Abstract
The bacterial chromosome must be compacted over 1000-fold to fit into its cellular compartment. How it is condensed, organized and ultimately segregated has been a puzzle for over half a century. Recent advances in live-cell imaging and genome-scale analyses have led to new insights into these problems. We argue that the key feature of compaction is orderly folding of DNA along adjacent segments, and that this organization provides easy and efficient access for protein-DNA transactions and plays a central role in driving segregation. Similar principles and common proteins are used in eukaryotes to condense and resolve sister chromatids at metaphase.
Introduction
The visualization and characterization of the genetic material in bacteria has had a bumpy and controversial history. In eukaryotes, the orderly segregation of sister chromatids in mitosis was described in awe-inspiring detail in the 1880's 1; in contrast, the bacterial chromosome, which tends to stain uniformly with basic dyes, was for many years believed to be unstructured. It was not until the 1930’s that light microscopists using DNA dyes with acid-treated cells convincingly demonstrated that the bacterial chromosome was concentrated in confined bodies with soft irregular outlines (Fig. 1A)2,3. These images changed the view of the bacterial chromosome from a formless material to a discrete structure that hinted at orderly and predictable behavior4. These cloud-like nuclear bodies were named nucleoids.
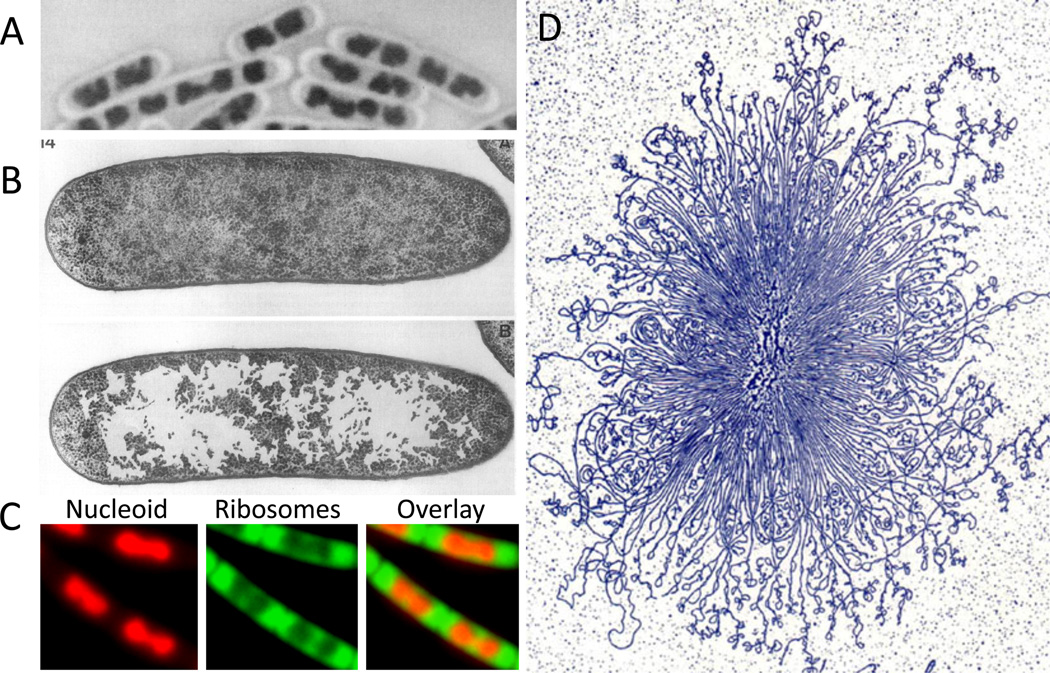
Figure 1. The bacterial nucleoid.
(A) B. subtilis nucleoid stained with Giemsa using acid-treated cells. (B) The nucleoid of growing E. coli in thin section after cryo-fixation followed by freeze-substitution. The upper and lower panels show the same section; in the lower panel, the ribosome-free spaces were enhanced by coloring by hand. (A) and (B) are adapted from Robinow and Kellenberger 4. (C) Nucleoid (stained with DAPI, colored red) and ribosomes (RplA-GFP, colored green) in live B. subtilis cells growing in rich media. Despite this commonly depicted cloud-like appearance of the bacterial chromosome, the morphology of the nucleoid varies among bacteria, and is influenced by growth rate and environmental conditions. For example, the nucleoid in C. crescentus, and in slow-growing E. coli and B. subtilis, appears more diffuse and occupies a greater proportion of the cell cytoplasm (not shown). (D) A gently isolated E. coli nucleoid bound by cytochrome C, spread on an EM grid, stained with uranyl acetate and visualized by transmission electron microscopy. Adapted from Physics in the twentieth century 140.
Cryoelectron microscopy of vitreous sections of nucleoids revealed structures with features similar to those observed using DNA dyes (Fig. 1B), with irregular and dispersed morphologies that occupied about half the intracellular space. Two striking features of these images were the presence of many corral-like projections that extended into the cytoplasm and the exclusion of the ribosomes from the nucleoid 4. Similar compartmentalization has since been observed using fluorescence microscopy 5 (Fig. 1C).
These images still provoke our thinking about the bacterial chromosome. We envision a nucleoid core and a DNA surface that interacts with proteins in the cytoplasm. Although proteins can penetrate into and reside within the interior of the nucleoid, most DNA transactions are thought to occur at its periphery.
In the early 1970’s, Pettijohn and colleagues developed methods to gently lyse Escherichia coli and obtain nucleoids for direct EM visualization 6–8, providing an enduring image of the bacterial chromosome as a collection of plectonemic (interwound) loops emanating from a dense core (Fig. 1D) suggested to be organized by proteins and RNA 6,7,9,10. The composition, organization, function (and even existence!) of the core remain important and outstanding issues in the field. These studies led to the rosette model of the bacterial chromosome in which interwound loops are organized by a nucleoid scaffold (Fig. 1D and Fig. 2A) creating a structure that resembles a bottlebrush. However, the molecular nature of this compact aggregate of DNA, its cellular localization and organization, and its local and global dynamics in living bacteria remained elusive.
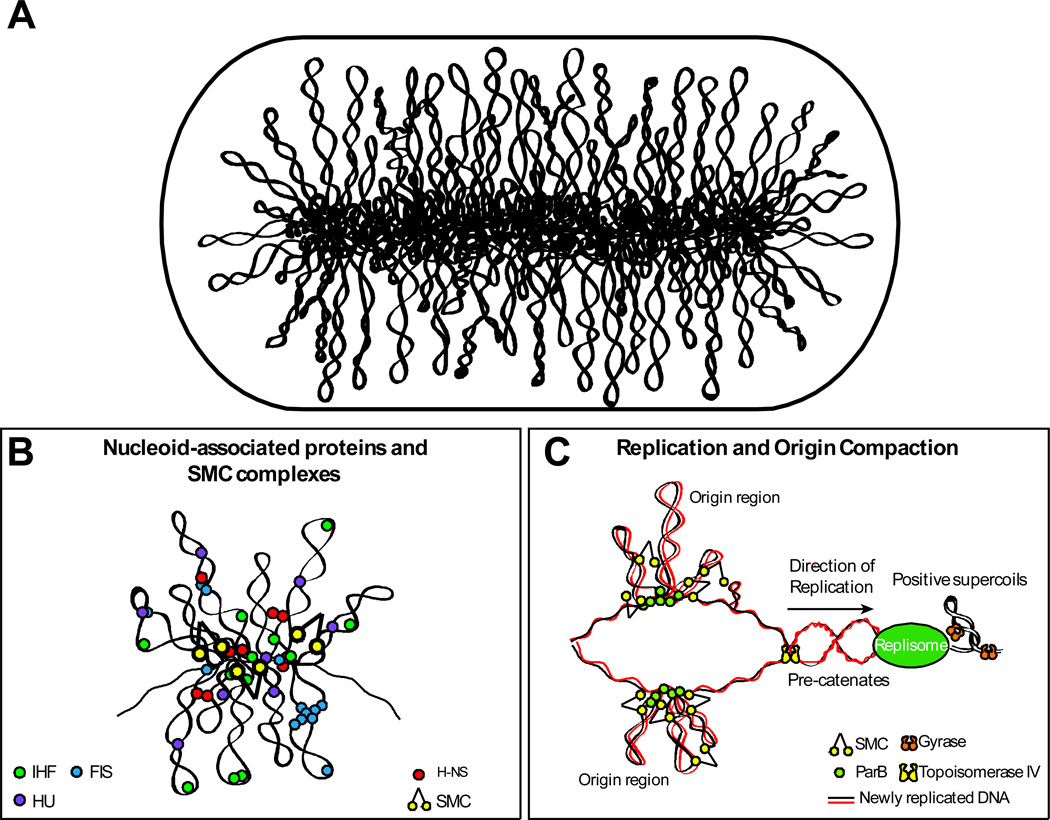
Figure 2. Topological organization of the bacterial chromosome.
