Abstract
Breast cancer is the most common cancer in women in developed countries and has a well-established genetic component. Germline mutations in a network of genes encoding BRCA1, BRCA2, and their interacting partners confer hereditary susceptibility to breast cancer. Abraxas directly interacts with the BRCA1 BRCT (BRCA1 carboxyl-terminal) repeats and contributes to BRCA1-dependent DNA damage responses, making Abraxas a candidate for yet unexplained disease susceptibility. Here, we have screened 125 Northern Finnish breast cancer families for coding region and splice-site Abraxas mutations and genotyped three tagging single-nucleotide polymorphisms within the gene from 991 unselected breast cancer cases and 868 female controls for common cancer-associated variants. A novel heterozygous alteration, c.1082G>A (Arg361Gln), that results in abrogated nuclear localization and DNA response activities was identified in three breast cancer families and in one additional familial case from an unselected breast cancer cohort, but not in healthy controls (P = 0.002). On the basis of its exclusive occurrence in familial cancers, disease cosegregation, evolutionary conservation, and disruption of critical BRCA1 functions, the recurrent Abraxas c.1082G>A mutation connects to cancer predisposition. These findings contribute to the concept of a BRCA-centered tumor suppressor network and provide the identity of Abraxas as a new breast cancer susceptibility gene.
INTRODUCTION
Breast cancer is the most common malignancy affecting women. The presence of a family history is the most important single risk factor, with the first-degree relatives having about a twofold increased risk over the general population. BRCA1 and BRCA2 are the two most important susceptibility genes in hereditary predisposition to breast and ovarian cancer. However, germline mutations in these tumor suppressors account for no more than 20% of the familial breast cancer cases worldwide (1). Most of the remaining cancer cases are predicted to involve mutations in moderate- and low-penetrance susceptibility genes together with influence from environmental factors, although the data from large multiple-case families strongly suggest that there might still be additional high-penetrance genes to be identified (2, 3). Improved knowledge on the functions of BRCA1 and BRCA2 in the DNA damage response pathway has been instrumental in the identification of a number of breast cancer susceptibility genes including PALB2, BRIP1, ATM, and CHEK2, all with protein products interacting directly or indirectly with BRCA1 and BRCA2 (4, 5).
BRCA1 plays multifaceted roles as a gatekeeper of genomic integrity. It is essential for efficient DNA repair by homologous recombination and execution of cell cycle checkpoints activated by DNA damage (6). The C-terminal region of BRCA1 contains two BRCT (BRCA1 C-terminal) repeats that are critical for its tumor suppressor function. These BRCA1 BRCT motifs are the most common site of clinical mis-sense mutations, which invariably disrupt phosphorylation-dependent interactions with direct binding partners (7, 8).
Abraxas (ABRA1, CCDC98, or FAM175A) serves as a central organizer of a large BRCA1 holoenzyme complex, directly binding via its phosphorylated C terminus to the BRCA1 BRCT motifs (9–11). This interaction links BRCA1 to a core protein complex dedicated to ubiquitin chain recognition and hydrolysis at DNA double-strand breaks (DSBs) (9, 12–16). Abraxas and the other members of this complex [RAP80, BRCC36, MERIT40/NBA1 (HUGO name BABAM1), and BRCC45] are required for DNA damage checkpoints and cellular resistance to ionizing radiation (IR).
A deleterious germline RAP80 variant (del81E) that abrogates ubiquitin binding, DSB targeting, and genome integrity has been reported in two breast cancer patients (17). Additionally, a recent genome-wide linkage consortium study suggested an association between the rare allele of single-nucleotide polymorphism (SNP) rs8170 in MERIT40/NBA1 and an increased propensity for hormone receptor–negative breast cancer, both in the general population and in BRCA1 mutation carriers (18). The same SNP has also been associated with susceptibility to serous epithelial ovarian cancer (19). These findings suggest that BRCA1, in association with the core Abraxas complex, has tumor-suppressive function in humans. Here, we provide evidence in support of this hypothesis by identifying a novel germline mutation in Abraxas that exclusively associates with familial cancer and abrogates BRCA1-dependent DNA repair function.
RESULTS
A genetic screen for Abraxas mutations identified a R361Q alteration that associates with familial breast cancer
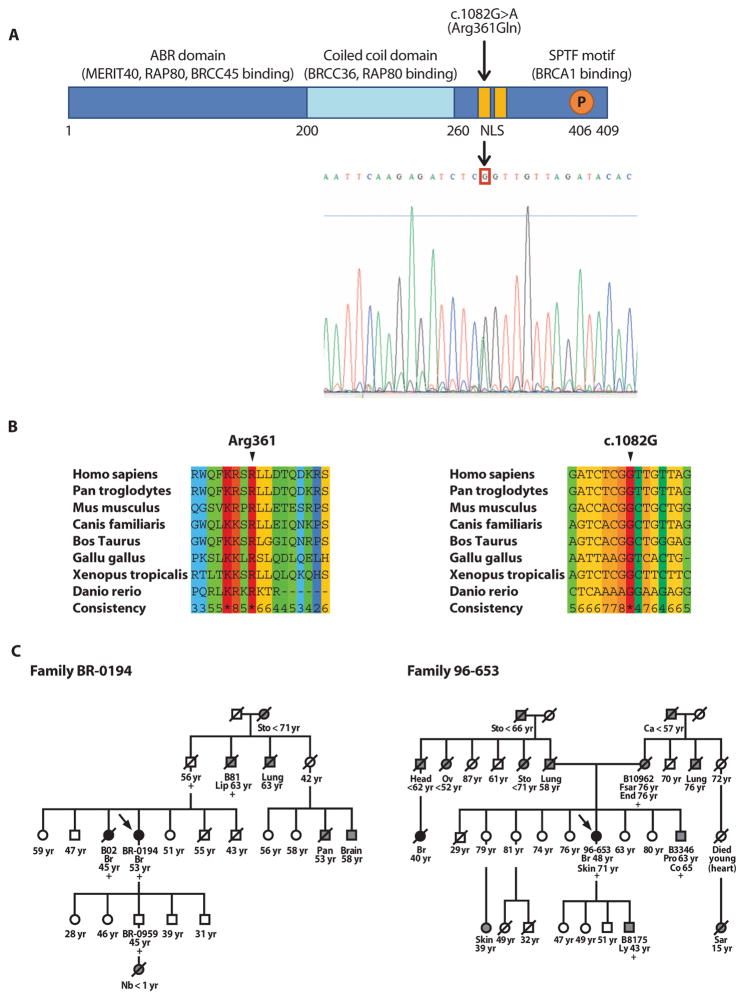
We screened 125 affected index patients of Northern Finnish breast cancer families for germline mutations in the Abraxas gene, revealing four intronic and six exonic variants (Table 1). Together, five of the changes have not been reported either in the National Center for Bio-technology Information (NCBI) SNP database (http://www.ncbi.nlm.nih.gov/SNP/) or by previous studies (20, 21). A computer simulation analysis using PolyPhen software indicated that of the observed changes, only c.1082G>A was likely to result in functional changes in the protein. ESEfinder 2.0 and NNSplice software analysis did not reveal any likely abnormalities. Alteration c.1082G>A results in Arg361Gln (R361Q), which changes the last residue of a putative bipartite nuclear localization signal (NLS) (Fig. 1A) (10). Both Abraxas protein and mRNA sequence alignments revealed absolute evolutionary conservation among vertebrates at the site of the wild-type sequence (Fig. 1B). None of the studied common SNPs within Abraxas showed a statistically significant association with cancer susceptibility (table S1).
