Abstract
Human MAP3K4 (MTK1) functions upstream of mitogen activated protein kinases (MAPKs). In this study we show MTK1 is required for human epidermal growth factor receptor 2/3 (HER2/HER3)-heregulin beta1 (HRG) induced cell migration in MCF-7 breast cancer cells. We demonstrate that HRG stimulation leads to association of MTK1 with activated HER3 in MCF-7 and T-47D breast cancer cells. Activated HER3 association with MTK1 is dependent on HER2 activation and is decreased by pre-treatment with the HER2 inhibitor, lapatinib. Moreover, we also identify the actin interacting region (AIR) on MTK1. Disruption of actin cytoskeletal polymerization with cytochalasin D inhibited HRG induced MTK1/HER3 association. Additionally, HRG stimulation leads to extracellular acidification that is independent of cellular proliferation. HRG induced extracellular acidification is significantly inhibited when MTK1 is knocked down in MCF-7 cells. Similarly, pre-treatment with lapatinib significantly decreased HRG induced extracellular acidification. Extracellular acidification is linked with cancer cell migration. We performed scratch assays that show HRG induced cell migration in MCF-7 cells. Knockdown of MTK1 significantly inhibited HRG induced cell migration. Furthermore, pre-treatment with lapatinib also significantly decreased cell migration. Cell migration is required for cancer cell metastasis, which is the major cause of cancer patient mortality. We identify MTK1 in the HER2/HER3-HRG mediated extracellular acidification and cell migration pathway in breast cancer cells.
Keywords: MTK1, MEKK4, HER3, HER2, Migration
1. Introduction
Mitogen activated protein kinases (MAPKs) are regulated by various extracellular stimuli resulting from a cascade of sequential phosphorylations. MAPKs, such as the extracellular signal-regulated kinases (ERKs), are phosphorylated by MEKs and MEKs are phosphorylated by MEKKs [1]. The MEKK family of MAP3Ks was cloned based on homology to the catalytic domain of the yeast MAP3K, Ste11 [1]. MEKK4 (MAP3K4) was cloned using cDNA isolated from mouse [2], while MTK1 (MAP3K4) was cloned using human cDNA [3] and the sequence homology between the two proteins is 88% amino acid identity and 92% amino acid homology. When Ssk2 was cloned from yeast [4] it became apparent that the MEKK4 and MTK1 amino acid sequences are more homologous to yeast Ssk2p than Stellp [5]. Ssk2p is regulated by osmotic stress [3]. In yeast lacking Ssk2p, MEKK4 rescues the loss of Ssk2p resulting in p38 MAPK activation indicating that MEKK4 compliments Sskp2 in yeast [3].
The heart is one of the first organs to develop and congenital malformations occur at a rate of about one in one hundred [6]. Mutation of lysine in the active site of MEKK4 produces a kinase inactive protein. Kinase inactive MEKK4 attenuates developmental epithelial to mesenchymal transformation in mouse atrioventricular canal and ventricular heart explants [7]. A knock-in mutation of kinase-inactive MEKK4 was introduced in mice and the pups die at birth from skeletal malformations and neural tube defects [8]. These findings emphasize the importance of MEKK4 kinase activity during development. In addition to kinase activity, MEKK4 protein expression is also important in development. MEKK4 is highly expressed in the developing neuroepithelium and MEKK4 knockout mice display neural tube defects resulting in exencephaly and spina bifida [9]. MEKK4 knockout mice also display a congenital malformation of the cerebral cortex and MEKK4 RNA interference impairs neuronal cell migration [10].
Human MAP3K4 catalytic activity is activated by binding of GADD45 to the amino-terminal domain of MTK1 [11]. In contrast when the amino- and carboxyl-terminal domains of MTKs associate, this interaction is auto-inhibitory, blocking kinase activity. GADD45 association with MTK1 causes dissociation of the MTK1 amino-terminal and carboxyl-terminal domains leading to dimerization, auto-phosphorylation and activation of MTK1 [12]. Human MAP3K4 (MTK1) and the mouse homolog (MEKK4) regulate MKK6, which is upstream of stress activated p38 MAPK [3,11,13]. In addition, stress induced activation of MEKK4 leads to activation of MEK4/7 and JNK [14].
Receptor tyrosine kinases (RTK’s) and the growth factors that regulate them, such as heregulin (HRG) are often over-expressed in breast cancer cells [15–18], leading to activation of ERK1/2 activity, cell cycle progression [19] and cell migration [20,21]. The human epidermal growth factor receptors (HER) 1–4 are required for cell proliferation and differentiation during development [22,23]. HER2 is an orphan receptor with no known ligand. HER2 can form a heterodimer with EGFR, HER3 or HER4 and is often over-expressed in breast cancer [24]. HER4 expression correlates with favorable prognosis, while EGFR, HER2 and HER3 correlate with poor prognosis in breast cancer patients [25]. The growth factor, heregulin, is a ligand for HER3 and HER4, however HER3 is not kinase active and requires hetero-dimerization with either EGFR, HER2 or HER4 for activity [26–28]. Furthermore, HER2/HER3 is the preferred heterodimer for heregulin and produces strong mitogenic signaling that is linked to cancer [28–31].
HER2 over expression in estrogen positive cells is associated with tamoxifen drug resistance in breast cancer [32–34]. The drugs trastuzumab and lapatinib show high efficacy with HER2 positive patients, however drug resistance still persists [35–39]. HER3 protein expression was shown to be up-regulated with lapatinib treatment, compensating for HER2 inhibition, and HER3 phosphorylation occurred by residual HER2 expression limiting the efficacy of lapatinib treatment [40]. Therefore, HER3 over-expression and recovery of phosphorylation appears to be a compensatory mechanism in response to drug targeting of HER2. HER3 requires the catalytic activity of other members of the HER family for phosphorylation. A unique feature of HER3 is the six YXXM binding motifs that when phosphorylated function as recruitment sites for the SH2 domain of p85 of phosphoinositide kinase 3 (PI3K) leading to increased cell motility, invasion and metastasis ([41].
Cell migration requires actin polymerization and intracellular coordination of actin binding proteins, which are regulated by HER2 and downstream signaling proteins [42]. For example, heregulin stimulation of breast cancer cells enhances the conversion of globular actin (g-actin) to filamentous actin (f-actin) increasing cell migration [43,44]. Additionally, HER3 is regulated by HRG stimulation through HER2 kinase activity, which links HER3 to actin cytoskeletal reorganization and cell migration. Ssk2p is an example of an actin binding protein and is a homolog of MTK1. Ssk2p has an actin interacting region (AIR) that is required for actin cytoskeleton recovery after osmotic stress [45]. Despite the evidence for Ssk2p involvement in actin cytoskeletal reorganization, a link between MTK1 and actin has not been established in mammalian cells. Furthermore, even though HER2 and HER3 are involved in actin reorganization and cell migration, MTK1 has not been identified in this signaling process.
Cancer cells have increased glycolytic metabolism leading to acid loading and excess protons are excreted by up-regulating proton transporters [46]. Heregulin stimulation of breast cancer cells leads to extra cellular acidification of media that is dependent on HER2/HER3 activity [47]. Additionally, extracellular acidification affects cell migration and invasion [48,49]. For instance, human melanoma cells treated with acidic media excrete proteases required for migration and are more invasive [50]. With regard to HER2/HER3 signaling, although many signaling proteins have been linked to these receptors it is not clear how HRG regulates proton transporters.