(A) Schematic representation of the bottlebrush model of the nucleoid. This diagram depicts the interwound supercoiled loops emanating from a dense core. The topologically isolated domains (microdomains) are on average 10 kb and therefore likely encompass several branched plectonemic loops. (B) Schematic representation of the small nucleoid-associated proteins and SMC. These proteins introduce DNA bends and also function in bridging chromosomal loci. (C) The diagram depicts replication fork progression and compaction of the origin region. Replication generates positive supercoils ahead of the fork, which can diffuse behind the replisome producing pre-catenanes. Positive supercoils are removed by DNA gyrase and pre-catenanes are unlinked by Topo IV. Newly replicated origin regions thought to be compacted by the SMC complexes that are recruited to the origin and by the action of small nucleoid-associated proteins (not shown).
Advances in fluorescence microscopy and live-cell imaging along with the development of genome-wide molecular and analytical approaches (see Box 1) are providing new and exciting insights into bacterial chromosome organization and dynamics. Here, we draw on recent studies to review our current understanding of two problems: how the chromosome is organized and compacted in the bacterial cell and how the replicated chromosomes are disentangled and segregated. We discuss these topics separately but, as you will see, they are intimately connected. Our guiding premise is that the orderly folding of the chromosome along adjacent DNA segments (called lengthwise condensation) in lock-step with its replication generates its higher order organization and functions as the driving force for bulk chromosome segregation. Throughout, we highlight which principles and molecular mechanisms are shared with eukaryotes, and which aspects are specific to the unique chromosomal dynamics of bacteria.
Fluorescence In situ hybridization (FISH): Visualization of individual genetic loci using fluorescently labeled locus-specific DNA probes with fixed and permeabilized cells.
Fluorescence Repressor-Operator System (FROS) and fluorescently tagged DNA-binding proteins: Visualization of individual genetic loci in live cells using fluorescent fusions to repressor proteins (LacI, TetR, or lambdaCI) and tandem operator (lacO, tetO, or λOL1) arrays 57,58,63,137 or fluorescent fusions to plasmid-encoded ParB proteins bound to parS sites 73. Plasmid parS sites do not resemble chromosomal parS sites.
Chromatin immunoprecipitation-on-chip (ChIP-chip)/-sequencing (ChIP-seq): Genome-wide identification of DNA binding sites for a DNA binding proteins under investigation.
Chromosome Conformation Capture (3C)/Carbon Copy (5C)/High Resolution (Hi-C): Examines global conformation of the chromosome by assessing the frequency that any two DNA loci can be cross-linked 69,138,139.
Compaction and organization of the bacterial chromosome
Most bacteria contain a single circular chromosome of 2–8 megabases (Mb) in size that replicates bi-directionally from a unique origin (oriC). If stretched out, this DNA molecule would be >1 mm in length, whereas the cellular compartment in which it resides is <1 µm in diameter. Accordingly, the bacterial chromosome must be linearly compacted more than 1,000-fold to fit inside the bacterial cell 11,12. The DNA is condensed in an orderly and hierarchical fashion and we describe this compaction and organization from its smallest unit to its largest domain.
Topological microdomains
The principal mechanism by which the bacterial chromosome is compacted is by negative DNA supercoiling. This underwinding of the DNA duplex generates plectonemic loops and branches like the ones observed by EM in the nucleoid spreads. Supercoiling condenses the chromosome but it also draws DNA in on itself, pulling it away from non-contiguous DNA (such as replicated sister DNA). Unlike plasmids, which can be relaxed by one single-stranded break, a number of nicks are required to completely relax the chromosome, suggesting that chromosomal DNA is organized into supercoiled domains that are topologically insulated from each other 7. Elegant molecular experiments that exploit supercoiling-sensitive activities (such as transcription and recombination) suggest that independent topological domains vary in size but are on average 10 kb 13–15. Thus, a 4 Mb genome would have approximately 400 topologically isolated domains. In the context of the highly schematized bottlebrush model for the nucleoid, these domains are the branched plectonemic loops that make up the bristles (Fig. 1D and Fig. 2A). In addition to their role in condensing the chromosome, these topological domains protect the chromosome from DNA relaxation, assist in decatenation of chromosomal links, and have been proposed to aid in the repair of double strand breaks by maintaining broken ends in close proximity 14.
For these interwound loops to be topologically insulated, they need boundary elements (so-called domainins) that restrict the free rotation of DNA. Domainins are thought to function by constraining loops and are likely to be concentrated at the nucleoid core. Many factors have been proposed to serve as domainins including abundant small nucleoid-associated proteins, structural maintenance of chromosome (SMC) condensin complexes, topoisomerases, RNA polymerase and even RNA 14,16,17 (Fig. 2B). Most of these factors and their roles in chromosome organization and compaction are discussed in greater detail below. Importantly, there do not appear to be specific sequence elements that define domain boundaries. Despite the image of a rosette with a static core that the nucleoid spreads evoke, the current view is that the interwound loops and their boundaries (and by extension, the domainins that define them) are highly dynamic, changing in response to DNA transactions that occur within and between them 13,14,16. Consistent with this idea, chromosomal loci display remarkable, albeit constrained, mobility within the nucleoid 18–20. Thus, we envision a dynamic nucleoid core or scaffold composed of a loose assemblage of domainins. This fluid scaffold provides structure and organization to the supercoiled loops without imposing rigidity.
Supercoiling homeostasis is principally governed by the opposing actions of DNA gyrase, which introduces negative supercoils, and topoisomerase I (Topo I) that relaxes them 21–23. Gyrase and Topo I localize and act throughout the chromosome 24,25. However, gyrase is also enriched ahead of replication forks and transciption bubbles where it alleviates the positive supercoils introduced by DNA unwinding. Positive supercoils ahead of replication fork that are not attended to by gyrase can diffuse backwards, generating entangled sister strands (called pre-catenanes)26,27 (Fig. 2C). Topoisomerase IV (Topo IV) is the principal enzyme responsible for removing these entanglements 28–30; as such, it plays a central role in segregating the replicated chromosomes. In eukaryotes, topoisomerase II performs an analogous function to Topo IV, removing entanglements generated by DNA replication to resolve sister chromatids during the early stages of mitosis31.
Unconstrained supercoils alone cannot account for the degree of compaction exhibited by the bacterial chromosome. Approximately half of the chromosome is thought to be constrained by small abundant DNA binding proteins 32 (Fig. 2B). These proteins are the bacterial equivalent of eukaryotic histones. Instead of wrapping DNA into nucleosomes, they bind specifically and non-specifically throughout the genome and facilitate chromosome compaction and organization by introducing bends in the DNA and by bridging chromosomal loci. Bending facilitates condensation of adjacent DNA segments while bridging stabilizes DNA loops 33–40. This latter activity suggests that these proteins function as domainins 32,37. In E. coli, the principal proteins in this class are HU, IHF, Fis, and H-NS. Other bacteria (such as Bacillus subtilis and Caulobacter crescentus) have a subset of this class of protein. Cells lacking these factors have defects in chromosome segregation. However, the nucleoid does not appear dramatically decondensed in their absence, raising the possibility that other factors may play a more significant role in constraining supercoiled domains.
The highly conserved SMC condensin complex is, perhaps, the best candidate to constrain plectonemic loops and to function as a dynamic nucleoid scaffold 41,42 (Fig. 2B). In eukaryotes, SMC complexes function in chromosome condensation, sister chromatid cohesion, recombination, and X-chromosome dosage compensation 43,44. The bacterial SMC complex is composed of the SMC protein, a kleisin (closure) subunit called ScpA, and third protein called ScpB 45–48. Structural and functional analogs of this complex (called MukB, MukF, and MukE) are found in E. coli49,50. Cells lacking any of the proteins in the condensin complex are inviable at 37°C. At lower temperature, bacteria survive without this complex but have decondensed nucleoids and severe defects in chromosome segregation 45,47,48,51. The mechanism by which these large complexes organize and compact DNA has remained enigmatic and is the subject of intense research. Biochemical studies indicate that SMC complexes or higher order multimers can bridge and constrain DNA loops 17,41,52. Our view is that this bridging activity works hand-in-hand with supercoiling and the small nucleoid-associated proteins to fold the chromosome along adjacent segments. If correct, then these complexes are likely to act locally on neighboring stretches of DNA, despite their large size. Understanding how this complex functions in vitro and in vivo lies at the heart of understanding how the bacterial chromosome is organized and compacted and how sisters are segregated.
