Abstract
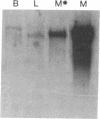
We have cloned the cDNA encoding glycogen phosphorylase (1,4-alpha-D-glucan:orthophosphate alpha-D-glucosyl-transferase, EC 2.4.1.1) from human liver. Blot-hybridization analysis using a large fragment of the cDNA to probe mRNA from rabbit brain, muscle, and liver tissues shows preferential hybridization to liver RNA. Determination of the entire nucleotide sequence of the liver message has allowed a comparison with the previously determined rabbit muscle phosphorylase sequence. Despite an amino acid identity of 80%, the two cDNAs exhibit a remarkable divergence in G+C content. In the muscle phosphorylase sequence, 86% of the nucleotides at the third codon position are either deoxyguanosine or deoxycytidine residues, while in the liver homolog the figure is only 60%, resulting in a strikingly different pattern of codon usage throughout most of the sequence. The liver phosphorylase cDNA appears to represent an evolutionary mosaic; the segment encoding the N-terminal 80 amino acids contains greater than 90% G+C at the third codon position. A survey of other published mammalian cDNA sequences reveals that the data for liver and muscle phosphorylases reflects a bias in codon usage patterns in liver and muscle coding sequences in general.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ashley P. L., MacDonald R. J. Kallikrein-related mRNAs of the rat submaxillary gland: nucleotide sequences of four distinct types including tonin. Biochemistry. 1985 Aug 13;24(17):4512–4520. doi: 10.1021/bi00338a005. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985 May 24;228(4702):953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Booth F. W., Watson P. A. Control of adaptations in protein levels in response to exercise. Fed Proc. 1985 Apr;44(7):2293–2300. [PubMed] [Google Scholar]
- David E. S., Crerar M. M. Quantitation of muscle glycogen phosphorylase mRNA and enzyme amounts in adult rat tissues. Biochim Biophys Acta. 1986 Jan 15;880(1):78–90. doi: 10.1016/0304-4165(86)90122-4. [DOI] [PubMed] [Google Scholar]
- Fletterick R. J., Madsen N. B. The structures and related functions of phosphorylase a. Annu Rev Biochem. 1980;49:31–61. doi: 10.1146/annurev.bi.49.070180.000335. [DOI] [PubMed] [Google Scholar]
- Gilbert W. Why genes in pieces? Nature. 1978 Feb 9;271(5645):501–501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Mercier R., Pavé A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980 Jan 11;8(1):r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HENION W. F., SUTHERLAND E. W. Immunological differences of phosphorylases. J Biol Chem. 1957 Jan;224(1):477–488. [PubMed] [Google Scholar]
- Hers H. G. The control of glycogen metabolism in the liver. Annu Rev Biochem. 1976;45:167–189. doi: 10.1146/annurev.bi.45.070176.001123. [DOI] [PubMed] [Google Scholar]
- Hwang P. K., See Y. P., Vincentini A. M., Powers M. A., Fletterick R. J., Crerar M. M. Comparative sequence analysis of rat, rabbit, and human muscle glycogen phosphorylase cDNAs. Eur J Biochem. 1985 Oct 15;152(2):267–274. doi: 10.1111/j.1432-1033.1985.tb09193.x. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985 Jan;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- Joh K., Mukai T., Yatsuki H., Hori K. Rat aldolase A messenger RNA: the nucleotide sequence and multiple mRNA species with different 5'-terminal regions. Gene. 1985;39(1):17–24. doi: 10.1016/0378-1119(85)90102-7. [DOI] [PubMed] [Google Scholar]