Proteins that function in the MTK1 pathway have not been fully characterized nor has the regulation of MTK1 kinase activity. Previously we have shown regulation of mouse MAP3K4 (MEKK4) to be through activation of the IFNγ cytokine receptor [51] and the GPCR for angiotensin II [52]. In this study we investigated whether MTK1 is also regulated by the activation of RTK’s in MCF-7 and T-47D epithelial breast cancer cells. We report the recruitment of MTK1 with only activated HER3 in response to HRG in both MCF-7 and T-47D cells. MTK1 is also required for HRG induced cell migration in MCF-7 breast cancer cells through the HER2/HER3 heterodimer. Additionally, HRG induces association of MTK1 with p85 of PI3K, likely via phosphoHER3. It has been reported that HRG stimulation leads to extracellular acidification [47] an event that is linked to cancer cell migration [46,53]. We demonstrate that knockdown of MTK1 inhibits HRG-induced extracellular acidification and cell migration. Furthermore, pre-treatment of MCF-7 cells with the HER2 kinase inhibitor lapatinib inhibits association of MTK1 and HER3. MTK1 also associates with actin through the actin interacting region (AIR) and disruption of the actin cytoskeleton using cytochalasin D inhibits MTK1 and HER3 association. Together, this report establishes MTK1 as an integral signaling protein downstream of activated HER2 and HER3, required for acidification of the extracellular environment and cell migration.
2. Materials and methods
2.1. Cell culture and treatments
HEK-293, T-47D and MDA-MB-231 cells were cultured in Dulbecco’s modified Eagles medium with high glucose (DMEM) pH 7.4, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. MCF-7 cells were maintained in the same media as T-47D cells and supplemented additionally with 10 μg/ml insulin. Prior to experimental procedures, cells were cultured in DMEM supplemented only with 1% penicillin–streptomycin for 16 h. Cells were stimulated with 10 nM heregulin-β1 (HRG) EGF-Domain (Millipore Cat # 01–201) for 12 min unless otherwise indicated, EGF 3.3 nM for 12 min, 0.3 M sorbitol for 30 min or vehicle (30% glycerol in 1× phosphate buffered saline pH 7.4) for 12 min. Pre-treatment with 250 nM lapatinib was performed during serum starvation for 16 h unless otherwise indicated. Cells were treated with 1 μg/ml Cytochalasin D for 30 min prior to addition of HRG.
2.2. Western blotting and antibodies
MCF-7 or T-47D cells were lysed in lysis buffer (70 mM β-glycerol phosphate, 1 mM EGTA, 1 mM dithiothreitol, 2 mM MgCl2 and 0.5% Triton X-100) with protease inhibitors: 0.5 mM phenylmethylsulfonyl fluoride, 127.4 KIU/ml aprotinin (Calbiochem Cat # 616399), 10 μM leupeptin and with 0.5 mM sodium orthovanadate. Proteins were resolved by 5%–12.5% gradient SDS-PAGE and transferred onto Protran 0.45 μm nitrocellulose blotting membrane (BioExpress Cat # F-3120-7). Membranes were blocked with 5% non-fat dry milk in 25 mM Tris–HCl, pH 7.4, 137 mM NaCl, 2.7 mM KCl and 0.15% Tween 20 (TBS-T). Immunostaining was performed in 5% non-fat dry milk in TBS-T and detected using chemiluminescence reagent (100 mM Tris pH 8.5, 250 mM luminol, 92 mM p-coumaric acid and 0.018% H2O2). Images were obtained using ChemiDoc™ XRS + (BIO-RAD) and quantification was performed with Image Lab Software. After the initial immunoblots were performed, the nitrocellulose membranes were stripped at 56 °C for 1 h using membrane stripping buffer (12.5 mM Tris pH 6.8, 2% SDS, 0.7% β-mercaptoethanol) to remove primary and secondary antibody. Membranes were re-imaged before additional immunoblots to ensure stripping was complete. Subsequent immunoblots were then performed the same way as described above. Antibodies were purchased from Cell Signaling (anti-mouse HRP-conjugated #7076S; anti-rabbit HRP-conjugated #7074S), Millipore (phosphotyrosine mouse monoclonal Clone 4G10 #05-321), Epitomics (EGFR #1902-1, HER2 #2064-1, HER3 #1186-1, HER4 #2218-1 and HER3 pY1289 #2526-1 rabbit monoclonal antibodies), Santa Cruz Biotechnology (PI3-Kinase p85α mouse monoclonal #sc-1637), Thermo Scientific (actin mouse monoclonal #MA1-744), Sigma (Anti-FLAG mouse monoclonal #F1804) and MTK1 antibodies used were developed as we previously described [52]. All commercial antibodies were used according to manufacturer recommendations.
2.3. Immunoprecipitation experiments
HEK-293, MCF-7, T-47D or MDA-MB-231 cells were stimulated with 10 nM HRG for 12 min, 0.3 M sorbitol for 30 min, 3.3 nM EGF for 12 min, 10 μg/ml insulin for 30 min or vehicle, unless otherwise indicated. The cells were then washed twice with ice cold 1× phosphate buffered saline pH 7.4. The cells were then lysed in lysis buffer and 2 mg of cell extract was immunoprecipitated for 1 h at 4 °C with 10 μg rabbit anti-MTK1 polyclonal antibody [52]; 4 μg mouse phosphotyrosine antibody (Millipore Clone 4G10 #05-321) or 20 μl anti-FLAG beads (Clontech #635686). Immune complexes recovered using protein A-Sepharose beads (Sigma Cat# P3391) were washed twice with ice cold lysis buffer and denatured with Laemmli sample buffer. Proteins were separated by 5–12.5% gradient SDS-PAGE and transferred to nitrocellulose membranes that were then subjected to immunoblot analysis as indicated.
2.4. Cell proliferation and media pH measurements
MCF-7 and T-47D cells were seeded in complete growth media at a density of 0.4 × 106 cells per well in 12-well plates (VWR # 62406–165) for proliferation assays and 0.7 × 106 cells per well for pH measurements. Cells were allowed to adhere for 16 h, followed by 4 h serum starvation in DMEM, 1% penicillin–streptomycin without fetal bovine serum. The cells were then stimulated with 10 nM HRG, 3.3 nM EGF, 250 nM lapatinib (lapatinib pre-treatment was started at time of serum starvation) or vehicle for 24 h in DMEM with 0.5% fetal bovine serum, 1.0% penicillin/streptomycin and 10 μg/ml human recombinant insulin in MCF-7 cells. Each condition was performed in triplicate and repeated a minimum of 3 times. After 24 h, the cells were counted using a TC-10 Bio-Rad automated cell counter and the media pH was recorded using a pH meter probe. Data shown for each condition represents n = 3. A two sided student t-test with standard deviation was used to perform statistical analyses on proliferation rates and media pH measurements. Statistical significance was determined by a p-value of ≤0.05.
2.5. siRNA knockdown experiments
MCF-7 cells were seeded (onto 10 cm cell culture plates, VWR # 353003) at a density of 5.0 × 106 cells and allowed to adhere for 16 h. The cells were then transfected with 434 pmol of siRNA specific for MTK1 (Thermo Scientific ON-TARGETplus SMARTpool, Human MAP3K4 (4216) Cat # L-003789-00-0005) or non-targeting (NS) (ON-TARGETplus Non-targeting Pool Cat # D-001810-10-05) as control using lipofectamine 2000 according to manufacture recommendations. The cells were transfected with siRNA a second time 24 h later using lipofectamine 2000 to enhance knockdown efficiency. Twenty four hours after the second transfection the cells were detached using 0.25% trypsin (Invitrogen # 15050–065). The cells were counted and seeded onto 12 well plates (VWR # 62406–165) at a density of 0.7 × 106 cells per well for pH measurements and scratch assays or at density of 0.4 × 106 cells per well for proliferation assays. MTK1 knockdown efficiency was determined by immunoblot analysis using 150 μg whole cell lysate at 120 hour post-secondary transfection. Each condition was performed in triplicate and repeated a minimum of 3 times. Data shown for each condition represents n = 3. Statistical analysis was performed as described above.