Macrodomains
The nucleoid is further organized into higher order structures called macrodomains, which are physically insulated from each other. These large regions (800 kb – 1 Mb in size) have been identified in E. coli but we suspect they are a common feature of many bacterial chromosomes. The higher order organization that defines a macrodomain does not appear to play a central role in the process of chromosome segregation but refines it and increase its fidelity 53 (see below). Organization of the chromosome into macrodomains was first recognized using fluorescence in situ hybridization (FISH) 54. Large regions of the genome spanning the origin (Ori) and terminus (Ter) exhibited spatially restricted localization patterns that were distinct from the rest of the chromosome, suggesting that loci in these regions cluster. Interestingly, a genetic assay based on recombination to assess the frequency of random collisions between different sites on the chromosome identified the same Ori and Ter macrodomains. This assay further delineated two additional insulated domains flanking the Ter macrodomain (called the Left and Right macrodomains) and two flexible or unstructured regions lying on either side of the Ori macrodomain 55. Consistent with the idea of structured macrodomains and unstructured flexible regions, time-lapse imaging of fluorescently labeled chromosomal loci were found to have different dynamic behaviors depending on their position in the chromosome 56. Loci in the unstructured regions displayed greater mobility than those within macrodomains.
The molecular mechanism underlying macrodomain organization is still unknown. However, recent evidence suggests that sequence-specific DNA binding proteins participate in this higher order organization. Bioinformatics analysis indentified a sequence motif (matS) that was highly overrepresented in the E. coli Ter macrodomain and virtually absent from the rest of the genome 53. This sequence element enabled the discovery of a DNA binding protein (MatP) that binds all 23 matS sites in vivo. Interestingly, MatP localizes as a focus that overlaps loci present in the Ter macrodomain, suggesting that it gathers or organizes matS sites. In this capacity it could act as a site-specific domainin; alternatively it could function in bundling interwound loops in the terminus region. In support of the idea that MatP is the Ter macrodomain organizer, loci in this domain become more mobile in cells lacking MatP and recombine with neighboring domains more frequently. The mechanism by which MatP organizes the terminus region; the identity of the proteins that specify the other macrodomains in E. coli; and whether or not similar domains exist in other bacterium are all active areas of investigation.
Cellular organization of the chromosome
Thus far, we have considered the organization and compaction of the chromosome without the spatial reference of the bacterial cell in which it resides. The development of methods to visualize individual chromosomal loci in live cells using fluorescence microscopy (see Box 1) 57–60 revealed a degree of spatial organization that had not been previously appreciated 60–64. This robust spatial organization reinforces our thinking about how the chromosome is compacted and informs our models for how newly replicated DNA is segregated.
The first cytological studies aimed at defining the subcellular localization of chromosomal loci were performed in B. subtilis62. Analysis of a locus adjacent to the origin revealed that replication initiates at or near mid-cell and that newly replicated origins rapidly segregate to the outer edges of the nucleoid. Subcellular localization of four chromosomal positions suggests that upon completion of replication the nucleoid adopts an organization in which the origins are present near opposite cell poles, the termini at mid-cell, and left and right chromosome arms lie between them 61 (Fig. 3A).
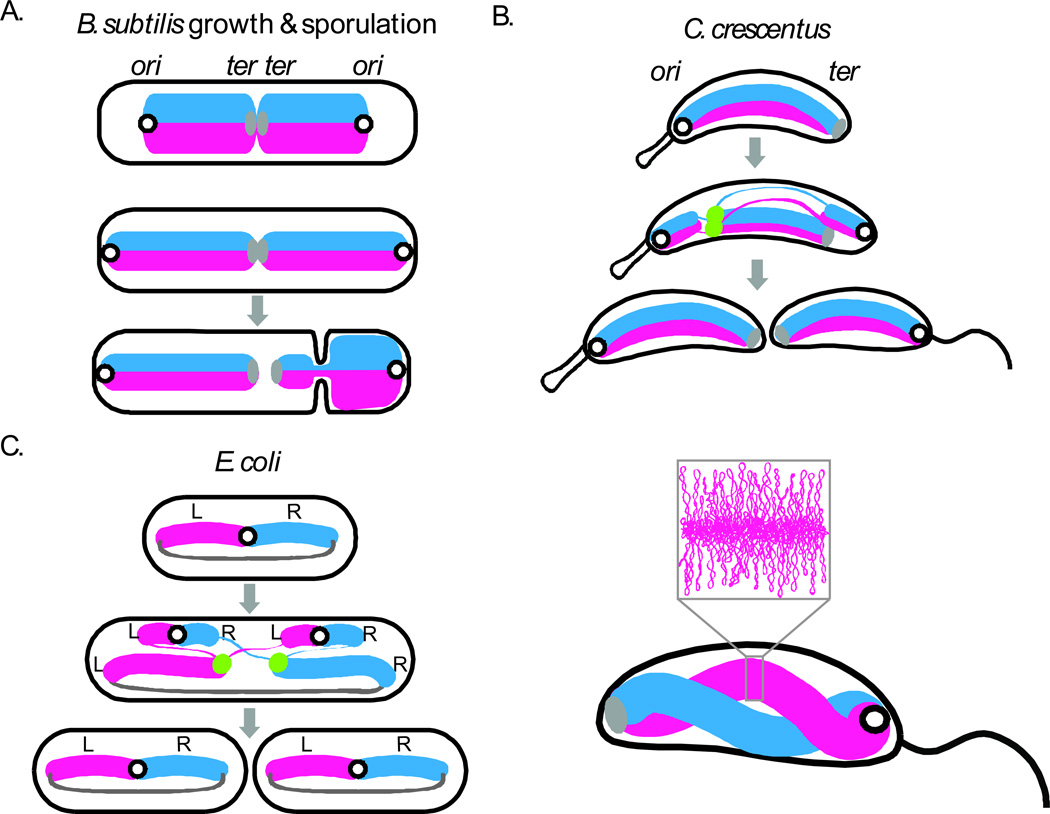
Figure 3. Spatial organization of bacterial chromosomes.
(A) Cellular organization of the chromosome in B. subtilis during growth and spore formation, C. crescentus (B) and slow growing E. coli (C) during vegetative cell cycles (gray arrows). During sporulation in B. subtilis, an asymmetric division traps about 25% of the chromosome in the smaller spore compartment 65,115. A DNA translocase (not shown) pumps the remaining 75% of the chromosome into spore after cytokinesis 125. The lower panel of (B) shows a model for the spatially separate but gently twisted arms of the C. crescentus chromosome based on chromosome conformation capture 69(new ref). The inset shows the plectonemic loops within one of the arms. Replication origins are illustrated as black open circles, termini as solid gray ovals (or lines in E. coli), the replisomes as green dots, the left (L) and right (R) replication arms as thick blue and pink lines, and newly replicated DNA as thinner lines. This figure is adapted from Jackson et al (2012)141.
How and when the origin moves to mid-cell to initiate a new round of replication and whether this movement impacts the overall organization of the chromosome remain to be addressed. Interestingly, using an elegant genetic assay 65, a similar “ori-ter ter-ori” nucleoid layout was identified in sporulating B. subtilis cells (Fig. 3A). During sporulation, the replicated chromosomes assemble into an elongated structure that extends from one cell pole to the other 62,66,67. Within this serpentine structure the replicated origins are located at opposite cell poles and the termini reside at mid-cell.
Analysis of chromosome organization in C. crescentus and E. coli followed the early cyological studies in B. subtilis and benefited from improved fluorescent proteins, methods to track chromosomal loci, and system-wide approaches. In C. crescentus, analysis of >100 separate loci revealed that the physical location of loci inside the cell recapitulates the genetic map 64. In cells that have not yet initiated replication, the origin and terminus are present at opposite poles, all other loci are organized along the long axis of the cell in an order that directly correlates with their position in the genome (Fig. 3B). Unlike the mid-cell initiation of replication in B. subtilis, replication initiates in C. crescentus at the origin-containing cell pole, and then one of the origins rapidly moves to the opposite pole. Replicated loci on the left and right arms follow suit. Thus, when replication is complete, the sister chromosomes have an ori-ter ter-ori organization (Fig. 3B). The linear organization of the chromosome arms suggests an orderly folding of adjacent DNA segments. However, the resolution of this cytological approach is not sufficient to assess whether or not the two chromosome arms are spatially resolved or entangled 68.
