 hours, and levels of murine TNF-α and IL-6 were measured by ELISA.
Acute Immunotoxicity Analysis
FBV mice were randomly divided into 12 groups (n=3). Each mouse received 15 μg siRNA in unprotected form (Scr or STAT3), as PEI polyplex (PEI/Scr or PEI/STAT3), as a mixture of pSi-Arg and PEI polyplex (pSi-APTES-Arg + PEI/Scr or pSi-APTES-Arg + PEI/STAT3), or in PCPS (PCPS/Scr or PCPS/STAT3). LPS (5 mg/kg) served as the positive control. Mice were dosed by tail vein injection, and blood samples were taken at 2 and 24 hours. Serum cytokine levels were measured with a multiplexed bead-based immunoassay from Millipore, which simultaneously evaluated the levels of: granulocyte/macrophage colony-stimulating factor (GM-CSF), interferon gamma (IFN-γ), interleukin-1 beta (IL-1β), IL-2, IL-6, IL-9, IL-10, IL-12 (p40 and p70), IL-15, IL-17, LPS-induced CXC chemokine (LIX), tumor necrosis factor-alpha (TNF-α), monokine induced by IFN-gamma (MIG), macrophage inflammatory protein 2 (MIP-2), MIP-1α, MIP-1β, macrophage colony-stimulating factor (M-CSF), regulated on activation normal T cell expressed and secreted (RANTES). The cytokines levels were read on the Luminex 200 System Multiplex Bio-Assay Analyzer, and quantified based on standard curves for each cytokine.
Sub-acute Toxicity Analysis
FVB mice received weekly treatment by tail vein injection for four weeks. The treatments included 15 μg siRNA in unprotected form (Scr or STAT3), as PEI polyplex (PEI/Scr or PEI/STAT3), as a mixture of pSi-Arg and PEI polyplex (pSi-APTES-Arg + PEI/Scr or pSi-APTES-Arg + PEI/STAT3), or in PCPS (PCPS/Scr or PCPS/STAT3). The animals were observed for signs of toxicity throughout the experiment. Water and food consumption were recorded weekly. Body weight of the mice was measured every three days. At the end of the 4-week treatment, blood samples were collected for cell count and biomarker analysis. The biomarkers included aspartate aminotransferase (AST), albumin (ALB), alanine aminotransferase (ALT), blood urea nitrogen (BUN), creatinine (CREA), Cl−, Na+ and K+, alkaline phosphatase (ALKP), and lactate dehydrogenase (LDH). To further evaluate potential organ damage by PCPS/siRNA, FVB mice were treated weekly with PCPS/scramble siRNA (75 μg of siRNA) or PCPS/STAT3 siRNA (15 μg or 75 μg) by tail vein injection for four weeks. All major organs were collected, and histological analyses were performed.
Histopathological Examination
Major organs including liver, spleen, kidney, lung, brain, and heart were harvested, fixed in formalin, and processed for histological evaluation by hematoxylin and eosin (H&E) staining. The tissues were fixed in formalin and embedded in paraffin. Tissue sections (4 μm) were analyzed to evaluate leukocyte infiltration, cell death, and other signs of organ damage. At least five random sections from each slide were examined.
Statistical Analysis
All quantitative data are expressed as mean ± standard deviations. Statistical analysis was performed with Student's t-test. Differences were considered statistically significant with p < 0.05 (*) and p < 0.01 (**).
Supplementary Material
Acknowledgements
The authors acknowledge financial support from the following sources: Department of Defense grants W81XWH-09-1-0212 and W81XWH-12-1-0414, National Institute of Health grants U54CA143837 and U54CA151668, the CPRIT grant RP121071 from the state of Texas, the Ernest Cockrell Jr. Distinguished Endowed Chair, the financial support from the National Natural Science Foundation of China (Nos. 21231007 and 21172274), National High-Tech Research and Development Programme of China (863 Program, Grant 2012AA020305), the Ministry of Education of China (Nos., 20100171110013 and 313058) and the Guangdong Provincial Natural Science Foundation (No. 9351027501000003).
Footnotes
The authors declare no competing financial interest.
All authors have given approval to the final version of this manuscript.