2.6. Scratch assay experiments
Scratch assays were performed in 12-well plates using MCF-7 cells at a density of 0.7 × 106 cells per well. The cells were allowed to adhere for 16 h followed by 4 h serum starvation in DMEM. The surface area of the cells was scratched using a 10 μl pipette tip (VWR # 89140–164) and the cells were then washed twice with 1× phosphate buffered saline pH 7.4. The cells were then stimulated with 10 nM HRG or vehicle, pretreated with or without 250 nM lapatinib (lapatinib pre-treatment was started at the time of serum starvation). Once scratches were performed and HRG stimulation was started, this was considered time zero and images were acquired using a 12.2 megapixel digital camera (GE model # W1200) visualized through a Leica Microsystems inverted microscope (model # DM IL) at 10× magnification. Additional images were taken at 24 and 48 h and measurements were taken from 24 h and 48 h and compared to time zero for each condition. Each condition was performed in triplicate and repeated a minimum of 3 times. Data shown for each condition represents n = 3. Statistical analysis was performed as described above.
3. Results
3.1. Association of a 180 kDa tyrosine phosphorylated protein with MTK1
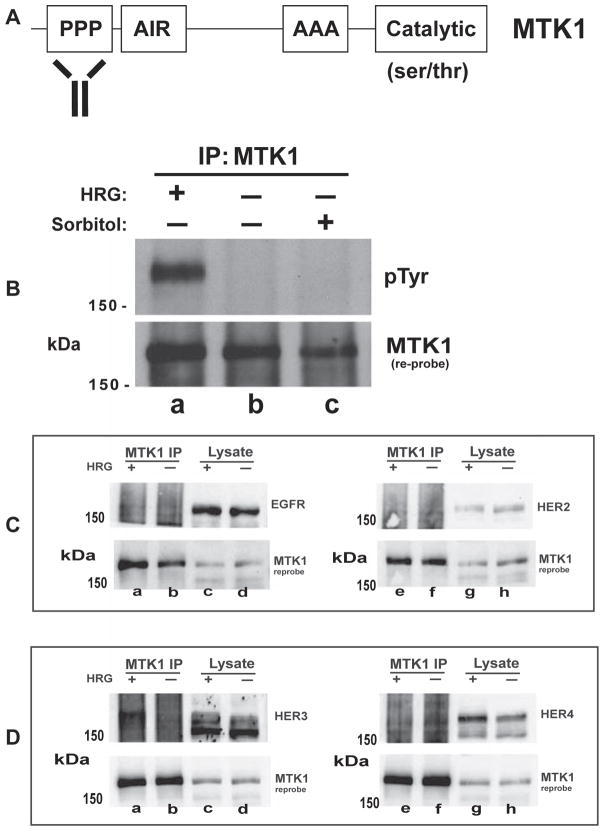
To investigate RTK regulation of MTK1 we selected MCF-7 and T-47D breast cancer epithelial cells, which express moderate levels of the ErbB RTK receptors 1–4 [26,54]. Heregulin β1 (HRG) was used to stimulate these cells because HER3 and HER4 bind HRG allowing homo and hetero dimerization between HER1-4 [55–57]. MCF-7 or T-47D cells were stimulated with HRG or sorbitol. Sorbitol was used to induce osmotic stress and activate MTK1, since MTK1 is known to function in the p38 MAP kinase pathway [11,14,58]. After stimulation, MTK1 was immunoprecipitated with MTK1 antibody that recognizes the amino-terminal proline-rich region of MTK1 (Fig. 1A). Immunoprecipitated MTK1 and associated proteins were resolved by SDS-PAGE, followed by transfer to nitrocellulose membrane and immunoblotting for tyrosine phosphorylation (pTyr). A tyrosine phosphorylated protein co-immunoprecipitated with MTK1 in response to HRG stimulation (Fig. 1B, top panel, lane a), which was not present in cells stimulated with vehicle or sorbitol (Fig. 1B, top panel, lanes b and c). The tyrosine phosphorylated protein had a molecular weight of 180 kDa, which is approximately the same molecular weight as MTK1. This result suggested that MTK1 may be phosphorylated on tyrosine in response to HRG stimulation, or that a HER family member co-precipitated with MTK1.
Fig. 1.
HER3 associates with MTK1 in response to HRG stimulation. A schematic representation of the proline-rich region (PPP), actin-interacting region (AIR), alanine-rich region (AAA), and catalytic domain of MTK1 is shown. The MTK1 antibody recognizes an amino terminal region (amino acids 18–139) of MTK1 (Panel A). T-47D cells were stimulated with 10 nM HRG for 12 min or 0.3 M sorbitol for 30 min as indicated (Panel B). MTK1 was immunoprecipitated and proteins were resolved by SDS-PAGE and immunoblotted using monoclonal antibody 4G10 (pTyr). T-47D cells were stimulated with HRG and MTK1 was immunoprecipitated as described above (Panels C & D). Immunoblots were performed using antibodies directed against EGFR and HER2 (Panel C) or HER3 and HER4 (Panel D). Membranes were stripped and immunoblotted with MTK1 antibody (B, C and D, bottom panels).
3.2. HER3 associates with MTK1 in response to HRG stimulation
We set out to investigate the identity of the 180 kDa protein. HER1–4 were candidate proteins for tyrosine phosphorylation and association with MTK1, since HER3 and HER4 bind HRG and can trigger homo and hetero-dimerization of these four receptors [18,59]. T-47D cells were stimulated with HRG and cell extracts were prepared in which MTK1 was immunoprecipitated in four different samples. The precipitated proteins were resolved by SDS-PAGE and immunoblotted for HER1–4 to identify which of the HER proteins associate with MTK1. HER3 was identified as associating with MTK1 in response to HRG stimulation (Fig. 1D, top panel, lane a). The fact that immunoblotted HER3 from the immunoprecipitation migrated slower than the proteins from the cell lysates suggested that phosphorylated HER3 preferentially interacted with MTK1. EGFR and HER2 did not associate with MTK1 in response to HRG or vehicle stimulation (Fig. 1C, top panel, lanes a & b, and lanes e & f). Moreover, HER4 did not associate with MTK1 in response to HRG (Fig. 1D, top panel, lanes e & f), even though HER4 binds HRG [60]. Immunoprecipitation of MTK1 was validated for each experimental condition by immunoblotting for MTK1 (Fig. 1C and D, bottom panels).
3.3. MTK1 is not tyrosine phosphorylated in response to HRG
Our results suggested that HER3 was the tyrosine phosphorylated protein that interacts with MTK1. However, the results did not exclude the possibility that MTK1 was phosphorylated on tyrosine especially since we were immunoprecipitating MTK1 and it has a similar molecular mass as the tyrosine phosphorylated protein. If MTK1 was tyrosine phosphorylated, we should be able to immunoprecipitate MTK1 with an antibody specific for phospho-tyrosine. We stimulated T-47D cells with HRG and performed an immunoprecipitation with monoclonal antibody 4G10 that recognizes tyrosine phosphorylation. A protein of 180 kDa was immunoprecipitated with the 4G10 monoclonal antibody and the protein was phosphorylated on tyrosine (Fig. 2A, top panel, lane a). Similarly, a 180 kDa tyrosine phosphorylated protein was present in the MTK1 immunoprecipitation (Fig. 2A, top panel, lane c) When normal rabbit IgG was used as a control during the immunoprecipitation, no tyrosine phosphorylated proteins were detected in response to HRG (Fig. 2A, top panel, lane e). The membrane was stripped and immunoblotted for MTK1. The results revealed that MTK1 was not present in the phosphotyrosine immunoprecipitation (Fig. 2A, bottom panel, lanes a & b), which strongly suggests that MTK1 is not tyrosine phosphorylated in response to HRG. As expected, MTK1 was present in the MTK1 immunoprecipitation (Fig. 2A, bottom panel, lanes c & d). Additionally, normal rabbit IgG did not immunoprecipitate MTK1 (Fig. 2A, bottom panel, lanes e & f).