Recent experiments in C. crescentus using a high throughput chromosome conformation capture assay (called 5C) (see Box 1) and computational modeling have addressed the disposition of the two arms and provided the first three-dimensional model of a bacterial chromosome 69 (Fig. 3B). Consistent with the fluorescence microscopy studies, the model suggests that in C. crescentus the right and left arms of the chromosome are symmetric and linearly organized along the ori-ter axis. Importantly, the two arms are spatially separated although they gently twist around each other approximately one and half times (Fig. 3B). Thus, it appears that this bacterium condenses its chromosome along its length, generating two bottlebrushes, one for each chromosome arm. This three-dimensional rendering of C. crescentus chromosome is reminiscent of the twisted nucleoid structures observed by fluorescence microscopy in B. subtilis70 and E. coli71.
Similar systematic and genome-wide cytological analyses were performed in E. coli using slow-growing cells with a eukaryotic-like cell cycle such that newborn progeny have a single copy of the chromosome (Fig. 3C). These studies revealed that the organization of the E. coli chromosome is strikingly different from C. crescentus. At birth, the origin localizes near mid-cell with the left and right chromosome arms in opposite cell halves 72–74. To complete the circle, the terminus region is in an elongated organization that spans the length of the cell to bridge the two arms. During the process of replication (and after its completion) the sister chromosomes are organized into a Left-ori-Right, Left-ori-Right conformation (Fig 3C), such that cell division recapitulates the original organization in the daughter cells. Despite the difference in global chromosome organization, like C. crescentus, the left and right chromosome arms are linearly organized and have approximately constant packing density 72–74. The observed Left-ori-Right organization is consistent with the low frequency of recombination between loci in the Left and Right macrodomains. However, the role of the ter region as a connector of the left and right arms appears to be at odds with a structured Ter macrodomain. How this macrodomain fits in the context of the cellular organization of the chromosome remains to be discovered. In summary, although there are fundamental differences in the arrangement of the chromosome in different bacteria, the emergent and unifying theme is that the DNA is linearly organized and condensed along its length with approximately constant packing density.
Chromosome segregation
The past two decades have revealed an amazing degree of spatial organization of the bacterial chromosome. Importantly, as we have discussed, this organization is generated in lock-step with replication and as part of the segregation process. With this backdrop, we now turn our attention to how the replicated sisters are segregated. Segregation of most bacterial chromosomes can be broken down into three discrete steps: separation of the newly replicated origins; bulk chromosome segregation; and resolution and transport of the replication termini at the division septum. A surprisingly small set of highly conserved proteins has been implicated in these steps. We discuss each step separately and in the context of the patterns of nucleoid organization described above and their recreation in the in the next generation.
Origin segregation
Unlike eukaryotic cells that have temporally distinct phases for DNA replication, chromosome condensation, and sister chromatid segregation, bacteria organize, compact, and segregate their chromosomes progressively as the sister chromosomes are generated 64,75,76 (with exceptions described below 77) . Accordingly, much attention has been focused on how the origins are segregated as origin re-positioning provides a path and a destination for the rest of the chromosome.
The mechanism by which the newly replicated origins are segregated has been the subject of speculation and investigation for more than half a century. The origin attachment model proposed by Jacob, Brenner, and Cuzin in 1963 was among the first and endured for more than three decades 78. This model posits that the two newly replicated origins are tethered to the cell envelope close to mid-cell and are separated by cell growth between them. It is now clear that cell elongation in rod-shaped bacteria is not restricted to zonal growth at mid-cell but occurs throughout the cell cylinder. Furthermore, the movement of the origins away from mid-cell is much faster than the rate of cell growth 18,64,79,80. Thus, this attractively simple model cannot account for origin segregation.
As opposed to passive segregation embodied in the origin attachment model, active partitioning systems were first identified on plasmids in the 1980’s 81. These partitioning systems are essential for stable maintenance of low-copy-number plasmids 82–86 and their molecular characterization continues to play an important role in our understanding of how chromosomal origins are segregated. Remarkably, over 65% of all sequenced bacterial genomes contain a chromosomally encoded partitioning (par) locus 87. These species include B. subtilis88,89, C. Crescentus90, and Vibrio cholerae91. By contrast, E. coli and its close relatives do not posses this system. Chromosomal par loci (like their plasmid counterparts) consist of two genes, parA and parB and a cis-acting DNA site called parS. This centromere-like DNA element is frequently present in multiple copies and they are almost always located in close proximity to the replication origin 87. Insertion of this 3-component partitioning module onto an unstable plasmid improves plasmid maintenance even in unrelated host bacteria (including E. coli) 84,92,93. Thus, this locus has all the information required to partition DNA harboring the parS sequence.
For years, bacterial cell biologists have searched for a mitotic apparatus akin to the machinery employed by eukaryotes to segregate sister chromatids. The Par system is likely to be the closest bacterial counterpart. However, instead of segregating fully replicated sister chromatids, this system helps separate newly replicated origins (Fig. 4A). ParB is a DNA binding protein that binds site-specifically to the parS sites generating a large nucleoprotein complex adjacent to the origin 89,94. ParA is an ATPase with non-specific DNA binding activity that acts on this centromeric complex 85,95. Instead of microtubules and motor proteins, the ParA motor uses the nucleoid (a veritable sea of nonspecific DNA) to pull the newly replicated origins toward opposite cell poles 96 (Fig. 4A). Elegant biochemical and cytological analyses have begun to uncover the mechanistic underpinnings of this simple segregation system (see Fig. 4A for details). We refer the interested reader to recent reviews on this topic 85,97,98.
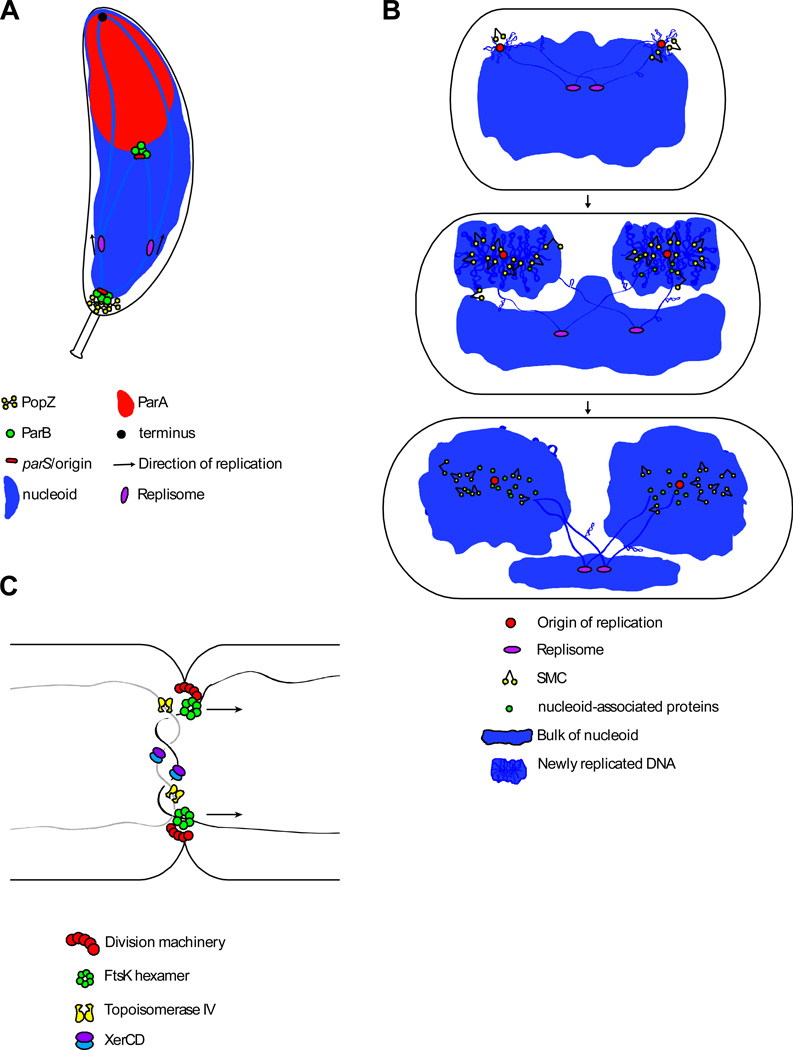
Figure 4. Chromosome segregation viewed in three steps.