Table 1.
Abraxas sequence variants observed in comprehensive mutation screening of familial breast cancer patients. The following sequence information was used: ENSG00000163322 (genomic DNA, with the correction that rs60946531 is c.*347C>T), ENST00000321945 (mRNA), and ENSP00000369857 (protein). c.1082G>A and c.1117G>A: unselected cases (USC) and controls (CT) comprised patients from both Northern Finland and Northern Savo in Eastern Finland. FC, familial cases; OR, odds ratio; CI, confidence interval; ND, not determined.
| Location | Nucleotide change | Effect on protein | rs number or reference | Frequency of heterozygotes, % (n/N)
|
P (OR; 95% CI) | ||
|---|---|---|---|---|---|---|---|
| FC | USC | CT | |||||
| Intron 2 | c.179-34_-38 delAATTA | — | — | 0.8 (1/125) | ND | 2.0 (2/100) | 0.6 (0.4; 0.4–4.4) |
| Intron 3 | c.216-44T>C | — | (21) | 2.4 (3/125) | ND | 5.3 (21/400) | 0.2 (0.4; 0.1–1.5) |
| Intron 4 | c.282+46T>A | — | — | 0.8 (1/125) | ND | — (—/400) | 0.2 (—) |
| Intron 7 | c.682-14A>G | — | — | 0.8 (1/125) | ND | 0.5 (2/400) | 0.6 (1.6; 0.1–17.8) |
| Exon 9 | c.1042G>A | Ala348Thr | rs12642536 (20, 21) | 52.8 (66/125) | ND | 51.1 (92/180) | 0.8 (1.1; 0.7–1.7) |
| Exon 9* | c.1082G>A | Arg361Gln | — | 2.4 (3/125) | 0.1 (1/991) | — (—/868) |
FC versus CT: 0.002 (—) FC versus USC: 0.005 (24.3; 2.5–235.9) |
| Exon 9 | c.1117G>A | Asp373Asn | rs13125836 (20, 21) | 12.8 (16/125) | 16.5 (163/990) | 13.9 (121/868) | FC versus CT: 0.8 (0.9; 0.5–1.6) USC versus CT: 0.1 (1.2; 0.9–1.6) |
| Exon 9† | c.*249delG | — | rs34610900 | 53.6 (67/125) | ND | 47.7 (42/88) | 0.4 (1.3; 0.7–2.2) |
| Exon 9† | c.*347C>T | — | rs60946531 rs6825184 |
0.8 (1/125) | ND | — (—/88) | 1.0 (—) |
| Exon 9† | c.*575A>G | — | — | 7.2 (9/125) | ND | 5.7 (5/88) | 0.8 (1.3; 0.4–3.9) |
Carriers of c.1082G>A (in bold) also harbored the c.1042G>A and c.*249delG SNPs.
3′ Untranslated region change.
Fig. 1.
Identification of Abraxas mutation c.1082G>A (Arg361Gln) in breast cancer index cases and cancer families. (A) Schematic diagram of the Abraxas protein and the site of Arg361Gln. Chromatogram of c.1082G>A (Arg361Gln) is displayed directly below the bipartite NLS 358-Lys-Arg-Ser-Arg-361 and 368-Lys-Arg-Ser-Lys-371 shown in yellow. (B) Evolutionary conservation of Abraxas c.1082G and the encoded codon Arg361. The conservation scoring was performed by PRALINE. The scoring scheme extends from 0 for the least conserved alignment position up to * for the most conserved alignment position. (C) Pedigrees of two Abraxas c.1082G>A mutation–positive breast cancer families where segregation analysis was possible (BR-0194 and 96-653). Black circles represent breast (Br) cancer cases; other cancer types [brain, colon (Co), endometrial (End), fibrosarcoma (Fsar), head, lip, lung, lymphoma (Ly), neuroblastoma (Nb), ovarian (Ov), pancreatic (Pan), prostate (Pro), sarcoma (Sar), skin, stomach (Sto); Ca, unknown] are marked with gray circles (females) or squares (males). Arrows point to index patients. A slashed symbol indicates a deceased individual. Sample identification codes are provided for all individuals where DNA specimens were available for mutation status analysis. Individuals are marked with a plus sign if mutation-positive. The age at diagnosis is indicated below the patients. The age at monitoring (or age at death), when known, is shown for the healthy individuals.
Abraxas c.1082G>A alteration was observed in 3 of 125 studied breast cancer families (2.4%) but was absent from 868 healthy female control individuals. The mutant allele was also identified in 1 of 991 breast cancer cases unselected for a family history of the disease (Table 1). The prevalence of c.1082G>A in familial compared with control cases, and also in familial compared with unselected breast cancer cases, was found to be significantly different (P = 0.002 and 0.005, respectively), which suggests that this variant is disease-associated and specifically correlated with familial cancer. In agreement, the only mutation-positive breast cancer patient in the unselected cohort also proved to have a familial cancer background (Fig. 1C, Family BR-0194).
Segregation analysis was performed in two of the mutation-positive families (BR-0194 and 96-653, Fig. 1C), showing cosegregation between Abraxas c.1082G>A and cancer phenotype. Because of a lack of suitable DNA samples, segregation analysis was not possible for the two remaining Abraxas mutation–positive breast cancer families (BR-02101 and 98-063). Of these, the index case of Family BR-02101 was diagnosed with both breast and endometrial cancer at the age of 37 and 49 years, respectively. She had a family history notable for stomach, lung, and prostate cancer, in addition to two maternal female cousins diagnosed with breast cancer at 48 and 54 years of age, respectively. The index case in the fourth family (98-063) was diagnosed with two ipsilateral primary breast tumors of different morphology (lobular and mucinous) at the age of 49 years. Her deceased sister displayed bilateral breast cancer at 53 and 71 years of age. The index patients of all four Abraxas mutation–positive breast cancer families tested negative for mutations in BRCA1, BRCA2, TP53, CDH1, and PALB2. The average age of disease onset of the confirmed five Abraxas mutation–positive breast cancer cases was 46 years (variation, 35 to 53 years), similar to those with Finnish BRCA1 (46 years; variation, 32 to 57 years) and BRCA2 (48 years; variation, 45 to 67 years) mutations (22). For comparison, the mean age of onset for the recurrent Finnish PALB2 c.1592delT was found to be 52.9 years (variation, 39 to 73 years), and the average age of onset for the unselected breast cancer cases was 57.9 years (variation, 23 to 92) (23).
Morphology studies of R361Q-positive breast cancers (Table 2) revealed a lobular phenotype in four of the five tumors (80%). A second primary disease of mucinous morphology was observed in one of the patients who initially presented with lobular disease. This was in marked contrast to the single case (20%) of the ductal phenotype, which is typically seen in about 75% of breast cancer cases (24). Immunohistochemistry was performed on sections from the same breast cancer cases noted above, and all four of the informative tumors revealed strong expression of both the estrogen receptor (ER) and the progesterone receptor (PR), as well as total absence of human epidermal growth factor receptor 2 (HER2) expression (Table 2). These phenotypic observations suggest that Abraxas mutation–positive breast cancer cases can deviate from the pattern of hormone receptor and HER2 negativity associated with BRCA1 tumors (25). Tumors from patients with mutations in BRCA2 and PALB2, which, like Abraxas, encode BRCA1-associated proteins, also frequently show hormone receptor positivity (23, 26).