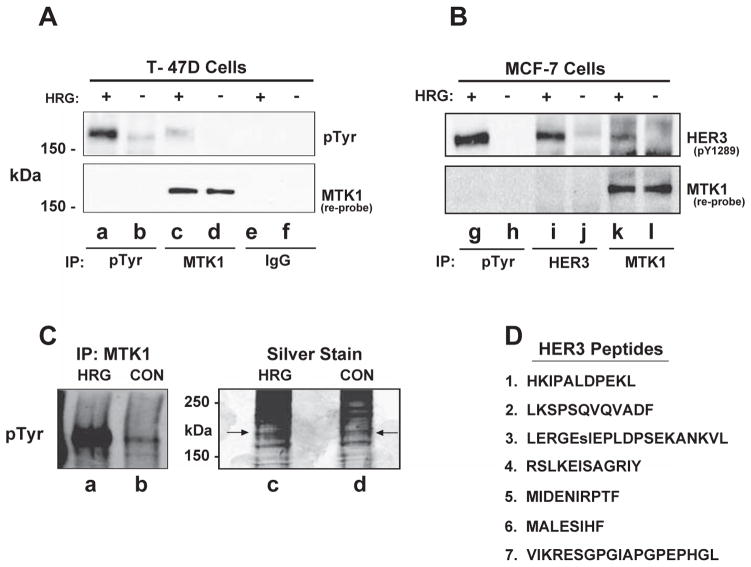
Fig. 2.
HER3 associates with MTK1 in both T-47D and MCF-7 cells. Cells were stimulated as described in Fig. 1. Cell lysates were incubated with antibody directed against phosphotyrosine (pTyr), MTK1, or normal rabbit IgG. Proteins were resolved by SDS-PAGE and immunoblotted with anti-phosphotyrosine antibody (A, top panel) or antibody specific for phosphotyrosine 1289 of HER3 (B, top panel). The membranes were re-probed for MTK1 (bottom panels). MCF-7 cell lysates were prepared as described above and a fraction of the immunoprecipitate (15%) was immunoblotted using anti-phosphotyrosine antibody (Panel C, lanes a & b), while the remaining immunoprecipitation was silver stained (lanes c & d). HER3 peptides were identified in the HRG stimulated sample (arrows) by LC–MS/MS with 99.9% confidence (D).
3.4. PhosphoHER3 associates with MTK1 in response to HRG
HER3 has a molecular mass of 180 kDa and among many phosphotyrosines there are several YXXM motifs that are phosphorylated in response to HRG [41]. There are commercial phospho-antibodies that recognize several of these phosphorylation sites, one of which is pY1289. In order to determine if MCF-7 cells behave similarly to T-47D cells, we used MCF-7 cells in our next experiment. We stimulated MCF-7 cells with HRG and performed an immunoprecipitation with monoclonal antibody 4G10. HER3 was immunoprecipitated with the 4G10 monoclonal antibody and was phosphorylated on Y1289 (Fig. 2B, top panel, lane g). Similarly when a HER3 antibody was used for immunoprecipitation, phosphoHER3 was detected with the pY1289 phospho antibody (Fig. 2B, top panel, lane i). Additionally, HER3 co-precipitated with MTK1 when the immunoprecipitation was performed with MTK1 antibody and HER3 was tyrosine phosphorylated on Y1289 (Fig. 2B, top panel, lane k). The membrane was stripped, immunoblotted for MTK1, and MTK1 was not detected in the phosphotyrosine immunoprecipitation (Fig. 2B, bottom panel, lanes g & h). These results demonstrate that HRG regulates the association of MTK1 with activated HER3 and that MTK1 is not phosphorylated on tyrosine in both T-47D and MCF-7 breast cancer cells.
Additionally, the University of Arizona Southwest Environmental Health Sciences Center (SWEHSC) proteomics core facility was used to further characterize the association between MTK1 and HER3 in response to HRG stimulation. A series of six MTK1 immunoprecipitations was performed in which 2 mg of cell extract was used for each immunoprecipitation. After washing the immunoprecipitations with lysis buffer, the samples were pooled and resolved in one lane by SDS-PAGE followed by silver staining. Silver stained proteins were aligned with a replicate experiment that was immunoblotted with phosphotyrosine antibody (Fig. 2C, lane a). The area of the gel aligning with the 180 kDa tyrosine phosphorylated protein was excised (Fig. 2C, lanes c & d). The proteins were digested with trypsin, separated and analyzed via Orbitrap LC–MS. HER3 was identified as associating with MTK1 in response to HRG with 26% of the HER3 protein sequenced and several of the peptides identified with a 99.9% degree of confidence (Fig. 2D, right panel). Several of these peptides were sequenced multiple times increasing confidence for positive identification of HER3. Proteomic analysis did not identify HER3 as associating with MTK1 in response to vehicle stimulation. This result is consistent with our immunoblot analysis which suggests that the association between HER3 and MTK1 is not constitutive, but requires HRG stimulation.
Although HER3 peptides were not identified from the MTK1 immunoprecipitation of vehicle stimulated MCF-7 cells, MTK1 peptides were identified because MTK1 and HER3 migrate at similar positions by SDS-PAGE (Supplemental Fig. 1). MTK1 peptides were identified from both the HRG and vehicle stimulated conditions. A total of 56% of the MTK1 protein was identified as tryptic peptides with a 99.9% degree of confidence for positive identification of MTK1. The identification of MTK1 peptides and not HER3 peptides from the vehicle stimulated sample indicates that the lack of HER3 peptides in this sample was not a technical artifact. In addition, there were no tyrosine phosphorylated peptides identified for MTK1 in either HRG or vehicle stimulated conditions which is consistent with the immunoblotting data.
A time course experiment was performed in which MCF-7 cells were stimulated with HRG for as long as 150 min. In other experiments, the tyrosine phosphorylation of the 180 kDa protein associating with MTK1 occurred within 30 s (data not shown). In longer time course experiments, the tyrosine phosphorylated protein that co-immunoprecipitated with MTK1 was observed from 5 for up to 150 min after HRG stimulation (Fig. 3A, upper panel, lanes b–j). In HRG washout experiments where HRG was washed out after each time point, the tyrosine phosphorylation was more transient (data not shown). These experiments demonstrate that constant HRG exposure results in continued HER3 activation and association with MTK1. This result may be significant in breast cancer cases in which HRG is over-expressed.
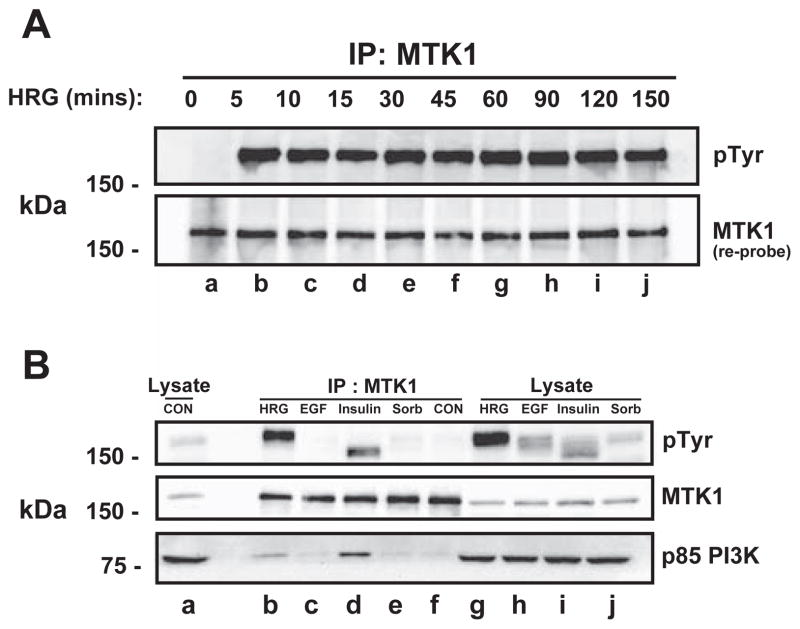
Fig. 3.