(A) Schematic model of Par-mediated origin segregation in C. crescentus99–101. The origin region is tethered to the cell pole through interactions of ParB/parS with the polar anchor PopZ. After initiation of replication, one of the sister origins is pulled toward the opposite pole through interactions between ParB/parS and ParA(ATP) bound non-specifically to the nucleoid. These interactions trigger hydrolysis of ParA(ATP) and release of ParA(ADP) from the nucleoid. The ParB/pars complex then binds to neighboring ParA(ATP) on the nucleoid (alternatively, another ParB in the nucleoprotein complex engaged a nearby ParA(ATP) prior to release of the first). Repeated cycles of binding, hydrolysis, and release results in movement of the ParB/pars complex toward the cell pole and a ParA-free nucleoid in its wake. This so-called diffusion-ratchet mechanism allows the ParB/parS complex to “surf” on top of the nucleoid toward the pole 96,98. In C. crescentus, an additional protein (TipN) located at the cell pole is required for Par-mediated segregation 99,100 (not shown). TipN localizes to the new cell pole where it likely functions to regenerate ParA(ATP) helping to set up a ParA(ATP) gradient on the nucleoid. In par-containing bacteria that do not anchor their origins at the cell pole, the partitioning system helps re-position the newly replicated origins at the polar edges of the nucleoid. (B) Schematic model of bulk chromosome segregation. After newly replicated origins are separated, lengthwise condensation mediated by supercoiling, small nucleoid-associated proteins and SMC, in lock-step with replication, drive disentanglement and segregation of the sister chromosomes. (C) Schematic of terminus segregation in E. coli. The replicated terminus is translocated to appropriate daughter cell by the FtsK DNA translocase, while Topoisomerase IV and XerCD resolve catenanes and chromosome dimers, respectively. FtsK specifically localizes at the division septum where it participates in cytokinesis and DNA segregation.
Interestingly, in some bacteria the Par system plays a role in both establishing and maintaining the cellular organization of the chromosome. For example, in C. crescentus, after the origin is replicated at the cell pole, one of the ParB-parS-origin complexes is pulled to the opposite cell pole in a ParA-dependent manner 99–102. When it gets there, ParB bound parS interacts with a polar anchoring protein called PopZ 103,104. Accordingly, the partitioning system together with PopZ helps regenerate the ori-ter linear organization of the chromosome.
Par-independent origin segregation
Despite the high degree of conservation among par loci, many bacteria, including E. coli, lack these partitioning modules. Nonetheless, the replicated origins in E. coli rapidly move away from mid-cell 60,63,76. Moreover, when this locus is deleted from par-containing species, in most cases there are only modest defects in chromosome segregation and the separation of replicated origins is impaired but not eliminated 79,88,105–107 Accordingly, these modules function to refine origin segregation and improve its efficiency but in most cases may not be the driver of it.
What then is the underlying mechanism by which origins are segregated? Orderly lengthwise condensation and resolution of the replicated origins (discussed below) could explain their separation but it does not account for the faster rate of origin movement compared to more distal chromosomal loci108. It is possible that factors that have yet to be discovered are responsible for origin re-positioning. However, an intriguing alternative model proposed by Kleckner and colleagues posits that origin regions are extruded toward the cell poles as a result of intranucleoid pushing forces. In this model, the replicated origins undergo condensation and resolution from each other but remain cohesed at specific origin-proximal sites (called snaps). Meanwhile, DNA replication and compaction continue unabated. The accumulation of these DNA bodies in the confined space of the bacterial cell generates internal pushing forces. When these forces exceed the strength of the snaps, cohesion is lost, resulting in the abrupt and rapid extrusion of the condensed origin regions toward opposite poles. Since replication likely initiates at the nucleoid periphery, the newly replicated DNA is naturally compartmentalized from the unreplicated chromosome. This helps prevent entanglements and provides an unimpeded path for the extruded origins. The molecular basis of the snaps is currently unknown. However, two closely spaced origin-proximal regions on the E. coli chromosome with snap-like properties have recently been described 76,77. This model requires further investigation and refinement but, if correct, could be broadly relevant both in bacteria that possess and in those that lack partitioning loci.
Origin segregation must be more highly orchestrated and nuanced than this compelling model suggests. One observation that highlights this is the strong positional bias in the segregation of the leading and lagging strands of newly replicated DNA in E. coli109. The chromosome arms generated by leading strand synthesis are more frequently located at the outer edges of the nucleoid while the lagging-strand-synthesized arms are present on the opposite side of the origins, close to mid-cell. How the leading and lagging strands are positioned on a particular side of the origin and whether this orientation is established before or after origin segregation is not known.
Bulk chromosome segregation
As highlighted throughout this review, our guiding premise is that the orderly folding of the replicated sisters along adjacent DNA segments serves as the principal driver of bulk chromosome segregation. This lengthwise condensation is mediated by the concerted action of supercoiling, small nucleoid-associated proteins, and SMC condensin complexes (Fig. 4B). Compaction of neighboring DNA segments draws replicated sisters away from each other, makes the newly generated DNA stiffer and thicker, and, with the help of Topo IV, eliminates pre-catenated entanglements 30,110,111.
This model is consistent with the linear organization of the bacterial chromosome within the cell and its near uniform packing density 64,74. It is also in line with time-lapse imaging of chromosomal loci during replication in E. coli, which shows that newly replicated sister loci are sequentially segregated and co-localize with neighboring genetic loci, thus suggesting that condensation and segregation proceed in a coupled manner. Finally, consistent with this condensation-resolution scheme, bulk chromosome segregation is impaired when the proteins and processes that function in chromosome compaction are compromised. Specifically, cells with defects in supercoiling or that lack the small nucleoid-associated proteins or components of the SMC complex have segregation defects characterized by the formation of anucleate cells 45,49,51,112–115. This model makes sense intuitively but is also supported by mathematical modeling of two flexible polymer rings: compaction of catenated rings in an orderly and locally controlled manner along their lengths is sufficient to eliminate entanglements between them provided a mechanism (such as TopoIV-mediated decatenation) exists to unlink the rings 110.
The folding of the chromosome in upon itself likely initiates at the origin and is propagated outward. Replicated DNA is then sequentially and progressively gathered into these condensed structures (Fig. 4B). Intriguingly, SMC and MukB complexes, are enriched at the origin of replication in E. coli, C. crescentus, and B. subtilis50,115–117. The mechanism by which it is concentrated at this site in E. coli and C. crescentus is unknown. However, In B. subtilis, the SMC complex is recruited to the origin by ParB bound to origin-proximal parS sites 115,117 and, like its eukaryotic counterpart, enriched at the highly transcribed rRNA genes, most of which reside in close proximity to the origin 118. Interestingly, the parS sites are principally clustered to the left of the origin in B. subtilis while the rRNA operons are present on the right arm. We imagine that origin-localized SMC plays an important role in “seeding” independent lengthwise condensation of the left and right chromosome arms.
A corollary to this model is that condensation seeded at the origin could also function to dictate the overall organization of the bacterial chromosome. In support of this idea, Danilova and colleagues 50 found that E. coli MukB mutants that successfully inherit a chromosome switch from a Left-ori-Right organization to an ori-ter organization. The absence of origin-localized condensin complexes is thought to be responsible for this switch 50. We suspect that SMC complexes work similarly in C. crescentus and sporulating B. subtilis. However, in these cases, the origin is anchored at the cell pole. Thus, as a result of this constraint on the origin, the Left and Right arms lie side-by-side in separately condensed bodies rather than on opposite sides of the origin.
Recently, a model for bulk chromosome segregation based on conformational entropy was proposed 119,120. In this model segregation is driven by the tendency of confined polymers to separate from each other in a cylindrical container. We favor a model in which lengthwise folding of the replicated sisters drives their separation because it is consistent with the linear organization of the chromosome and the segregation defects observed in cells lacking compaction proteins, and it is applicable to all bacterial cells, regardless of their shape. However, we suspect that rod-shape bacteria take advantage of their carefully constructed geometry and in these organisms entropic sorting forces could facilitate the condensation-resolution process discussed here.
Segregation of the terminus region
In principle, condensation-resolution should be sufficient to segregate replicated sister chromosomes prior to cytokinesis. However, bacteria have evolved a septal localized DNA translocase to ensure efficient segregation of the terminus and to attend to the particular challenges of replicating a circular chromosome (Fig. 4C).