Supporting Information Available: SEM images, release profiles, Western blots, qRT-PCR data, viability evluation and immunotoxicity testing. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Makley LN, Gestwicki JE. Expanding the Number of ‘Druggable’ Targets: Non-Enzymes and Protein-Protein Interactions. Chem. Biol. Drug Des. 2013;81:22–32. doi: 10.1111/cbdd.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ, Jenkins NA, Copeland NG. New Genes Involved in Cancer Identified by Retroviral Tagging. Nat. Genet. 2002;32:166–174. doi: 10.1038/ng949. [DOI] [PubMed] [Google Scholar]
- 3.Li J, Shen H, Himmel KL, Dupuy AJ, Largaespada DA, Nakamura T, Shaughnessy JD, Jr., Jenkins NA, Copeland NG. Leukaemia Disease Genes: Large-Scale Cloning and Pathway Predictions. Nat. Genet. 1999;23:348–353. doi: 10.1038/15531. [DOI] [PubMed] [Google Scholar]
- 4.Schlabach MR, Luo J, Solimini NL, Hu G, Xu Q, Li MZ, Zhao Z, Smogorzewska A, Sowa ME, Ang XL, et al. Cancer Proliferation Gene Discovery through Functional Genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis Elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Shen H, Sun T, Ferrari M. Nanovector Delivery of siRNA for Cancer Ttherapy. Cancer Gene Ther. 2012;19:367–373. doi: 10.1038/cgt.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumura Y, Maeda H. A New Concept for Macromolecular Therapeutics in Cancer Chemotherapy: Mechanism of Tumoritropic Accumulation of Proteins and the Antitumor Agent Smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 8.Maeda H. The Enhanced Permeability and Retention (EPR) Effect in Tumor Vasculature: The Key Role of Tumor-Selective Macromolecular Drug Targeting. Adv. Enzyme Regul. 2001;41:189–207. doi: 10.1016/s0065-2571(00)00013-3. [DOI] [PubMed] [Google Scholar]
- 9. wwww.clinicaltrials.com.
- 10.Davis ME, Zuckerman JE, Choi CH, Seligson D, Tolcher A, Alabi CA, Yen Y, Heidel JD, Ribas A. Evidence of RNAi in Humans from Systemically Administered siRNA via Targeted Nanoparticles. Nature. 2010;464:1067–1070. doi: 10.1038/nature08956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aliabadi HM, Landry B, Sun C, Tang T, Uludag H. Supramolecular Assemblies in Functional siRNA Delivery: Where Do We Stand? Biomaterials. 2012;33:2546–2569. doi: 10.1016/j.biomaterials.2011.11.079. [DOI] [PubMed] [Google Scholar]
- 12.Godin B, Chiappini C, Srinivasan S, Alexander JF, Yokoi K, Ferrari M, Decuzzi P, Liu X. Discoidal Porous Silicon Particles: Fabrication and Biodistribution in Breast Cancer Bearing Mice. Adv. Funct. Mater. 2012;22:4225–4235. doi: 10.1002/adfm.201200869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Decuzzi P, Godin B, Tanaka T, Lee SY, Chiappini C, Liu X, Ferrari M. Size and Shape Effects in the Biodistribution of Intravascularly Injected Particles. J. Control Release. 2010;141:320–327. doi: 10.1016/j.jconrel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 14.van de Ven AL, Kim P, Haley O, Fakhoury JR, Adriani G, Schmulen J, Moloney P, Hussain F, Ferrari M, Liu X, et al. Rapid Tumoritropic Accumulation of Systemically Injected Plateloid Particles and Their Biodistribution. J. Control Release. 2011;158:148–155. doi: 10.1016/j.jconrel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Huang Y, Mai J, Zhang G, Guo X, Xia X, Koay EJ, Qin G, Erm DR, Li Q, et al. Multistage Vectored siRNA Targeting Ataxia-Telangiectasia Mutated for Breast Cancer Therapy. Small. 2013;9:1799–1808. doi: 10.1002/smll.201201510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen H, Rodriguez-Aguayo C, Xu R, Gonzalez-Villasana V, Mai J, Huang Y, Zhang G, Guo X, Bai L, Qin G, et al. Enhancing Chemotherapy Response with Sustained EphA2 Silencing Using Multistage Vector Delivery. Clin. Cancer Res. 2013;19:1806–1815. doi: 10.1158/1078-0432.CCR-12-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, You J, Zhang G, Ziemys A, Li Q, Bai L, Deng X, Erm DR, Liu X, Li C, et al. Cooperative, Nanoparticle-Enabled Thermal Therapy of Breast Cancer. Adv. Healthcare Mater. 2012;1:84–89. doi: 10.1002/adhm.201100005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonawane ND, Szoka FC, Jr., Verkman AS. Chloride Accumulation and Swelling in Endosomes Enhances DNA Transfer by Polyamine-DNA Polyplexes. J. Biol. Chem. 2003;278:44826–44831. doi: 10.1074/jbc.M308643200. [DOI] [PubMed] [Google Scholar]