Sustained MTK1 and HER3 association. MCF-7 cells were stimulated with 10 nM HRG for the indicated times. MTK1 was immunoprecipitated and proteins were resolved by SDS-PAGE, then immunoblotted with anti-phosphotyrosine antibody (A, top panel). The membrane was re-robed with MTK1 antibody (bottom panel). T-47D cells were stimulated with 10 nM HRG for 12 min, 3.3 nM EGF for 12 min, 10 μg/ml insulin for 15 min or 0.3 M sorbitol for 30 min followed by immunoprecipitation of MTK1. The proteins were resolved by SDS-PAGE and immunoblotted with the indicated antibodies.
3.5. HER3 and MTK1 association is specific to HRG stimulation
In order to determine whether other receptor tyrosine kinases interacted with MTK1, T-47D cells were stimulated with EGF or insulin and then MTK1 was immunoprecipitated as described above. As a positive control, cells were stimulated with HRG. Immunoblot analysis using 4G10 mouse monoclonal antibody revealed a protein of approximately 150 kDa that was tyrosine phosphorylated in response to insulin (Fig. 3B, top panel, lane d). No tyrosine phosphorylated proteins immunoprecipitated with MTK1 after stimulation with EGF, sorbitol or vehicle (Fig. 3B, top panel, lanes c, e & f). Since EGF can activate the EGFR and HER2, the lack of tyrosine phosphorylated proteins indicates that neither EGFR nor HER2 interact with MTK1, while HRG selectively activates HER3 to interact with MTK1 (lane b). The ~150 kDa tyrosine phosphorylated protein that immunoprecipitated with MTK1 in response to insulin was later identified as insulin receptor substrate-1 (IRS-1; data not shown). The membrane was stripped and immunoblotted for MTK1 (Fig. 3B, middle panel). To establish stimulation efficacy, lysates from vehicle, HRG, EGF, insulin and sorbitol were immunoblotted with G410 mouse monoclonal antibody (Fig. 3B, top panel lanes a & g–j).
3.6. p85 of PI3K associates with MTK1
HER3 has several YXXM motifs which, when phosphorylated, function as recruitment sites for the SH2 domain of the p85 regulatory subunit of PI3K [41]. Therefore, it was possible that p85 would co-precipitate with MTK1 in response to HRG. p85 of PI3K was identified as immunoprecipitating with MTK1 in response to HRG stimulation (Fig. 3B, bottom panel, lane b). Additionally, p85 was also shown to immunoprecipitate with MTK1 in response to insulin stimulation, likely through an interaction with IRS (Fig. 3B, bottom panel, lane d). These results demonstrate that the HER3/MTK1 complex also includes PI3K suggesting that the HER3/MTK1 complex utilizes 3′ phosphorylated inositol phosphates as part of the signaling mechanism.
3.7. HER2 is required for co-immunoprecipitation of phosphorylated HER3 with MTK1
HER3 does not have intrinsic kinase activity and is phosphorylated by hetero-dimerization with either HER1, 2 or 4 [28]. HER2, HER3 and HER4 are tyrosine phosphorylated in MCF-7 cells stimulated with HRG (data not shown). Thus, HER2 and HER4 are possible candidates for phosphorylating HER3, since HER2/HER3 and HER3/HER4 heterodimers are formed in response to HRG. Since HER2 and HER3 are preferred heterodimers [31], HER2 was hypothesized as the most likely candidate for phosphorylation of HER3. MCF-7 cells were pretreated with lapatinib to inhibit the kinase activity of HER2 and then stimulated with HRG followed by immunoprecipitation of MTK1. Lapatinib has a Ki of 13 nM for inhibition of HER2 [61]. Lapatinib decreased the interaction between HER3 and MTK1 in a dose dependent manner as evidenced by decreased tyrosine phosphorylation of HER3 (Fig. 4A). These results indicate that HER2 catalytic activity is required for HER3 phosphorylation and subsequent interaction with MTK1 in response to HRG. The membrane was stripped and immunoblotted for MTK1 (Fig. 4A, bottom panel). Lysates from the same experiment show tyrosine phosphorylation of HER3 is decreased in response to lapatinib (Fig. 4B, top panel, lanes c–e).
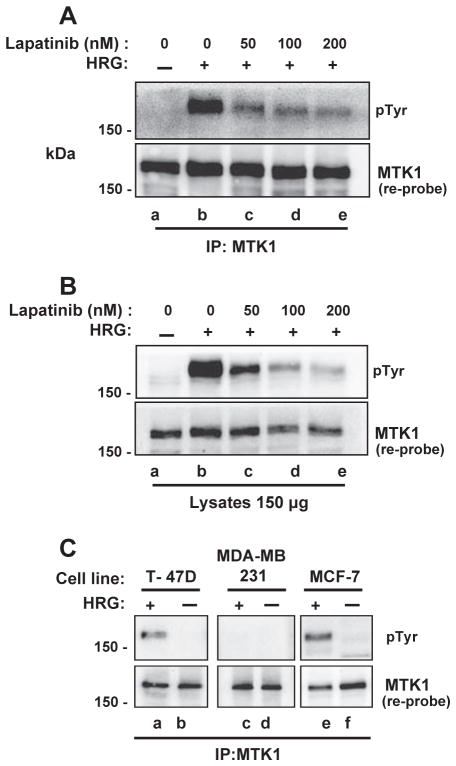
Fig. 4.
HER2 kinase activity is required for MTK1/HER3 association. MCF-7 cells were pre-treated with 0, 50, 100 or 200 nM lapatinib for 1 h followed by 10 nM HRG stimulation for 12 min. Phosphotyrosine immunoblot analysis of MTK1 immunoprecipitations show 50 nM lapatinib attenuates MTK1/HER3 association (Panel A, lane c). Cell extracts (150 μg) that were used for the immunoprecipitation were resolved by SDS-PAGE and immunoblotted for phosphotyrosine (B, top panel). HER2 negative, but HER3 positive MDA-MB-231 cells were stimulated with 10 nM HRG followed by MTK1 immunoprecipitations and compared to T-47D and MCF-7 cells. Phosphotyrosine immunoblot analysis shows HER2 is required for MTK1/HER3 association in response to HRG (Panel C, lane c). Membranes were stripped and immunoblotted for MTK1 (A, B and C, bottom panels).
MDA-MB-231 breast cancer cells were used to further characterize HER2 involvement in the formation of the HER3-MTK1 protein complex in response to HRG. MDA-MB-231 cells are a triple negative cell line and do not express estrogen receptor, progesterone receptor or HER2, but do express HER3 [62]. If HER2 is required for HER3 phosphorylation and association with MTK1, then we should not see the association of HER3 with MTK1 in MDA-MB-231 cells. T-47D, MDA-MB-231 and MCF-7 cells were stimulated with HRG followed by immunoprecipitation of MTK1. Immunoblot analysis using 4G10 mouse monoclonal antibody revealed that a 180 kDa tyrosine phosphorylated protein immunoprecipitated with MTK1 in T-47D and MCF-7 cells dependent on HRG (Fig. 4C, top panel, lanes a & e). No tyrosine phosphorylated proteins immunoprecipitated with MTK1 from the MDA-MB-231 cells (Fig. 4C, top panel, lanes c & d), which strongly suggests that HER2 is required for formation of the HER3 MTK1 complex. The membranes were stripped and immunoblotted for MTK1 to demonstrate that MTK1 is expressed in MDA-MB-231 cells (Fig. 4C, bottom panel).