Replicating a circular chromosome generates two topological challenges: catenanes and dimers. When pre-catenanes are not removed by Topo IV during replication the replicated sisters remain linked to each other forming interlocked rings 26,27. In addition, as a result of homologous recombination between sisters during replication repair and an uneven number of crossovers, ~15% of the population ends up with conjoined sister chromosomes as a single circle 121. Thus, to complete sister chromosome segregation, TopoIV must remove the catenanes and a recombinase (called XerCD in E. coli and RipX and CodV in B. subtilis) must convert the chromosome dimers into monomers 122–124. Decatenation and dimer resolution are coordinated and facilitated by a DNA translocase (called FtsK in E. coli and C. crescentus, and SpoIIIE in B. subtilis). These membrane-anchored ATPases associate with the cell division apparatus at mid-cell and take advantage of strand-specific base-composition skew in the DNA to translocate the chromosome arms towards the replication termini and the site of dimer resolution (Fig 4C). These translocases are employed when DNA is present at the septum as a result of mis-segregation, chromosome dimers or catenanes. Interestingly, the B. subtilis DNA translocase is also used during sporulation to pump ~75% of the chromosome into the developing spore 65,115,125 (Fig 3A). Translocation during vegetative growth and sporulation brings the termini to mid-cell where XerCD and TopoIV can catalyze their unlinking and complete the segregation process 126–130 (Fig. 4C).
Concluding remarks and future directions
Live cell imaging and genome scale molecular approaches have taken the disembodied image of the bacterial nucleoid - plectonemes emanating from a central core - and provided a context in which to interpret it. The linear organization of the chromosome with its uniform packing density and the ordered layering of chromosomal loci during replication provide a clearer picture of the nucleoid and suggests a plausible and compelling mechanism for its segregation.
Defining how origins are segregated with and without a Par system and the mechanism by which SMC compacts DNA and are outstanding issues that will be addressed in the near future. Defining SMC action will inform (and be informed by) studies on eukaryotic SMC complexes. However, an equally important and challenging question is how the different compaction and segregation factors interface during the replication-segregation cycle. Intriguing hints of interconnections have been described over the past decade. In B. subtilis, the ParA protein appears to regulate replication initiation 131, while ParB bound to parS recruits SMC to the origin to facilitate compaction and segregation 115,117. Moreover, SMC (MukB) in E. coli has been found to interact with TopoIV and stimulate its activity in vitro 132. To round out this picture, the FtsK translocase also interacts with TopoIV 133, facilitating terminus separation and cell division. Understanding how these factors (and others) work together will provide a more complete picture of how the chromosome is organized and accurately segregated with such high fidelity.
The other major challenge for the future is understanding how chromosome condensation and segregation are influenced by the physical-chemical properties of the cell and basic cellular processes. We have touched upon a possible role for confinement but have not mentioned the crowded and metabolically active cytoplasm in which the nucleoid resides. Molecular crowding can contribute to chromosome compaction directly by creating a phase separation between the DNA and the rest of the cytoplasm, and indirectly by enhancing the interactions between the chromosome and DNA binding proteins 134,135. How crowding and confinement influence organization and segregation remain to be elucidated. As well, the chromosome is constantly being pushed and bullied by the replication and transcription machineries as well as recombination and repair proteins. These activities clearly influence chromosome dynamics. Interestingly, inhibition of transcription leads to a dramatic decondensation of the nucleoid, the molecular basis of which remains unknown 136. Understanding the interplay between the condensation and segregation machineries in the context of the crowded and metabolically active cell is, of course, a long way off but is a goal worthy of our efforts.
Acknowledgements
We thank members of the Rudner lab for advice and encouragement, John Marko and Suckjoon Jun for invaluable discussions, Nancy Kleckner for sharing unpublished data, and Becky Ward for expert editing. Support for this work comes from the National Institute of Health Grant GM086466, X.W. is a long-term fellow of the Human Frontier Science Program, P.M.L. is a Helen Hay Whitney fellow.
References
- 1.Baker JR. The cell-theory: a restatement, history and critique. Part V. The multiplication of nuclei. Q J Microsc Sci. 1955;96:449–481. [Google Scholar]
- 2.Piekarski G. Zytologische Untersuchungen an Paratyphusund Coli Bakterien. Arch. Mikrobiol. 1937;8:428–429. [Google Scholar]
- 3.Stille B. Zytologische Untersuchungen an Bakterien mit Hilfe der Feulgenschen Nuclealreaktion. Arch. Mikrobiol. 1937;8:124–148. [Google Scholar]
- 4.Robinow C, Kellenberger E. The bacterial nucleoid revisited. Microbiol Rev. 1994;58:211–232. doi: 10.1128/mr.58.2.211-232.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis PJ, Thaker SD, Errington J. Compartmentalization of transcription and translation in Bacillus subtilis. EmboJ. 2000;19:710–718. doi: 10.1093/emboj/19.4.710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delius H, Worcel A. Letter: Electron microscopic visualization of the folded chromosome of Escherichia coli. J Mol Biol. 1974;82:107–109. doi: 10.1016/0022-2836(74)90577-4. [DOI] [PubMed] [Google Scholar]
- 7.Worcel A, Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972;71:127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]
- 8.Kavenoff R, Ryder OA. Electron microscopy of membrane-associated folded chromosomes of Escherichia coli. Chromosoma. 1976;55:13–25. doi: 10.1007/BF00288323. [DOI] [PubMed] [Google Scholar]
- 9.Kavenoff R, Bowen BC. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma. 1976;59:89–101. doi: 10.1007/BF00328479. [DOI] [PubMed] [Google Scholar]
- 10.Pettijohn DE, Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Holmes VF, Cozzarelli NR. Closing the ring: links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci U S A. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trun NJ, Marko JF. Architecture of a bacterial chromosome. American Society for Microbiology News. 1998;64 [Google Scholar]
- 13.Higgins NP, Yang X, Fu Q, Roth JR. Surveying a supercoil domain by using the gamma delta resolution system in Salmonella typhimurium. J Bacteriol. 1996;178:2825–2835. doi: 10.1128/jb.178.10.2825-2835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Postow L, Hardy CD, Arsuaga J, Cozzarelli NR. Topological domain structure of the Escherichia coli chromosome. Genes Dev. 2004;18:1766–1779. doi: 10.1101/gad.1207504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stein RA, Deng S, Higgins NP. Measuring chromosome dynamics on different time scales using resolvases with varying half-lives. Mol Microbiol. 2005;56:1049–1061. doi: 10.1111/j.1365-2958.2005.04588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng S, Stein RA, Higgins NP. Organization of supercoil domains and their reorganization by transcription. Mol Microbiol. 2005;57:1511–1521. doi: 10.1111/j.1365-2958.2005.04796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Y, Petrushenko ZM, Rybenkov VV. MukB acts as a macromolecular clamp in DNA condensation. Nat Struct Mol Biol. 2008;15:411–418. doi: 10.1038/nsmb.1410. [DOI] [PubMed] [Google Scholar]
- 18.Fiebig A, Keren K, Theriot JA. Fine-scale time-lapse analysis of the biphasic, dynamic behaviour of the two Vibrio cholerae chromosomes. Mol Microbiol. 2006;60:1164–1178. doi: 10.1111/j.1365-2958.2006.05175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber SC, Spakowitz AJ, Theriot JA. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc Natl Acad Sci U S A. 2012;109:7338–7343. doi: 10.1073/pnas.1119505109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 22.Luttinger A. The twisted 'life' of DNA in the cell: bacterial topoisomerases. Mol Microbiol. 1995;15:601–606. doi: 10.1111/j.1365-2958.1995.tb02369.x. [DOI] [PubMed] [Google Scholar]
- 23.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Hsu YH, Chung MW, Li TK. Distribution of gyrase and topoisomerase IV on bacterial nucleoid: implications for nucleoid organization. Nucleic Acids Res. 2006;34:3128–3138. doi: 10.1093/nar/gkl392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tadesse S, Graumann PL. Differential and dynamic localization of topoisomerases in Bacillus subtilis. J Bacteriol. 2006;188:3002–3011. doi: 10.1128/JB.188.8.3002-3011.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khodursky AB, et al. Analysis of topoisomerase function in bacterial replication fork movement: use of DNA microarrays. Proc Natl Acad Sci U S A. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peter BJ, Ullsperger C, Hiasa H, Marians KJ, Cozzarelli NR. The structure of supercoiled intermediates in DNA replication. Cell. 1998;94:819–827. doi: 10.1016/s0092-8674(00)81740-7. [DOI] [PubMed] [Google Scholar]