- Kagawa Y., Nojima H., Nukiwa N., Ishizuka M., Nakajima T., Yasuhara T., Tanaka T., Oshima T. High guanine plus cytosine content in the third letter of codons of an extreme thermophile. DNA sequence of the isopropylmalate dehydrogenase of Thermus thermophilus. J Biol Chem. 1984 Mar 10;259(5):2956–2960. [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kruk B., Kaciuba-Uściłko H., Nazar K., Greenleaf J. E., Kozłowski S. Hypothalamic, rectal, and muscle temperatures in exercising dogs: effect of cooling. J Appl Physiol (1985) 1985 May;58(5):1444–1448. doi: 10.1152/jappl.1985.58.5.1444. [DOI] [PubMed] [Google Scholar]
- MacDonald R. J., Stary S. J., Swift G. H. Two similar but nonallelic rat pancreatic trypsinogens. Nucleotide sequences of the cloned cDNAs. J Biol Chem. 1982 Aug 25;257(16):9724–9732. [PubMed] [Google Scholar]
- MacDonald R. J., Swift G. H., Quinto C., Swain W., Pictet R. L., Nikovits W., Rutter W. J. Primary structure of two distinct rat pancreatic preproelastases determined by sequence analysis of the complete cloned messenger ribonucleic acid sequences. Biochemistry. 1982 Mar 16;21(6):1453–1463. doi: 10.1021/bi00535a053. [DOI] [PubMed] [Google Scholar]
- Max E. E. New twist to DNA methylation. Nature. 1984 Jul 12;310(5973):100–100. doi: 10.1038/310100a0. [DOI] [PubMed] [Google Scholar]
- Paolella G., Santamaria R., Izzo P., Costanzo P., Salvatore F. Isolation and nucleotide sequence of a full-length cDNA coding for aldolase B from human liver. Nucleic Acids Res. 1984 Oct 11;12(19):7401–7410. doi: 10.1093/nar/12.19.7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinto C., Quiroga M., Swain W. F., Nikovits W. C., Jr, Standring D. N., Pictet R. L., Valenzuela P., Rutter W. J. Rat preprocarboxypeptidase A: cDNA sequence and preliminary characterization of the gene. Proc Natl Acad Sci U S A. 1982 Jan;79(1):31–35. doi: 10.1073/pnas.79.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schane H. P. Molecular weight estimation of rat uterine phosphorylase. Anal Biochem. 1965 May;11(2):371–374. doi: 10.1016/0003-2697(65)90025-4. [DOI] [PubMed] [Google Scholar]
- Swift G. H., Dagorn J. C., Ashley P. L., Cummings S. W., MacDonald R. J. Rat pancreatic kallikrein mRNA: nucleotide sequence and amino acid sequence of the encoded preproenzyme. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7263–7267. doi: 10.1073/pnas.79.23.7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T. C., Goldstein J. L., Brown M. S., Russell D. W. The LDL receptor gene: a mosaic of exons shared with different proteins. Science. 1985 May 17;228(4701):815–822. doi: 10.1126/science.2988123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titani K., Koide A., Hermann J., Ericsson L. H., Kumar S., Wade R. D., Walsh K. A., Neurath H., Fischer E. H. Complete amino acid sequence of rabbit muscle glycogen phosphorylase. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4762–4766. doi: 10.1073/pnas.74.11.4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolan D. R., Amsden A. B., Putney S. D., Urdea M. S., Penhoet E. E. The complete nucleotide sequence for rabbit muscle aldolase A messenger RNA. J Biol Chem. 1984 Jan 25;259(2):1127–1131. [PubMed] [Google Scholar]
- Tsutsumi K., Mukai T., Tsutsumi R., Hidaka S., Arai Y., Hori K., Ishikawa K. Structure and genomic organization of the rat aldolase B gene. J Mol Biol. 1985 Jan 20;181(2):153–160. doi: 10.1016/0022-2836(85)90081-6. [DOI] [PubMed] [Google Scholar]
- WOSILAIT W. D., SUTHERLAND E. W. The relationship of epinephrine and glucagon to liver phosphorylase. II. Enzymatic inactivation of liver phosphorylase. J Biol Chem. 1956 Jan;218(1):469–481. [PubMed] [Google Scholar]
- Winter G., Koch G. L., Hartley B. S., Barker D. G. The amino acid sequence of the tyrosyl-tRNA synthetase from Bacillus stearothermophilus. Eur J Biochem. 1983 May 2;132(2):383–387. doi: 10.1111/j.1432-1033.1983.tb07374.x. [DOI] [PubMed] [Google Scholar]