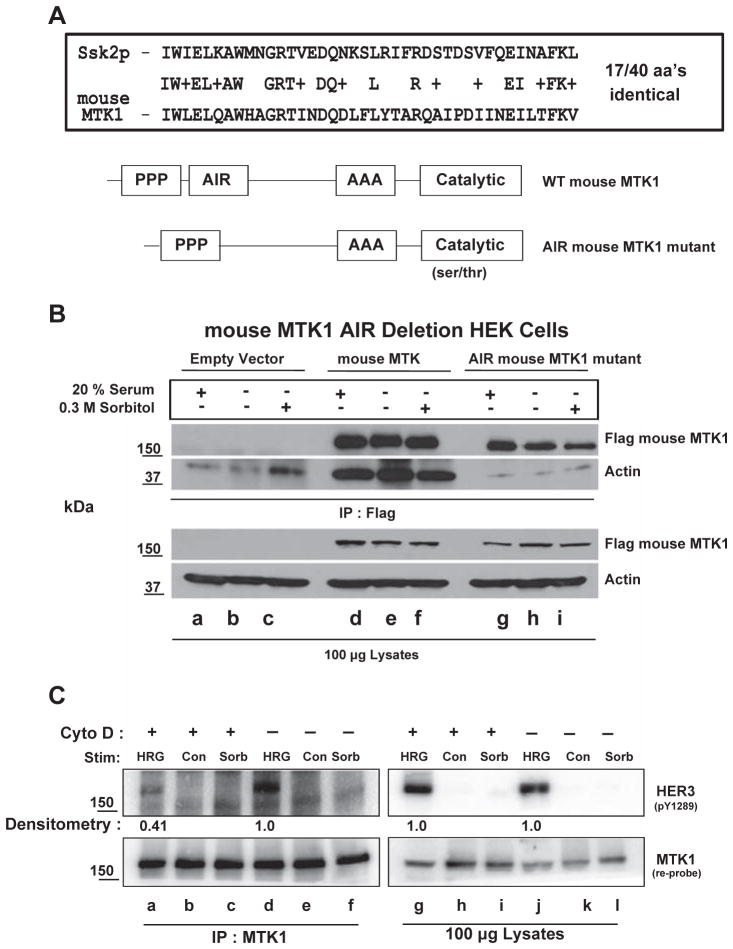
3.8. Actin interacting region (AIR) of MEKK4 is required for actin association with MEKK4
Human MTK1 and the mouse homolog, MEKK4, have similar homology to the yeast protein, Ssk2p [5]. Ssk2p has an actin interacting region (AIR) that is required for actin cytoskeleton recovery after osmotic stress [45]. Alignment of the AIR of Ssk2p with MEKK4 shows 17/40 amino acids are identical between these proteins and 25/40 are highly conserved, suggesting that MEKK4/MTK1 might interact with actin (Fig. 5A). The AIR of MEKK4 consisting of amino acids 256–295 was deleted from the MEKK4 cDNA. Flag-tagged wild-type MEKK4 and flag-tagged AIR MEKK4 were transfected into HEK 293 cells. After 48 h, the HEK cells were serum starved for 4 h followed by stimulation with 20% fetal bovine serum or sorbitol as possible inducers of the MTK1-actin interaction. MEKK4 was immunoprecipitated with a flag antibody-coupled to beads that specifically recognizes the flag epitope. Actin did not associate with MEKK4 when the AIR was deleted (Fig. 5B, second panel, lanes g–i), while actin constitutively associated with wild-type MEKK4 independent of stimulation (Fig. 5B, second panel, lanes d–f). These results demonstrate that MEKK4/MTK1 behave like Ssk2p and interact with actin within a unique 40 amino acid region.
Fig. 5.
Actin associates with MTK1 and inhibition of actin polymerization with cytochalasin D inhibits MTK1/HER3 association. Comparison of mouse MTK1 and the actin interacting region (AIR) of the yeast homolog Ssk2p shows 17/40 amino acids are identical and 25/40 are conserved (Panel A). Amino acids 256–295 were deleted from the mouse MTK1 cDNA (AIR mutant). HEK-293 cells were transfected with empty vector, flag-tagged wild-type mouse MTK1 or the flag-tagged AIR mutant mouse MTK1 cDNA. After 48 h, the cells were stimulated with 20% fetal bovine serum or 0.3 M sorbitol. Flag antibody-beads were used for immunoprecipitation for flag-tagged proteins. Proteins were resolved by SDS-PAGE and immunoblotted with antibody that recognizes the FLAG epitope (B, top panel) or actin (lower panel). Cell lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. MCF-7 cells were pre-treated with 1 μg/ml cytochalasin D (Cyto D) for 30 min followed by 10 nM HRG for 12 min or 0.3 M sorbitol for 30 min. MTK1 was then immunoprecipitated from each condition (C, lanes a–f). Antibody directed against phosphotyrosine 1289 of HER3 was used to immunoblot (top panel). The membrane was re-probed for MTK1 (bottom panel). Cell lysates were resolved by SDS-PAGE and immunoblotted as indicated. Cytochalasin D had no effect on the phosphorylation of HER3 (lanes g & j), but diminished interaction between MTK1 and HER3 (lanes a & d).
3.9. Disruption of the actin cytoskeleton significantly decreases the association between HER3 and MTK1
HRG enhances the conversion of g-actin to f-actin leading to increased cell migration [43]. Since HER3 is regulated by HRG, the association between MTK1 and HER3 that we observed may be facilitated by actin in response to HRG. To test this hypothesis, MCF-7 cells were pre-treated with cytochalasin D to disrupt actin polymerization and the formation of f-actin. The cells were then stimulated with HRG or sorbitol followed by immunoprecipitation of MTK1 and pY1289 HER3 immunoblots. In the presence of cytochalasin D, the interaction between HER3 and MTK1 was diminished by 60% (Fig. 5C, top panel, compare lanes a & d). Interestingly, phosphorylation of HER3 remained the same in the HRG stimulated lysates (Fig. 5C, top panel, lanes g & j). These data suggest that f-actin is required for the association between activated HER3 and MTK1 in response to HRG, but f-actin is not required for HRG-dependent activation of HER3.
3.10. MTK1 is required for HRG-induced extracellular acidification
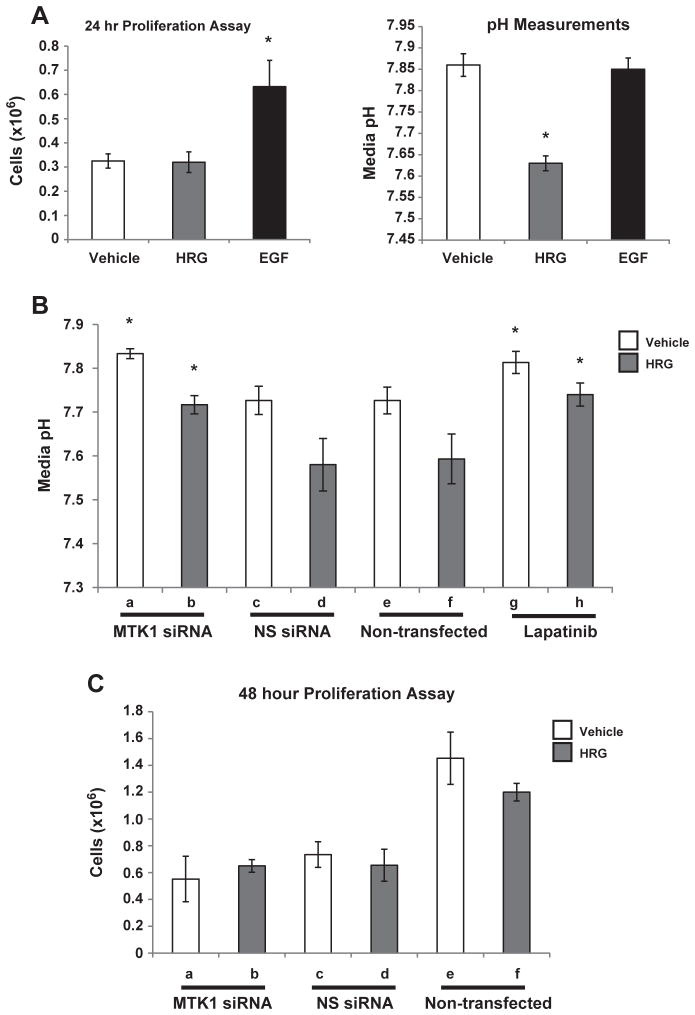
Since HRG is known to induce proliferation, invasion, migration and metastasis [63–68], it was necessary to identify the specific functional role of HRG in MCF-7 breast cancer cells. Assays were performed to determine if HRG stimulation leads to proliferation of MCF-7 cells. MCF-7 cells were stimulated with HRG or EGF and after 24 h the cells were counted using a Bio-Rad TC-10 automated cell counter in the presence of trypan blue. EGF, unlike HRG, induced cell proliferation (Fig. 6A, left panel). There was no difference in cell viability between each condition (data not shown). However, there was a noticeable difference in the color of the DMEM media after 24 h of HRG stimulation, which was not observed with EGF stimulation. Media from the HRG stimulated cells appeared orange, which is an indicator of increased acidity or decreased pH. Measurements of the media pH revealed that HRG stimulation led to extracellular acidification of the cell culture media when compared to vehicle (Fig. 6A, right panel). EGF stimulation did not induce extracellular acidification (Fig. 6A, right panel). Similarly, T-47D cells stimulated with HRG also had extracellular acidification independent of cell proliferation (data not shown).