- 19.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-Nucleotide RNAs Mediate RNA Interference in Cultured Mammalian Cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 20.Seavey MM, Dobrzanski P. The Many Faces of Janus kinase. Biochem. Pharmacol. 2012;83:1136–1145. doi: 10.1016/j.bcp.2011.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, et al. The JAK2/STAT3 Signaling Pathway is Required for Growth of CD44(+)CD24(−) Stem Cell-Like Breast Cancer Cells in Human Tumors. J. Clin. Invest. 2011;121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen J, Chen X, Hendershot L, Prywes R. ER Stress Regulation of ATF6 Localization by Dissociation of BiP/GRP78 Binding and Unmasking of Golgi Localization Signals. Dev. Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 23.Qased AB, Yi H, Liang N, Ma S, Qiao S, Liu X. MicroRNA-18a Upregulates Autophagy and Ataxia Telangiectasia Mutated Gene Expression in HCT116 Colon Cancer Cells. Mol. Med. Report. 2013;7:559–564. doi: 10.3892/mmr.2012.1214. [DOI] [PubMed] [Google Scholar]
- 24.Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and In Vitro Propagation of Tumorigenic Breast Cancer Cells with Stem/Progenitor Cell Properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- 25.Tateda K, Matsumoto T, Miyazaki S, Yamaguchi K. Lipopolysaccharide-Induced Lethality and Cytokine Production in Aged Mice. Infect. Immun. 1996;64:769–774. doi: 10.1128/iai.64.3.769-774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Hajj M, Becker MW, Wicha M, Weissman I, Clarke MF. Therapeutic Implications of Cancer Stem Cells. Curr. Opin. Genet. Dev. 2004;14:43–47. doi: 10.1016/j.gde.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. Let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 28.Hunter AC. Molecular Hurdles in Polyfectin Design and Mechanistic Background to Polycation Induced Cytotoxicity. Adv. Drug Deliv. Rev. 2006;58:1523–1531. doi: 10.1016/j.addr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I. Sequence-Dependent Stimulation of the Mammalian Innate Immune Response by Synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 30.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, et al. Sequence-Specific Potent Induction of IFN-alpha by Short Interfering RNA in Plasmacytoid Dendritic Cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 31.Thomas M, Klibanov AM. Conjugation to Gold Nanoparticles Enhances Polyethylenimine's Transfer of Plasmid DNA into Mammalian Cells. Proc. Natl. Acad. Sci. U S A. 2003;100:9138–9143. doi: 10.1073/pnas.1233634100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 33.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ananta JS, Godin B, Sethi R, Moriggi L, Liu X, Serda RE, Krishnamurthy R, Muthupillai R, Bolskar RD, Helm L, et al. Geometrical Confinement of Gadolinium-Based Contrast Agents in Nanoporous Particles Enhances T1 Contrast. Nat. Nanotechnol. 2010;5:815–821. doi: 10.1038/nnano.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanaka T, Mangala LS, Vivas-Mejia PE, Nieves-Alicea R, Mann AP, Mora E, Han HD, Shahzad MM, Liu X, Bhavane R, et al. Sustained Small Interfering RNA Delivery by Mesoporous Silicon Particles. Cancer Res. 2010;70:3687–3696. doi: 10.1158/0008-5472.CAN-09-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.