Table 2.
Presentation of breast cancer, other tumors, and cellular dysplasia in patients heterozygous for germline Abraxas c.1082G>A. TNM, tumor-node-metastasis; NA, data not available.
| Patient | Breast cancer
|
Other cancer | Dysplasia | |||
|---|---|---|---|---|---|---|
| Morphology | Receptor status* | TNM | Age at diagnosis/metastasis (years) | |||
| BR-0194 | Lobular | ER++, PR+++, HER2− | T2N1M0 | 53/bone, 62 | — | Endometrial leiomyoma |
| B02 | Lobular | ER+++, PR+++, HER2− | T3–4N1M0 | 45/bone, 48; brain, 51 | — | — |
| BR-02101 | Ductal | ER+++, PR+++, HER2− | T2N0M0 | 35/— | Endometrial, 48 years | — |
| 98-063 | Lobular/mucinous | ER+++, PR+++, HER2− | T1N0M0 | 49/— | — | Colon tubular adenoma (low-grade dysplasia) |
| 96-653 | Lobular | NA | T1N0M0 | 48/— | Skin, 71 years (lentigo maligna) | Colon tubular adenoma (low-grade dysplasia) |
Positive staining for the ER and PR is defined as nuclear immunostaining in 1 to 10% (+), 10 to 50% (++), or >50% (+++) of the tumor cells, whereas a minus (−) indicates negative staining. Positive staining for HER2 is defined as membranous immunostaining of the tumor cells at levels + (faint positivity), ++ (moderate positivity), or +++ (strong, circumferential positivity), whereas HER2− indicates a completely negative staining.
BRCA1 and BRCA2 mutated cancers often display loss of heterozygosity (LOH), whereas loss of the wild-type allele at other BRCA-associated breast cancer susceptibility genes is less common (27, 28). To test whether LOH had occurred in tumors of individuals heterozygous for the Abraxas mutation (a total of six R361Q-positive tumors), we extracted genomic DNA from pure or highly enriched tumor cell populations obtained by laser-capture microdissection from formalin-fixed, paraffin-embedded tumor tissue sections, and we amplified and sequenced an Abraxas gene segment surrounding the c.1082G>A mutation by polymerase chain reaction (PCR). The existence of the c.1082G>A mutation was confirmed in all studied tumors, whereas no evidence of loss of the wild-type allele was ever seen (table S2).
Abraxas R361Q mutation impairs nuclear localization and disrupts BRCA1 DNA damage response function
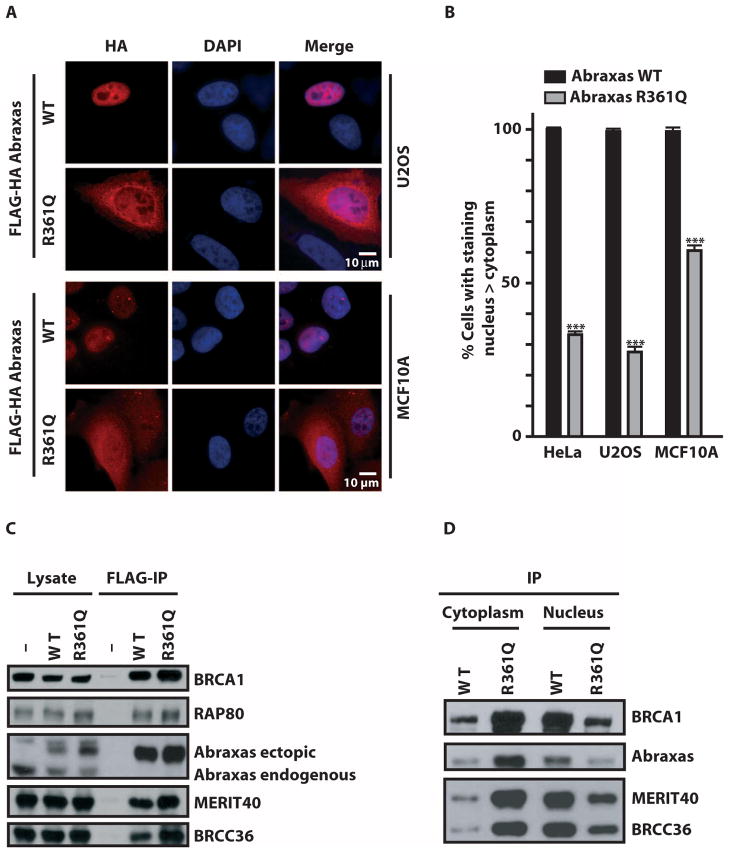
To assess the impact of R361Q on Abraxas subcellular localization, we stably expressed epitope-tagged wild-type or R361Q mutant Abraxas in three different cell lines at near-endogenous levels and examined subcellular localization by immunofluorescence (IF). Wild-type Abraxas demonstrated predominantly nuclear localization by IF, whereas Abraxas R361Q was primarily cytoplasmic (Fig. 2, A and B). Coimmunoprecipitation experiments revealed that R361Q maintained interactions with BRCA1 and other core components of the holoenzyme complex (Fig. 2C). Notably, Abraxas R361Q preferentially immunoprecipitated with BRCA1 and other interacting partners in the cytoplasm, whereas wild-type Abraxas primarily displayed these interactions in the nucleus (Fig. 2D and fig. S1).
Fig. 2.
Abraxas R361Q disrupts nuclear localization and recruitment to DNA DSBs. (A) IF of HA-tagged Abraxas wild-type (WT) and Abraxas R361Q transiently transfected in U2OS (top) and MCF10A (bottom) cells demonstrates cytoplasmic localization for the Abraxas mutant R361Q. DAPI, 4′,6-diamidino-2-phenylindole. (B) Quantification of cells presenting a predominantly nuclear staining in HeLa, U2OS, and MCF10A stably expressing FLAG-HA Abraxas WT or R361Q. Error bars represent means ± SD. P values were calculated by an unpaired t test. ***P < 0.0001. (C) Immunoblot (IB) of complexes after FLAG immunoprecipitation (IP) of FLAG-HA–tagged Abraxas from whole-cell lysates of 293T cells (-) or 293T cell lines stably expressing either Abraxas WT or R361Q. The location of endogenous and ectopic Abraxas protein is indicated. (D) IB of FLAG-HA–tagged Abraxas complexes after FLAG-IP from cytoplasmic and nuclear fractions of HeLa S3 cell lines that maintain stable expression of either Abraxas WT or Abraxas R361Q.