- 28.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J Biol Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 29.Zechiedrich EL, Khodursky AB, Cozzarelli NR. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 31.Shintomi K, Hirano T. Sister chromatid resolution: a cohesin releasing network and beyond. Chromosoma. 2010;119:459–467. doi: 10.1007/s00412-010-0271-z. [DOI] [PubMed] [Google Scholar]
- 32.Hardy CD, Cozzarelli NR. A genetic selection for supercoiling mutants of Escherichia coli reveals proteins implicated in chromosome structure. Mol Microbiol. 2005;57:1636–1652. doi: 10.1111/j.1365-2958.2005.04799.x. [DOI] [PubMed] [Google Scholar]
- 33.Azam TA, Hiraga S, Ishihama A. Two types of localization of the DNA-binding proteins within the Escherichia coli nucleoid. Genes Cells. 2000;5:613–626. doi: 10.1046/j.1365-2443.2000.00350.x. [DOI] [PubMed] [Google Scholar]
- 34.Azam TA, Ishihama A. Twelve species of the nucleoid-associated protein from Escherichia coli. Sequence recognition specificity and DNA binding affinity. J Biol Chem. 1999;274:33105–33113. doi: 10.1074/jbc.274.46.33105. [DOI] [PubMed] [Google Scholar]
- 35.Dame RT, Wyman C, Goosen N. H-NS mediated compaction of DNA visualised by atomic force microscopy. Nucleic Acids Res. 2000;28:3504–3510. doi: 10.1093/nar/28.18.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. H-NS promotes looped domain formation in the bacterial chromosome. Curr Biol. 2007;17:R913–R914. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Skoko D, et al. Mechanism of chromosome compaction and looping by the Escherichia coli nucleoid protein Fis. J Mol Biol. 2006;364:777–798. doi: 10.1016/j.jmb.2006.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 39.Luijsterburg MS, Noom MC, Wuite GJ, Dame RT. The architectural role of nucleoid-associated proteins in the organization of bacterial chromatin: a molecular perspective. J Struct Biol. 2006;156:262–272. doi: 10.1016/j.jsb.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Luijsterburg MS, White MF, van Driel R, Dame RT. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit Rev Biochem Mol Biol. 2008;43:393–418. doi: 10.1080/10409230802528488. [DOI] [PubMed] [Google Scholar]
- 41.Hirano M, Hirano T. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 42.Hirano T. At the heart of the chromosome: SMC proteins in action. Nat Rev Mol Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- 43.Haering CH, Nasmyth K. Building and breaking bridges between sister chromatids. Bioessays. 2003;25:1178–1191. doi: 10.1002/bies.10361. [DOI] [PubMed] [Google Scholar]
- 44.Nasmyth K, Haering CH. The structure and function of SMC and kleisin complexes. Annu Rev Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- 45.Britton RA, Lin DC, Grossman AD. Characterization of a prokaryotic SMC protein involved in chromosome partitioning. Genes Dev. 1998;12:1254–1259. doi: 10.1101/gad.12.9.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirano M, Hirano T. Positive and negative regulation of SMC-DNA interactions by ATP and accessory proteins. EMBO J. 2004;23:2664–2673. doi: 10.1038/sj.emboj.7600264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soppa J, et al. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- 49.Niki H, Jaffe A, Imamura R, Ogura T, Hiraga S. The new gene mukB codes for a 177 kd protein with coiled-coil domains involved in chromosome partitioning of E. coli. EmboJ. 1991;10:183–193. doi: 10.1002/j.1460-2075.1991.tb07935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danilova O, Reyes-Lamothe R, Pinskaya M, Sherratt D, Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jensen RB, Shapiro L. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc Natl Acad Sci U S A. 1999;96:10661–10666. doi: 10.1073/pnas.96.19.10661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrushenko ZM, Cui Y, She W, Rybenkov VV. Mechanics of DNA bridging by bacterial condensin MukBEF in vitro and in singulo. EmboJ. 2010;29:1126–1135. doi: 10.1038/emboj.2009.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mercier R, et al. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 54.Niki H, Yamaichi Y, Hiraga S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000;14:212–223. [PMC free article] [PubMed] [Google Scholar]
- 55.Valens M, Penaud S, Rossignol M, Cornet F, Boccard F. Macrodomain organization of the Escherichia coli chromosome. EMBOJ. 2004;23:4330–4341. doi: 10.1038/sj.emboj.7600434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- 57.Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- 58.Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- 59.Robinett CC, et al. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Sergueev K, Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol. 2002;46:985–996. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- 61.Teleman AA, Graumann PL, Lin DC, Grossman AD, Losick R. Chromosome arrangement within a bacterium. Curr Biol. 1998;8:1102–1109. doi: 10.1016/s0960-9822(98)70464-6. [DOI] [PubMed] [Google Scholar]
- 62.Webb CD, et al. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 63.Lau IF, et al. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol Microbiol. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 64.Viollier PH, et al. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc Natl Acad Sci USA. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu LJ, Errington J. Use of asymmetric cell division and spoIIIE mutants to probe chromosome orientation and organization in Bacillus subtilis. Mol Microbiol. 1998;27:777–786. doi: 10.1046/j.1365-2958.1998.00724.x. [DOI] [PubMed] [Google Scholar]
- 66.Ben-Yehuda S, Rudner DZ, Losick R. RacA, a bacterial protein that anchors chromosomes to the cell poles. Science. 2003;299:532–536. doi: 10.1126/science.1079914. [DOI] [PubMed] [Google Scholar]
- 67.Lin DC, Levin PA, Grossman AD. Bipolar localization of a chromosome partition protein in Bacillus subtilis. Proc Natl Acad Sci U S A. 1997;94:4721–4726. doi: 10.1073/pnas.94.9.4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Breier AM, Cozzarelli NR. Linear ordering and dynamic segregation of the bacterial chromosome. Proc Natl Acad Sci U S A. 2004;101:9175–9176. doi: 10.1073/pnas.0403722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Umbarger MA, et al. The three-dimensional architecture of a bacterial genome and its alteration by genetic perturbation. Mol Cell. 2011;44:252–264. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Berlatzky IA, Rouvinski A, Ben-Yehuda S. Spatial organization of a replicating bacterial chromosome. Proc Natl Acad Sci U S A. 2008;105:14136–14140. doi: 10.1073/pnas.0804982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hadizadeh Yazdi N, Guet CC, Johnson RC, Marko JF. Variation of the folding and dynamics of the Escherichia coli chromosome with growth conditions. Mol Microbiol. 2012 doi: 10.1111/mmi.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Liu X, Possoz C, Sherratt DJ. The two Escherichia coli chromosome arms locate to separate cell halves. Genes Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nielsen HJ, Ottesen JR, Youngren B, Austin SJ, Hansen FG. The Escherichia coli chromosome is organized with the left and right chromosome arms in separate cell halves. Mol Microbiol. 2006;62:331–338. doi: 10.1111/j.1365-2958.2006.05346.x. [DOI] [PubMed] [Google Scholar]
- 74.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci U S A. 2010;107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lesterlin C, Gigant E, Boccard F, Espeli O. Sister chromatid interactions in bacteria revealed by a site-specific recombination assay. EMBOJ. 2012;31:3468–3479. doi: 10.1038/emboj.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 77.Joshi MC, et al. Escherichia coli sister chromosome separation includes an abrupt global transition with concomitant release of late-splitting intersister snaps. Proc Natl Acad Sci U S A. 2011;108:2765–2770. doi: 10.1073/pnas.1019593108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jacob F, Brenner S, Cuzin F. On the regulation of DNA replication in bacteria. Cold Spring Harb Symp Quant Biol. 1963;28:329–348. [Google Scholar]
- 79.Webb CD, et al. Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol. 1998;28:883–892. doi: 10.1046/j.1365-2958.1998.00808.x. [DOI] [PubMed] [Google Scholar]
- 80.Wang X, Sherratt DJ. Independent segregation of the two arms of the Escherichia coli ori region requires neither RNA synthesis nor MreB dynamics. J Bacteriol. 2010;192:6143–6153. doi: 10.1128/JB.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abeles AL, Friedman SA, Austin SJ. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 82.Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci U S A. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sengupta M, Nielsen HJ, Youngren B, Austin S. P1 plasmid segregation: accurate redistribution by dynamic plasmid pairing and separation. J Bacteriol. 2010;192:1175–1183. doi: 10.1128/JB.01245-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ebersbach G, et al. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol Microbiol. 2006;61:1428–1442. doi: 10.1111/j.1365-2958.2006.05322.x. [DOI] [PubMed] [Google Scholar]