Fig. 6.
MTK1 is required for HRG induced extracellular acidification. MCF-7 cells were stimulated with 10 nM HRG or 3.3 nM EGF, after 24 h the cells were counted (Panel A, left graph) and the pH of the media was measured (right graph). MTK1 siRNA knockdown was used to determine whether MTK1 is required for HRG induced extracellular acidification. MCF-7 cells were transfected as described in the Materials and methods section. The cells were then stimulated with 10 nM HRG for 24 h followed by pH measurements of the media (Panel B). Non-transfected or cells pre-treated with 250 nM lapatinib were also stimulated with HRG followed by pH measurements (Panel B). MTK1 knockdown did not have an effect on cell proliferation compared to NS knockdown (Panel C). Statistical analysis was performed using a two-sided student t-test with standard deviations and a minimum of three replicates, p-value of ≤0.05 was considered statistically significant and each experiment was repeated a minimum of three independent times.
3.11. MTK1 knockdown inhibits HRG induced extracellular acidification
HRG induced extracellular acidification and association of tyrosine phosphorylated HER3 with MTK1. These results suggest that MTK1 is required for HRG-induced proton excretion from the cell. To test this hypothesis MCF-7 cells were double transfected with siRNA that specifically targets MTK1 and non-specific (NS) siRNA as a control (double transfections were performed to increase transfection efficiency). The cells were stimulated with HRG and after 24 h, media was collected and pH measurements were recorded with a pH meter probe. Knockdown of MTK1 inhibited HRG induced extracellular acidification when compared to NS siRNA and non-transfected cells (Fig. 6B, lanes b, d & f). In addition, pre-treatment of MCF-7 cells with lapatinib for 30 min prior to stimulation with HRG resulted in inhibition of HRG-induced extracellular acidification (Fig. 6B, lanes g & h) indicating a requirement for HER2 in the acidification response. Together, these results show that HER2 and MTK1 are required for HRG-induced extracellular acidification.
Knockdown of MTK1 could affect the proliferation rate resulting in fewer cells excreting protons. Therefore, proliferation assays were performed to rule out whether MTK1 knockdown had an effect on proliferation rates. MCF-7 cells were transfected with siRNA, as described previously and stimulated with HRG. After 48 h the cells were counted using a Bio-Rad TC-10 automated cell counter. Knockdown of MTK1 did not have an effect on the proliferation rate when compared to NS siRNA knockdown (Fig. 6C, lanes a–d). Knockdown of MTK1 did not have an effect on viability either (data not shown). Since all of the experimental conditions shown in Fig. 6C started with the same amount of cells, it appears that transfection of MCF-7 cells did have an effect on proliferation rate when compared to non-transfected cells (Fig. 6C, lanes e & f). However, the decreased proliferation rate was independent of MTK1 siRNA but dependent on transfection, likely reflecting the difficulty associated with transfecting MCF-7 cells.
3.12. MTK1 is required for HRG induced cell migration
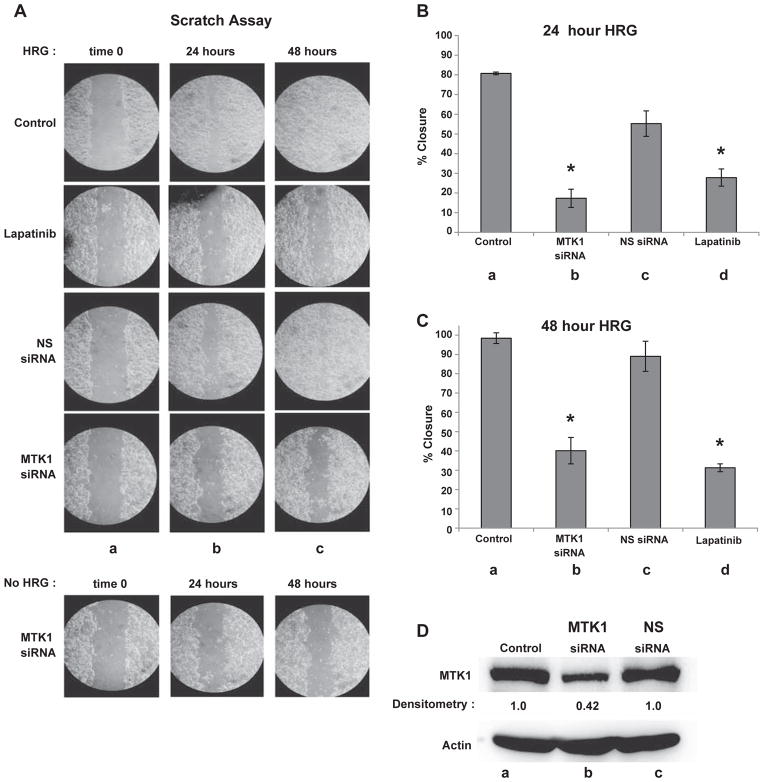
Cancer cells have increased glycolytic metabolism leading to acid loading and excess protons are excreted by up-regulating proton transporters [46]. Migration is a possible functional response to HRG stimulation because acid excretion creates micro extracellular environments that are favorable for cell migration [53]. HRG is also linked to intracellular actin cytoskeletal reorganization that leads to increased migration [43]. Furthermore, MTK1 may have a role in HRG induced cell migration since MTK1 knockdown blocks extracellular acidification. To determine whether MTK1 is required for cell migration, HRG-induced scratch assays were performed with and without MTK1 knockdown. MCF-7 cells were transfected with MTK1 and NS siRNA as described above. MCF-7 cell transfections were performed using 10 cm culture dishes and then the cells were transferred to a 12-well plate after a 24 h post-transfection recovery period. The surface of the well was scratched followed by HRG stimulation and digital image acquisition at 0, 24 and 48 hour post scratch. After 24 h of HRG stimulation, non-transfected cells closed the scratched area by 80% (Fig. 7A, column b). Cells pretreated with lapatinib only closed the scratched area by 27%. Similarly, cells transfected with MTK1 siRNA closed the scratched area by approximately 17% while the cells transfected with NS siRNA had closed the scratched area by approximately 60% (Fig. 7A, column b). Additionally, MTK1 knockdown or lapatinib pretreatment continued to severely impair cell migration into the scratched area after 48 h (Fig. 7A, column c), while the cells transfected with NS siRNA had almost completely migrated into the scratched area, much like the non-transfected cells (Fig. 7A, top row). Statistical analysis revealed that MTK1 knockdown had a mean of 17.3% ± 4.6 cell migration (Fig. 7B, lane b) and cells pretreated with lapatinib had a mean of 27.8% ± 4.4 cell migration (Fig. 7B, lane d) compared to NS knockdown mean of 55.3% ± 6.4 (Fig. 7B, lane c). Similar results were observed at 48 h with MTK1 knockdown cells migrating by a mean of 40.1% ± 6.8 (Fig. 7C, lane b) and lapatinib treated cells migrating by a mean of 31.3% ± 2 (Fig. 7C, lane d) compared to NS knockdown migrating by a mean of 89% ± 7.8 (Fig. 7C, lane c). Very little cell migration was observed in cells not stimulated with HRG. Therefore, scratch assays of cells transfected with MTK1 siRNA or NS siRNA, and non-transfected cells all looked similar (only MTK1 siRNA is shown, Fig. 7A bottom panel). MTK1 protein expression levels were measured 48 h after scratch by immunoblot analysis. MTK1 protein expression was knocked down by 58% compared to NS siRNA (Fig. 7D, top panel, lanes b & c). Actin was used as a loading control (Fig. 7D, bottom panel). Together these results demonstrate that HRG-induced cell migration requires MTK1.