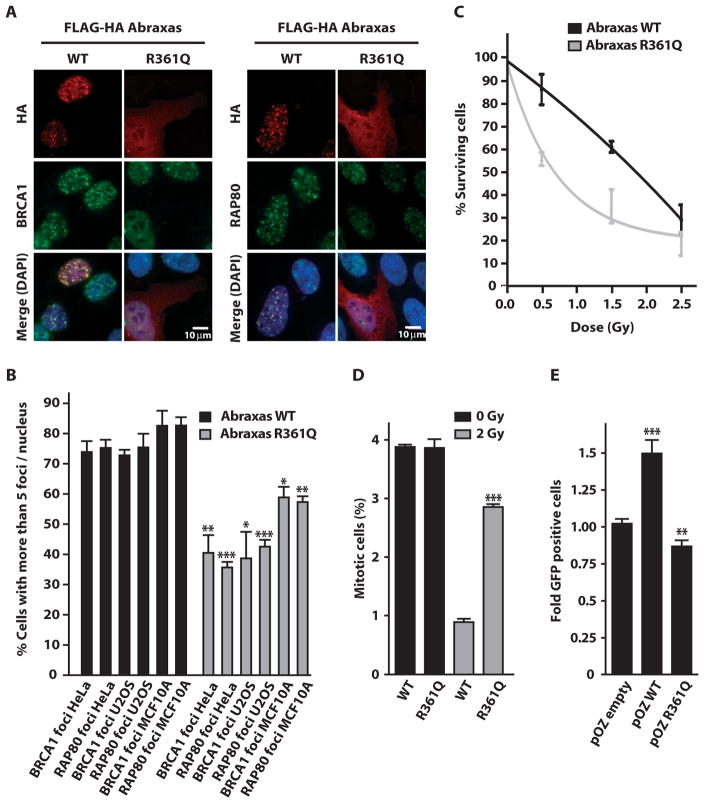
To determine the impact of R361Q on DNA damage response functions, we examined recruitment of wild-type Abraxas or R361Q to DSBs. R361Q recruitment to a specific site of nuclease-induced DSBs (29) was strongly reduced in comparison to wild-type Abraxas, suggesting deficiency in DNA damage response functions due to impaired nuclear accumulation (fig. S2A). Similar results were observed at IR-induced foci (IRIF) in three different cell types (Fig. 3, A and B). Consistent with a dominant-negative activity, R361Q expression significantly reduced both BRCA1 and RAP80 IRIF formation and resulted in IR hypersensitivity in three different cell lines (Fig. 3, A to C, and fig. S2, C and D). In addition, R361Q expression partially disrupted the G2 checkpoint in response to IR and reduced the efficiency of homology-directed DSB repair (Fig. 3, D and E). These results suggest that reduced nuclear accumulation by Abraxas R361Q negatively affects DSB localization of its interacting partners. In support of this concept, inhibition of nuclear export by leptomycin B treatment increased nuclear Abraxas R361Q and restored IRIF for Abraxas R361Q, BRCA1, and RAP80 (fig. S3, A and B). Conversely, Abraxas R361Q expression did not aberrantly affect subcellular localization of β-catenin, further emphasizing the specificity for its impact on BRCA1 and RAP80 (fig. S4). Abraxas, in addition to BRIP1 (FANCJ) and RAP80 (17, 27, 30), is now the third BRCA1 BRCT–interacting partner with germline human breast cancer–associated mutations shown to exert dominant-negative effects on BRCA1 DNA repair function. These findings suggest that the nature of the Abraxas R361Q mutation may obviate the need for LOH in tumors from affected patients.
Fig. 3.
Abraxas R361Q impairs BRCA1 and RAP80 DNA damage response functions. (A) IF in U2OS cells at 7 hours after 10 Gy IR demonstrates decreased BRCA1 and RAP80 IRIF in cells expressing Abraxas R361Q. (B) Quantification of the percentage of HeLa, U2OS, and MCF10A cells expressing Abraxas WT or R361Q with greater than five BRCA1 or RAP80 foci per nucleus. The mean was calculated from more than 200 cells for each condition in triplicate. Error bars represent means ± SD. P values were calculated by an unpaired t test. *P = 0.0181; **P = 0.0056; ***P < 0.0001. (C) Clonogenic survival assay of U2OS cells stably expressing Abraxas WT or R361Q after exposure to the indicated doses of γ-radiation. Points represent the average of three independent experiments run in triplicate. Error bars represent means ± SD. (D) Evaluation of the IR-induced G2-M checkpoint in transiently transfected 293T cells. Mitotic cells were detected by FACS using an antibody against phospho–histone H3. The mean mitotic cell population was calculated from three independent experiments in which, in each sample, 10,000 cells were examined. Error bars represent SD. P values were calculated by Student’s t test assuming unequal variances. ***P < 0.0001. (E) Evaluation of the homology-directed DNA repair of a nuclease-induced DSB after transfection of a DR-GFP reporter cell line with the indicated plasmids. GFP-positive cells indicate the presence of homology-directed DNA repair. Data are representative of four independent experiments. Error bars represent means ± SD. P values were calculated by an unpaired t test. **P = 0.0028; ***P < 0.0001.
DISCUSSION
Here, we report the identity of a novel, recurrent, constitutional mutation in the Abraxas gene in familial breast cancer. Abraxas R361Q demonstrated exclusive association with cancer, segregation with disease within families, and loss of biological function in the DNA damage response. The impaired DNA damage response function extends beyond Abraxas itself, because Abraxas R361Q exerted a dominant-negative influence on BRCA1 and RAP80 by diminishing their accumulation at IR-induced DNA damage foci. In light of this finding, it is interesting that breast cancer–associated mutation RAP80delE81 displayed similar dominant-negative properties with respect to BRCA1 localization to IR-induced foci (17).
These observations complement existing knowledge of breast cancer–associated mutations within genes encoding proteins present in other BRCA1-containing protein complexes, reinforcing the concept of a BRCA-centered tumor suppressor network dedicated to the maintenance of genomic integrity (4, 31). Moreover, they establish ubiquitin recognition at DNA damage sites as a bona fide tumor suppression function of BRCA1-associated protein complexes. The BRCA1 protein complex containing Abraxas and RAP80 is unique in comparison to BRCA1 complexes that contain breast cancer suppressors PALB2, BRCA2, and BRIP1. RAP80 targets BRCA1 and Abraxas to ubiquitinated chromatin extending for a distance away from DSBs, whereas BRCA1 complexes containing BRCA2 and PALB2 or BRIP1 are thought to directly interact with DNA intermediates during homologous recombination–dependent DNA repair (32). Mutation in any of these genes results in severely reduced homologous recombination, whereas cells deficient for Abraxas or RAP80 display elevated use of homology-directed DNA repair mechanisms (33, 34). Abraxas R361Q cells displayed slightly reduced homology-directed repair of a nuclease-induced DSB (Fig. 3E), suggesting that Abraxas R361Q expression impairs BRCA1-dependent DNA repair differently than would an Abraxas-null allele. It is also unclear whether the IR hypersensitivity displayed in three different Abraxas R361Q–expressing cell lines was a result of the slight reduction in homology-directed DNA repair or alternatively due to defective G2 checkpoint signaling (Fig. 3D). Therefore, it would be interesting to determine whether cancers expressing Abraxas or RAP80 dominant-negative mutant alleles would exhibit similar responses to chemotherapy regimens as tumors harboring mutations in other genes within the BRCA network. The development of various Abraxas- or RAP80-deficient tumor models will be important to test such predictions.
In addition to breast cancer, Abraxas R361Q families displayed some relatively rare cancer types. Recently, a genome-wide association study associated a novel variant, rs1494961, located near Abraxas, with genetic susceptibility to upper aero-digestive tract cancers (35). In our current study, both lung and lip cancer and lymphoma of the throat occurred in the two Abraxas c.1082G>A families shown in Fig. 1C. In addition, Family BR-0194 had a case of neuroblastoma; mutations in anotherBRCA1-associatedgene, BARD1, have recently been connected to this disease (36).