- 85.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 86.Schumacher MA. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 87.Livny J, Yamaichi Y, Waldor MK. Distribution of centromere-like parS sites in bacteria: insights from comparative genomics. J Bacteriol. 2007;189:8693–8703. doi: 10.1128/JB.01239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ireton K, Gunther NW, Grossman AD. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin DC, Grossman AD. Identification and characterization of a bacterial chromosome partitioning site. Cell. 1998;92:675–685. doi: 10.1016/s0092-8674(00)81135-6. [DOI] [PubMed] [Google Scholar]
- 90.Mohl DA, Gober JW. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 91.Heidelberg JF, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Godfrin-Estevenon AM, Pasta F, Lane D. The parAB gene products of Pseudomonas putida exhibit partition activity in both P. putida and Escherichia coli. Mol Microbiol. 2002;43:39–49. doi: 10.1046/j.1365-2958.2002.02735.x. [DOI] [PubMed] [Google Scholar]
- 94.Murray H, Ferreira H, Errington J. The bacterial chromosome segregation protein Spo0J spreads along DNA from parS nucleation sites. Mol Microbiol. 2006;61:1352–1361. doi: 10.1111/j.1365-2958.2006.05316.x. [DOI] [PubMed] [Google Scholar]
- 95.Leonard TA, Butler PJ, Lowe J. Bacterial chromosome segregation: structure and DNA binding of the Soj dimer--a conserved biological switch. EmboJ. 2005;24:270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vecchiarelli AG, et al. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mierzejewska J, Jagura-Burdzy G. Prokaryotic ParA-ParB-parS system links bacterial chromosome segregation with the cell cycle. Plasmid. 2012;67:1–14. doi: 10.1016/j.plasmid.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 98.Szardenings F, Guymer D, Gerdes K. ParA ATPases can move and position DNA and subcellular structures. Curr Opin Microbiol. 2011;14:712–718. doi: 10.1016/j.mib.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 99.Ptacin JL, et al. A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol. 2010;12:791–798. doi: 10.1038/ncb2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Shebelut CW, Guberman JM, van Teeffelen S, Yakhnina AA, Gitai Z. Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci U S A. 2010;107:14194–14198. doi: 10.1073/pnas.1005274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Toro E, Hong SH, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci U S A. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in caulobacter. Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bowman GR, et al. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell. 2008;134:945–955. doi: 10.1016/j.cell.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fogel MA, Waldor MK. A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 2006;20:3269–3282. doi: 10.1101/gad.1496506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. J Bacteriol. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lewis RA, Bignell CR, Zeng W, Jones AC, Thomas CM. Chromosome loss from par mutants of Pseudomonas putida depends on growth medium and phase of growth. Microbiology. 2002;148:537–548. doi: 10.1099/00221287-148-2-537. [DOI] [PubMed] [Google Scholar]
- 108.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008;455:1248–1250. doi: 10.1038/nature07282. [DOI] [PubMed] [Google Scholar]
- 110.Marko JF. Linking topology of tethered polymer rings with applications to chromosome segregation and estimation of the knotting length. Phys Rev E Stat Nonlin Soft Matter Phys. 2009;79:051905. doi: 10.1103/PhysRevE.79.051905. [DOI] [PubMed] [Google Scholar]
- 111.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 112.Painbeni E, Caroff M, Rouviere-Yaniv J. Alterations of the outer membrane composition in Escherichia coli lacking the histone-like protein HU. Proc Natl Acad Sci U S A. 1997;94:6712–6717. doi: 10.1073/pnas.94.13.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moriya S, et al. A Bacillus subtilis gene-encoding protein homologous to eukaryotic SMC motor protein is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 114.Schwartz MA, Shapiro L. An SMC ATPase mutant disrupts chromosome segregation in Caulobacter. Mol Microbiol. 2011;82:1359–1374. doi: 10.1111/j.1365-2958.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sullivan NL, Marquis KA, Rudner DZ. Recruitment of SMC by ParB-parS organizes the origin region and promotes efficient chromosome segregation. Cell. 2009;137:697–707. doi: 10.1016/j.cell.2009.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jensen RB, Shapiro L. Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J Bacteriol. 2003;185:3068–3075. doi: 10.1128/JB.185.10.3068-3075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gruber S, Errington J. Recruitment of condensin to replication origin regions by ParB/SpoOJ promotes chromosome segregation in B. subtilis. Cell. 2009;137:685–696. doi: 10.1016/j.cell.2009.02.035. [DOI] [PubMed] [Google Scholar]
- 118.Jarvis ED, et al. Chromosomal organization of rRNA operons in Bacillus subtilis. Genetics. 1988;120:625–635. doi: 10.1093/genetics/120.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jun S, Wright A. Entropy as the driver of chromosome segregation. Nat Rev Micro. 2010;8:600–607. doi: 10.1038/nrmicro2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Steiner WW, Kuempel PL. Sister chromatid exchange frequencies in Escherichia coli analyzed by recombination at the dif resolvase site. J Bacteriol. 1998;180:6269–6275. doi: 10.1128/jb.180.23.6269-6275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lemon KP, Kurtser I, Grossman AD. Effects of replication termination mutants on chromosome partitioning in Bacillus subtilis. Proc Natl Acad Sci U S A. 2001;98:212–217. doi: 10.1073/pnas.011506098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kaimer C, Schenk K, Graumann PL. Two DNA translocases synergistically affect chromosome dimer resolution in Bacillus subtilis. J Bacteriol. 2011;193:1334–1340. doi: 10.1128/JB.00918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cortez D, et al. Evidence for a Xer/dif system for chromosome resolution in archaea. PLoS Genet. 2010;6:e1001166. doi: 10.1371/journal.pgen.1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu LJ, Errington J. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 126.Duggin IG, Wake RG, Bell SD, Hill TM. The replication fork trap and termination of chromosome replication. Mol Microbiol. 2008;70:1323–1333. doi: 10.1111/j.1365-2958.2008.06500.x. [DOI] [PubMed] [Google Scholar]
- 127.Grainge I, et al. Unlinking chromosome catenanes in vivo by site-specific recombination. EmboJ. 2007;26:4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grainge I, Lesterlin C, Sherratt DJ. Activation of XerCD-dif recombination by the FtsK DNA translocase. Nucleic Acids Res. 2011;39:5140–5148. doi: 10.1093/nar/gkr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sivanathan V, et al. KOPS-guided DNA translocation by FtsK safeguards Escherichia coli chromosome segregation. Mol Microbiol. 2009;71:1031–1042. doi: 10.1111/j.1365-2958.2008.06586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Steiner W, Liu G, Donachie WD, Kuempel P. The cytoplasmic domain of FtsK protein is required for resolution of chromosome dimers. Mol Microbiol. 1999;31:579–583. doi: 10.1046/j.1365-2958.1999.01198.x. [DOI] [PubMed] [Google Scholar]
- 131.Murray H, Errington J. Dynamic control of the DNA replication initiation protein DnaA by Soj/ParA. Cell. 2008;135:74–84. doi: 10.1016/j.cell.2008.07.044. [DOI] [PubMed] [Google Scholar]
- 132.Hayama R, Marians KJ. Physical and functional interaction between the condensin MukB and the decatenase topoisomerase IV in Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:18826–18831. doi: 10.1073/pnas.1008140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Espeli O, Levine C, Hassing H, Marians KJ. Temporal regulation of topoisomerase IV activity in E. coli. Mol Cell. 2003;11:189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- 134.Iborra FJ. Can visco-elastic phase separation, macromolecular crowding and colloidal physics explain nuclear organisation? Theor Biol Med Model. 2007;4 doi: 10.1186/1742-4682-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou HX, Rivas G, Minton AP. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu Rev Biophys. 2008;37:375–397. doi: 10.1146/annurev.biophys.37.032807.125817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cabrera JE, Jin DJ. The distribution of RNA polymerase in Escherichia coli is dynamic and sensitive to environmental cues. Mol Microbiol. 2003;50:1493–1505. doi: 10.1046/j.1365-2958.2003.03805.x. [DOI] [PubMed] [Google Scholar]
- 137.Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- 138.Lieberman-Aiden E, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 140.Suplee C. Physics in the twentieth Century. Harry N; Abrams, NY: 1999. [Google Scholar]
- 141.Jackson D, Wang X, Rudner DZ. Spatio-temporal organization of replication in bacteria and eukaryotes (nucleoids and nuclei) Cold Spring Harb Perspect Biol. 2012;4:a010389. doi: 10.1101/cshperspect.a010389. [DOI] [PMC free article] [PubMed] [Google Scholar]