In conclusion, we have identified a coding variant of the Abraxas gene with a significantly different distribution in the familial cancer cases compared to the studied controls. This alteration is found in 2.4% of the studied Northern Finnish familial breast cancer cases and predominantly associates to a lobular tumor phenotype. On the basis of its exclusive occurrence in familial cancer cases, disease cosegregation, evolutionary conservation, and disruption of critical BRCA1 DNA damage response functions, the recurrent mutation connects to cancer predisposition. Similar to BRCA1 and BRCA2, mutations in Abraxas appear to be involved in susceptibility to certain other malignancy types beyond breast cancer. The present results warrant investigation of Abraxas as a new cancer susceptibility gene in other populations as well. The identification of additional Abraxas mutation–positive families would provide the means for more reliable cancer risk assessments and evaluation of the potential use of Abraxas mutation testing in clinical diagnostics.
MATERIALS AND METHODS
Familial and unselected breast cancer cases
Mutation screening of Abraxas was performed on blood DNA samples obtained from 125 breast and breast-ovarian cancer families originating from Northern Finland (23). One index patient from each family was chosen according to the youngest age of breast cancer onset. Inclusion criteria for the 73 high-risk families were as follows: (i) three or more cases of breast cancer, potentially in combination with single ovarian cancer in first- or second-degree relatives, or (ii) two cases of breast or breast and ovarian cancer in first- or second-degree relatives, of which at least one with early disease onset (<35 years), bilateral breast cancer, or multiple primary tumors including breast or ovarian cancer in the same individual. The remaining 52 families were indicative of moderate disease susceptibility, presenting either two cases of breast cancer in first- or second-degree relatives, or breast cancer under the age of 35 (2 cases). Together, 15 of the studied index cases had previously been tested positive for known breast cancer–associated germline mutations in BRCA1 or BRCA2 (11 cases), TP53 (1 case), and PALB2 (3 cases). All of the biological specimens and clinical information of the familial breast cancer cases investigated were collected at the Oulu University Hospital, with the informed consent of the patients.
For the Abraxas c.1082G>A genotyping and tagging SNP (tagSNP) analysis, DNAs from an unselected cohort of breast cancer patients (n = 991) were collected without selection for a family history of breast cancer. This sample set consisted of 544 Northern Finnish cases operated at the Oulu University Hospital during the years 2000 to 2007 and 447 patients with invasive breast cancer from the Kuopio Breast Cancer Project (KBCP), originating from the province of Northern Savo in Eastern Finland and diagnosed at the Kuopio University Hospital between 1990 and 1995 (37).
Informed consent to participate in the study has been obtained from each patient, and the studies have been approved by the Finnish Ministry of Social Affairs and Health, and appropriate ethical committees of each of the participating University Hospitals.
Control cases
Together, 868 Finnish female control cases were used for genotyping and tagSNP analysis. The Northern Finnish control samples (where the number of studied individuals varied between 88 and 506, depending on the specific analysis) derived from anonymous cancer-free Finnish Red Cross blood donors (age ≥45 years) originating from the same geographical region as the studied cancer patient cohort. The age- and area of residence–matched KBCP cohort consisted of DNA from 362 control subjects selected from the National Population Register during the same time period as the unselected breast cancer patients (37). All control individuals were cancer-free at the time of donation of the blood sample. There was no follow-up on donor health status.
Mutation and tagSNP analysis
The entire coding region and exon-intron boundaries of the Abraxas gene were screened for germline mutations either by conformation-sensitive gel electrophoresis (CSGE, exons 2 to 8) or by direct sequencing (exons 1 and 9). Samples with deviating CSGE patterns or those directly sequenced were analyzed with the Li-Cor IR2 4200-S DNA Analysis system (Li-Cor Inc.) using the SequiTherm EXEL II DNA Sequencing Kit-LC (Epicentre Technologies) or with ABI3730 (ABI Perkin Elmer) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Oligonucleotides were designed with Primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi, table S3).
Genotyping of Abraxas c.1082G>A and tagSNPs rs12499395, rs12649417, and rs13125836 was done with MassARRAY mass spectrometer (Sequenom Inc.) and iPLEX Gold (Sequenom) on 384-well plate format (used primers in table S4). TagSNPs were selected with the HapMap Genome Browser release 2 (Phase 3, NCBI build 36, bdSNP b126) as of 23 February 2010 (http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap3r2_B36/). TagSNPs for region chr4:84593218-84633217 were picked out for the CEU population with the Tagger multimarker algorithm with r2 cutoff at 0.8 and minor allele frequency (MAF) cutoff at 0.05. MassARRAY was used for spectra acquisitions from the SpectroCHIP (Sequenom). Data analysis and genotype calling were done with Typer Analyzer software version 4.0.3.18 (Sequenom). Each 384-well plate contained a minimum of eight nontemplate controls. For the c.1082G>A mutation, DNAs from three heterozygous mutation carriers were used as positive controls on each plate. For quality control, duplicate analysis was done for 6.5% of the samples from Oulu and for 6.7% of the samples from Kuopio.
Statistical and bioinformatic analysis
Carrier frequencies between patients and healthy controls were compared with Pearson χ2 or Fisher’s exact test (two-sided, SPSS version 17.0 for Windows). All alterations were checked with NNSplice software for potential effects on splicing (http://www.fruitfly.org/seq_tools/splice.html). Arg361Gln was tested for possible pathogenicity with PolyPhen software (http://genetics.bwh.harvard.edu/pph). For tagSNP data, the overall association as well as the Hardy-Weinberg equilibrium, allele-specific P, odds ratio, and confidence interval were computed with Cochran-Armitage trend test.
Tumor ER, PR, and HER2 immunohistochemistry
Formalin-fixed, paraffin-embedded tissue sections (4 μm) were de-paraffinized and rehydrated in graded alcohols. Heat-induced epitope retrieval was performed with a digital pressure cooker in citrate buffer (pH 6.0) before application of the appropriate polyclonal (or monoclonal) antibody. The primary antibody was detected with the Envision+ system (K4011, DakoCytomation) that uses horseradish peroxidase–labeled polymer conjugated to goat anti-rabbit immuno-globulin antibodies. The immune complexes were identified using a peroxidase reaction with 3,3′-diaminobenzidine-plus as chromogen. Slides were counterstained with Mayer’s hematoxylin. Antibodies against ER (monoclonal, clone 1D5), PR (monoclonal, clone PgR636), and HER2 (polyclonal) were all purchased from Dako and used at 1:100, 1:200, and 1:1000, respectively.
Antibodies used in the immunofluorescence and immunoprecipitation analysis for functional assessment of the germline Abraxas R361Q alteration
BRCA1 was detected by immunoblotting (IB) with mouse monoclonal antibody MS110 at a 1:10 dilution, and by IF with a rabbit polyclonal antibody 07-434 (Millipore) at 1:500. A RAP80 rabbit polyclonal antibody was used for IB at 1:500 and IF at 1:100 dilutions. BRCC36 was detected by IB with a rabbit polyclonal antibody (12). MERIT40 was detected by IB at 1:1000 with a rabbit polyclonal antibody (14). Hemagglutinin (HA)–tagged proteins were detected by IB at 1:1000 and for IF at 1:1000 dilutions with mouse monoclonal antibody HA.11 (Covance).
Cells
HeLa, 293T, MCF10A, and U2OS cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco) with 10% calf serum. Transient transfections were performed with LipoD293 (SignaGen Laboratories) according to the manufacturer’s protocols.
Cell fractionation and immunoprecipitation
293T cells were lysed in NETN-150 buffer [150 mM tris-HCl (pH 7.4), 1 mM EDTA, 0.05% Nonidet P-40, 0.5 mM phenylmethylsulfonyl fluoride, and 5 mM β-mercaptoethanol] for the production of whole-cell extracts. Nuclear and cytoplasmic fractions were obtained from cells by Dounce homogenization of cells treated with hypotonic buffer [10 mM KCl, 10 mM tris-HCl (pH 7.4), 1.5 mM MgCl2]. Nuclei were lysed in NETN-150 buffer at 4°C for 45 min. All subsequent steps were performed in NETN-150 buffer. The quality of our fractionation was evaluated by the detection of specific markers: γ-tubulin for the cytoplasmic fraction and proliferating cell nuclear antigen for the nuclear compartment.
DNA damage induction
All radiation exposures were performed with a Gammacell 40 irradiator (Nordion International), which used cesium-137 as the radiation source.
IF analysis
U2OS cells were cultured on glass coverslips and transiently transfected with LipoD293 (SignaGen Laboratories) according to the manufacturer’s protocols. Cells were treated with 10 Gy IR and allowed to recover for the indicated times in a 37°C incubator. Cells were washed with phosphate-buffered saline (PBS) and then fixed in 3% paraformaldehyde/2% sucrose containing solution for 10 min at room temperature. Cells were subsequently permeabilized with 0.5% Triton solution for 5 min at 4°C and then incubated with the appropriate primary antibody for 20 min at 37°C. Cells were then washed with PBST (PBS–Tween 20) and incubated with secondary antibody for 20 min at 37°C. Nuclei were then stained by incubating the cells in a PBS solution containing Hoechst 33258 dye (1 μg/ml). After four washes in PBST, coverslips were mounted onto glass slides with Vectashield mounting media (Vector Labs) and visualized with a Nikon Eclipse 80i fluorescence microscope.
G2 checkpoint assay
The G2 checkpoint assay was performed by assessing the percentage of mitotic cells at 0 Gy and at 1 hour after 2 Gy IR as previously described (14). A rabbit polyclonal antibody against phosphorylated histone H3 (Upstate Biotechnology) was used to detect mitotic cells.
Homology-directed DNA repair assay
Homology-directed DNA repair assay was performed as described (34). U2OS cells containing stable integration of the direct repeat green fluorescent protein (DR-GFP) reporter locus were transiently transfected with I-SceI and Abraxas plasmids and then scored for homology-directed DNA repair by fluorescence-activated cell sorting (FACS) for GFP-positive cells 72 hours after transfection.
Plasmids
The FLAG-HA–tagged version of Abraxas R361Q was created from Addgene vector 27495.
Supplementary Material
Table S1. Association analysis of three Abraxas tagSNPs with breast cancer.
Table S2. LOH analysis of laser-capture microdissected tumor cell DNA of Abraxas c.1082G>A germline mutation carriers.
Table S3. Abraxas primers and PCR amplicon details.
Table S4. iPLEX primers used for Abraxas c.1082G>A and tagSNP genotyping.
Fig. S1. Abraxas R361Q is preferentially immunoprecipitated with its interacting partners in the cytoplasm.
Fig. S2. Expression of Abraxas R361Q affects DNA damage response.
Fig. S3. Nuclear retention of Abraxas R361Q restores localization and recruitment to DSB.
Fig. S4. Cytoplasmic localization of Abraxas R361Q does not aberrantly activate β-catenin.
Acknowledgments
We thank A. Mustonen, A. Jukkola-Vuorinen, M. Grip, V. Kataja, and nurses K. Mononen and O. Kajula for their help in sample and data collection and in patient contacts. The contribution of H. Erkko and technical assistance by M. Otsukka, H. Konola, R. Vuento, and E. Myöhänen are gratefully acknowledged. We thank all the patients and their family members for volunteering to participate in these studies, as well as the Finnish Red Cross Blood Service for help with collection of population control blood samples. We are also grateful to M. Jasin (Memorial Sloan-Kettering Cancer Center) for providing the DR-GFP cell line and I-SceI expression plasmid.
Funding: R.W. gratefully acknowledges the financial support from the Finnish Cancer Foundation, the Academy of Finland (grant number 122715 and Center of Excellence grant number 251314), CIMO, the University of Oulu, the Oulu University Hospital, the Cancer Fund of Northern Finland, the Sigrid Jusélius Foundation, the Cancer Fund of Savo, the Ida Montini Foundation, and the Tyyni Tani Foundation. The Kuopio group acknowledges the support from the Finnish Cancer Society, the Academy of Finland (grant number 127220), the special Government Funding (EVO) of Kuopio University Hospital (grant numbers 5654112 and 5501), and the strategic funding of the University of Eastern Finland. R.A.G. gratefully acknowledges funding from the National Cancer Institute (1R01CA138835-01), an American Cancer Society Research Scholar Grant, and funds from the Abramson Family Cancer Research Institute. J.P.-F. was supported in part by a Doctoral Research Award from the Canadian Institutes for Health Research.
Footnotes
Author contributions: R.W. and R.A.G. designed and oversaw the study with helpful input from K.P. S.S. performed the mutation analysis and the statistical and bioinformatic assessment of the initial genetic findings obtained from the familial and unselected breast cancer cohorts and controls from Oulu. K.P. was responsible for the sequencing and loss of heterozygosity analysis on archival patient tissue material, with some early-phase mutation analysis help from S.S. J.M.H., A.M., and V.-M.K. contributed with the KBCP cohort, and J.M.H. and A.M. performed the tagSNP association and haplotype analysis as well as iPLEX c.1082G (a mutation testing in both unselected cases and controls from Kuopio). A.M., K.P., and J.M.H. performed the statistical analysis of the obtained tagSNP data. R.W. gathered the extended genealogy and cancer registry data for the mutation-positive families, and S.K. was responsible for obtaining suitable archival tissue samples as well as laser-capture dissection of tumor material for the LOH analysis and for getting detailed clinicopathology data of the studied cancer cases. J.P.-F. and B.A. performed the functional studies. S.S. prepared the initial manuscript draft with critical input from K.P., R.W., and R.A.G. R.W., R.A.G., K.P., B.A., S.S., and J.P.-F. prepared the advanced manuscript, and all authors contributed to critical review of the paper.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Anglian Breast Cancer Study Group. Prevalence and penetrance of BRCA1 and BRCA2 mutations in a population-based series of breast cancer cases. Br J Cancer. 2000;83:1301–1308. doi: 10.1054/bjoc.2000.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, Struewing JP, Morrison J, Field H, Luben R, Wareham N, Ahmed S, Healey CS, Bowma R, Meyer KB, Haiman CA, Kolonel LK, Henderson BE, Le Marchand L, Brennan P, Sangrajrang S, Gaborieau V, Odefrey F, Shen CY, Wu PE, Wang HC, Eccles D, Evans DG, Peto J, Fletcher O, Johnson N, Seal S, Stratton MR, Rahman N, Chenevix-Trench G, Bojesen SE, Nordestgaard BG, Axelsson CK, Garcia-Closas M, Bogdanova N, Schürmann P, Dörk T, Tollenaar RA, Jacobi CE, Devilee P, Klijn JG, Sigurdson AJ, Doody MM, Alexander BH, Zhang J, Cox A, Brock IW, MacPherson G, Reed MW, Couch FJ, Goode EL, Olson JE, Meijers-Heijboer H, van den Ouweland A, Uitterlinden A, Rivadeneira F, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Hopper JL, McCredie M, Southey M, Giles GG, Schroen C, Justenhoven C, Brauch H, Hamann U, Ko YD, Spurdle AB, Beesley J, Chen X, Mannermaa A, Kosma VM, Kataja V, Hartikainen J, Day NE, Cox DR, Ponder BA SEARCH collaborators, kConFab AOCS Management Group. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 4.Walsh T, King MC. Ten genes for inherited breast cancer. Cancer Cell. 2007;11:103–105. doi: 10.1016/j.ccr.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 5.Shuen AY, Foulkes WD. Inherited mutations in breast cancer genes—Risk and response. J Mammary Gland Biol Neoplasia. 2011;16:3–15. doi: 10.1007/s10911-011-9213-5. [DOI] [PubMed] [Google Scholar]
- 6.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 7.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 8.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 9.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain–binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 12.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B, Hurov K, Hofmann K, Elledge SJ. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009;23:729–739. doi: 10.1101/gad.1770309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao G, Patterson-Fortin J, Messick TE, Feng D, Shanbhag N, Wang Y, Greenberg RA. MERIT40 controls BRCA1–Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009;23:740–754. doi: 10.1101/gad.1739609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 16.Feng L, Huang J, Chen J. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009;23:719–728. doi: 10.1101/gad.1770609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nikkilä J, Coleman KA, Morrissey D, Pylkäs K, Erkko H, Messick TE, Karppinen SM, Amelina A, Winqvist R, Greenberg RA. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009;28:1843–1852. doi: 10.1038/onc.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antoniou AC, Wang X, Fredericksen ZS, McGuffog L, Tarrell R, Sinilnikova OM, Healey S, Morrison J, Kartsonaki C, Lesnick T, Ghoussaini M, Barrowdale, EMBRACE D, Peock S, Cook M, Oliver C, Frost D, Eccles D, Evans DG, Eeles R, Izatt L, Chu C, Douglas F, Paterson J, Stoppa-Lyonnet D, Houdayer C, Mazoyer S, Giraud S, Lasset C, Remenieras A, Caron O, Hardouin A, Berthet P, Hogervorst FB, Rookus MA, Jager A, van den Ouweland A, Hoogerbrugge N, van der Luijt RB, Meijers-Heijboer H, Gómez García EB, HEBON, Devilee P, Vreeswijk MP, Lubinski J, Jakubowska A, Gronwald J, Huzarski T, Byrski T, Górski B, Cybulski C, Spurdle AB, Holland H, Goldgar DE, John EM, Hopper JL, Southey M, Buys SS, Daly MB, Terry MB, Schmutzler RK, Wappenschmidt B, Engel C, Meindl A, Preisler-Adams S, Arnold N, Niederacher D, Sutter C, Domchek SM, Nathanson KL, Rebbeck T, Blum JL, Piedmonte M, Rodriguez GC, Wakeley K, Boggess JF, Basil J, Blank SV, Friedman E, Kaufman B, Laitman Y, Milgrom R, Andrulis IL, Glendon G, Ozcelik H, Kirchhoff T, Vijai J, Gaudet MM, Altshuler D, Guiducci C, SWE-BRCA, Loman N, Harbst K, Rantala J, Ehrencrona H, Gerdes AM, Thomassen M, Sunde L, Peterlongo P, Manoukian S, Bonanni B, Viel A, Radice P, Caldes T, de la Hoya M, Singer CF, Fink-Retter A, Greene MH, Mai PL, Loud JT, Guidugli L, Lindor NM, Hansen TV, Nielsen FC, Blanco I, Lazaro C, Garber J, Ramus SJ, Gayther SA, Phelan C, Narod S, Szabo CI, SQUAD MOD, Benitez J, Osorio A, Nevanlinna H, Heikkinen T, Caligo MA, Beattie MS, Hamann U, Godwin AK, Montagna M, Casella C, Neuhausen SL, Karlan BY, Tung N, Toland AE, Weitzel J, Olopade O, Simard J, Soucy P, Rubinstein WS, Arason A, Rennert G, Martin NG, Montgomery GW, Chang-Claude J, Flesch-Janys D, Brauch; H, GENICA, Severi G, Baglietto L, Cox A, Cross SS, Miron P, Gerty SM, Tapper W, Yannoukakos D, Fountzilas G, Fasching PA, Beckmann MW, Dos Santos Silva I, Peto J, Lambrechts D, Paridaens R, Rüdiger T, Försti A, Winqvist R, Pylkäs K, Diasio RB, Lee AM, Eckel-Passow J, Vachon C, Blows F, Driver K, Dunning A, Pharoah PP, Offit K, Pankratz VS, Hakonarson H, Chenevix-Trench G, Easton DF, Couch FJ kConFab GEMO Study Collaborators. A locus on 19p13 modifies risk of breast cancer in BRCA1 mutation carriers and is associated with hormone receptor–negative breast cancer in the general population. Nat Genet. 2010;42:885–892. doi: 10.1038/ng.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, Sher T, Gentry-Maharaj A, Wozniak E, Tsai YY, Weidhaas J, Paik D, Van Den Berg DJ, Stram DO, Pearce CL, Wu AH, Brewster W, Anton-Culver H, Ziogas A, Narod SA, Levine DA, Kaye SB, Brown R, Paul J, Flanagan J, Sieh W, McGuire V, Whittemore AS, Campbell I, Gore ME, Lissowska J, Yang HP, Medrek K, Gronwald J, Lubinski J, Jakubowska A, Le ND, Cook LS, Kelemen LE, Brook-Wilson A, Massuger LF, Kiemeney LA, Aben KK, van Altena AM, Houlston R, Tomlinson I, Palmieri RT, Moorman PG, Schildkraut J, Iversen ES, Phelan C, Vierkant RA, Cunningham JM, Goode EL, Fridley BL, Kruger-Kjaer S, Blaeker J, Hogdall E, Hogdall C, Gross J, Karlan BY, Ness RB, Edwards RP, Odunsi K, Moyisch KB, Baker JA, Modugno F, Heikkinenen T, Butzow R, Nevanlinna H, Leminen A, Bogdanova N, Antonenkova N, Doerk T, Hillemanns P, Dürst M, Runnebaum I, Thompson PJ, Carney ME, Goodman MT, Lurie G, Wang-Gohrke S, Hein R, Chang-Claude J, Rossing MA, Cushing-Haugen KL, Doherty J, Chen C, Rafnar T, Besenbacher S, Sulem P, Stefansson K, Birrer MJ, Terry KL, Hernandez D, Cramer DW, Vergote I, Amant F, Lambrechts D, Despierre E, Fasching PA, Beckmann MW, Thiel FC, Ekici AB, Chen X, Johnatty SE, Webb PM, Beesley J, Chanock S, Garcia-Closas M, Sellers T, Easton DF, Berchuck A, Chenevix-Trench G, Pharoah PD, Gayther SA Australian Ovarian Cancer Study Group; Australian Cancer Study (Ovarian Cancer); Ovarian Cancer Association Consortium. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osorio A, Barroso A, García MJ, Martínez-Delgado B, Urioste M, Benítez J. Evaluation of the BRCA1 interacting genes RAP80 and CCDC98 in familial breast cancer susceptibility. Breast Cancer Res Treat. 2009;113:371–376. doi: 10.1007/s10549-008-9933-4. [DOI] [PubMed] [Google Scholar]
- 21.Novak DJ, Sabbaghian N, Maillet P, Chappuis PO, Foulkes WD, Tischkowitz M. Analysis of the genes coding for the BRCA1-interacting proteins, RAP80 and Abraxas (CCDC98), in high-risk, non-BRCA1/2, multiethnic breast cancer cases. Breast Cancer Res Treat. 2009;117:453–459. doi: 10.1007/s10549-008-0134-y. [DOI] [PubMed] [Google Scholar]
- 22.Sarantaus L, Huusko P, Eerola H, Launonen V, Vehmanen P, Rapakko K, Gillanders E, Syrjäkoski K, Kainu T, Vahteristo P, Krahe R, Pääkkönen K, Hartikainen J, Blomqvist C, Löppönen T, Holli K, Ryynänen M, Butzow R, Borg A, Wasteson Arver B, Holmberg E, Mannermaa A, Kere J, Kallioniemi OP, Winqvist R, Nevanlinna H. Multiple founder effects and geographical clustering of BRCA1 and BRCA2 families in Finland. Eur J Hum Genet. 2000;8:757–763. doi: 10.1038/sj.ejhg.5200529. [DOI] [PubMed] [Google Scholar]
- 23.Erkko H, Xia B, Nikkilä J, Schleutker J, Syrjäkoski K, Mannermaa A, Kallioniemi A, Pylkäs K, Karppinen SM, Rapakko K, Miron A, Sheng Q, Li G, Mattila H, Bell DW, Haber DA, Grip M, Reiman M, Jukkola-Vuorinen A, Mustonen A, Kere J, Aaltonen LA, Kosma VM, Kataja V, Soini Y, Drapkin RI, Livingston DM, Winqvist R. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 24.Li CI, Anderson BO, Daling JR, Moe RE. Trends in incidence rates of invasive lobular and ductal breast carcinoma. JAMA. 2003;289:1421–1424. doi: 10.1001/jama.289.11.1421. [DOI] [PubMed] [Google Scholar]
- 25.Mavaddat N, Antoniou AC, Easton DF, Garcia-Closas M. Genetic susceptibility to breast cancer. Mol Oncol. 2010;4:174–191. doi: 10.1016/j.molonc.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borg A. Molecular and pathological characterization of inherited breast cancer. Semin Cancer Biol. 2001;11:375–385. doi: 10.1006/scbi.2001.0393. [DOI] [PubMed] [Google Scholar]
- 27.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 28.Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71:2222–2229. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K, North B, McGuffog L, Evans DG, Eccles D, Easton DF, Stratton MR, Rahman N Breast Cancer Susceptibility Collaboration (UK) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet. 2006;38:1239–1241. doi: 10.1038/ng1902. [DOI] [PubMed] [Google Scholar]
- 31.Greenberg RA. Recognition of DNA double strand breaks by the BRCA1 tumor suppressor network. Chromosoma. 2008;117:305–317. doi: 10.1007/s00412-008-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Messick TE, Greenberg RA. The ubiquitin landscape at DNA double-strand breaks. J Cell Biol. 2009;187:319–326. doi: 10.1083/jcb.200908074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Scully R, Sobhian B, Xie A, Shestakova E, Livingston DM. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011;25:685–700. doi: 10.1101/gad.2011011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman KA, Greenberg RA. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J Biol Chem. 2011;286:13669–13680. doi: 10.1074/jbc.M110.213728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McKay JD, Truong T, Gaborieau V, Chabrier A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Bucur A, Bencko V, Holcatova I, Janout V, Foretova L, Lagiou P, Trichopoulos D, Benhamou S, Bouchardy C, Ahrens W, Merletti F, Richiardi L, Talamini R, Barzan L, Kjaerheim K, Macfarlane GJ, Macfarlane TV, Simonato L, Canova C, Agudo A, Castellsagué X, Lowry R, Conway DI, McKinney PA, Healy CM, Toner ME, Znaor A, Curado MP, Koifman S, Menezes A, Wünsch-Filho V, Neto JE, Garrote LF, Boccia S, Cadoni G, Arzani D, Olshan AF, Weissler MC, Funkhouser WK, Luo J, Lubiński J, Trubicka J, Lener M, Oszutowska D, Schwartz SM, Chen C, Fish S, Doody DR, Muscat JE, Lazarus P, Gallagher CJ, Chang SC, Zhang ZF, Wei Q, Sturgis EM, Wang LE, Franceschi S, Herrero R, Kelsey KT, McClean MD, Marsit CJ, Nelson HH, Romkes M, Buch S, Nukui T, Zhong S, Lacko M, Manni JJ, Peters WH, Hung RJ, McLaughlin J, Vatten L, Njølstad I, Goodman GE, Field JK, Liloglou T, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, González CA, Quirós JR, Martínez C, Navarro C, Ardanaz E, Larrañaga N, Khaw KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjønneland A, Kumle M, Riboli E, Välk K, Vooder T, Metspalu A, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Blanché H, Gut IG, Galan P, Heath S, Hashibe M, Hayes RB, Boffetta P, Lathrop M, Brennan P. A genome-wide association study of upper aerodigestive tract cancers conducted within the INHANCE consortium. PLoS Genet. 2011;7:e1001333. doi: 10.1371/journal.pgen.1001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capasso M, Devoto M, Hou C, Asgharzadeh S, Glessner JT, Attiyeh EF, Mosse YP, Kim C, Diskin SJ, Cole KA, Bosse K, Diamond M, Laudenslager M, Winter C, Bradfield JP, Scott RH, Jagannathan J, Garris M, McConville C, London WB, Seeger RC, Grant SF, Li H, Rahman N, Rappaport E, Hakonarson H, Maris JM. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–723. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hartikainen JM, Tuhkanen H, Kataja V, Eskelinen M, Uusitupa M, Kosma VM, Mannermaa A. Refinement of the 22q12-q13 breast cancer–associated region: Evidence of TMPRSS6 as a candidate gene in an eastern Finnish population. Clin Cancer Res. 2006;12:1454–1462. doi: 10.1158/1078-0432.CCR-05-1417. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Association analysis of three Abraxas tagSNPs with breast cancer.
Table S2. LOH analysis of laser-capture microdissected tumor cell DNA of Abraxas c.1082G>A germline mutation carriers.
Table S3. Abraxas primers and PCR amplicon details.
Table S4. iPLEX primers used for Abraxas c.1082G>A and tagSNP genotyping.
Fig. S1. Abraxas R361Q is preferentially immunoprecipitated with its interacting partners in the cytoplasm.
Fig. S2. Expression of Abraxas R361Q affects DNA damage response.
Fig. S3. Nuclear retention of Abraxas R361Q restores localization and recruitment to DSB.
Fig. S4. Cytoplasmic localization of Abraxas R361Q does not aberrantly activate β-catenin.