Fig. 7.
MTK1 is required for HRG induced cell migration. MCF-7 cells were transfected as described previously. The cells were serum starved for 4 h, then scratched followed by HRG stimulation. Digital images were acquired at 0, 24 and 48 hour post scratch (Panel A). Statistical analysis revealed MTK1 knockdown cells migrated by a mean of 17.3% ± 4.6 (Panel B, lane b) and cells pretreated with lapatinib had a mean of 27.8% ± 4.4 cell migration (Panel B, lane d) compared to a NS knockdown mean of 55.3% ± 6.4 (Panel B, lane c). Similar results were observed at 48 h with MTK1 knockdown migrating by a mean of 40.1% ± 6.8 (Panel C, lane b) and lapatinib treated cells migrating by a mean of 31.3% ± 2 (Panel C, lane d) compared to NS knockdown by a mean of 89% ± 7.8 (Panel C, lane c). Statistical analysis was performed as described in fig. 6. To measure knockdown efficiency of MTK1, cell extracts (150 μg) were resolved by SDS-PAGE, followed by MTK1 immunoblot analysis and densitometry measurements using BioRad Image Lab software to detect extent of MTK1 knockdown (Panel D, lane b), actin was used as a loading control (bottom panel).
4. Discussion
Prior studies have used ectopic expression of MEKK4/MTK1 in yeast [3], over-expression of wild-type or dominant-negative mutants in mammalian cells [2,7,11,13,69], or manipulation of MEKK4 genomic DNA [8,10,70] as a means to study MEKK4/MTK1 function. In this study we have focused on the scaffolding properties of endogenous MTK1. We identify a heregulin-dependent recruitment of tyrosine phosphorylated HER3 to MTK1. HER2 kinase activity is required for the formation of the HER3/MTK1 heterodimer, which is stable and involves only tyrosine phosphorylated HER3. These results have clinical implications in patients prescribed drugs like trastuzumab and lapatinib to inhibit HER2 activity, since these drugs will more than likely indirectly inhibit the activity of the phosphoHER3/MTK1 heterodimer. We demonstrate that extracellular acidification and cell migration require MTK1. Furthermore, lapatinib inhibited extracellular acidification and cell migration. Thus, both normal and malignant biological processes dependent on extracellular acidification and cell migration would likely be inhibited in patients treated with trastuzumab or lapatinib.
Although it is not clear how these proteins interact, it is clear that tyrosine phosphorylated HER3 preferentially associates with MTK1 as demonstrated when the hyper-phosphorylated, slower migrating form of HER3 co-immunoprecipitates with MTK1 (Fig. 1D, top panel, lane a). Consistent with the immunoblotting results, multiple peptides were identified by LC–MS/MS from heregulin-stimulated HER3, but no peptides were detected from the control sample, again demonstrating that only activated HER3 interacts with MTK1. These results strongly suggest that a specific or multiple phosphotyrosines mediate the interaction between phosphoHER3 and MTK1. MTK1 has no known SH2 domains so a direct interaction between MTK1 and phosphoHER3 seems unlikely. However, Shc, PI3K, and Grb7 have SH2 domains and are examples of proteins that interact with specific phosphotyrosines on HER3 [71] and these proteins could mediate the interaction between MTK1 and phosphoHER3.
The HER2/HER3 heterodimer has been reported as an oncogenic unit that drives breast tumor cell proliferation. Although HER2 is the only catalytically active kinase within the heterodimer, HER3 is required for breast tumor formation [40,72,73]. Thus even though HER3 lacks tyrosine kinase activity and depends on HER2 for phosphorylation, the proteins that “decorate” phosphoHER3 are also mediators of HER2/HER3 tumorigenicity. We show for the first time that HRG stimulation recruits MTK1 to HER3, which is sustained for up to 150 min indicating that sustained exposure to HRG results in continuous signaling through MTK1 and does not result in receptor down-regulation, consistent with previous findings demonstrating lack of receptor down-regulation with HER2 and HER3 [74]. The ability of HER2/HER3 to continue signaling through MTK1 in the presence of HRG may have clinical implications for patients with HRG-driven tumors as opposed to tumors with HER2 gene amplification. In tumors with over-expression of HRG, an autocrine or paracrine loop may play a role in cellular transformation and tumor progression that does not require HER2 over-expression. Identification of the HER3 regulated MTK1 pathway may provide a new molecular target for clinical intervention under conditions of normal levels of HER2/HER3 expression.
We also investigated whether MTK1 interacts with other RTKs. Although HRG also binds to HER4 [75], phosphoHER4 does not appear to interact with MTK1 (Fig. 1D). EGF is also an activator of HER2 [76], but stimulation of MCF-7 cells did not result in the recruitment of EGFR or HER2, or any other tyrosine phosphorylated proteins that would migrate at the position of 150 kDa or greater (Fig. 3, lane c). Insulin stimulation of MCF-7 cells led to the recruitment of a 150 kDa tyrosine phosphorylated protein (Fig. 3B, lane d). We identified this protein as the insulin receptor substrate-1. When the insulin receptor is activated, IRS-1 associates with the insulin receptor and is tyrosine phosphorylated. IRS-1 helps attenuate insulin signaling in healthy individuals [77]. However, IRS-1 is also associated with breast cancer and is characterized as a transforming oncogene [78]. The interaction of MTK1 with HER3 and IRS-1 strongly links MTK1 to RTK signaling. Thus, the interaction between MTK1 and IRS-1 may be a potential new avenue to explore in breast cancer therapy. Finally, stimulating MCF-7 cells with sorbitol did not result in the recruitment of phosphoHER3 with MTK1 indicating that the interaction between MTK1 and phosphoHER3 is not related to osmotic stress (Fig. 3B, lane e). In summary, the recruitment of MTK1 with phosphoHER3 appears to be unique to HRG binding to the HER2/HER3 heterodimer identifying an entirely new signaling pathway downstream of HER3.
The carboxyl-terminus of HER3 contains six YXXM motifs that when phosphorylated allow association of p85 of PI3K [41,79]. Mutation of the YXXM motifs from tyrosine to phenylalanine inhibits HRG-induced migration suggesting that recruitment of PI3K with HER3 is important in mediating cell migration [80]. Furthermore, inhibition of PI3K with PIK-75 inhibits cell motility and invasion [80]. We report that knockdown of MTK1 also blocks HRG-induced migration (Fig. 7). It is tempting to speculate that a single mole of phosphoHER3 and MTK1 functions as a signaling complex to recruit as many as six moles of PI3K, leading to amplification of the cell migration signal. Thus even though MTK1 knockdown was not highly efficient, loss of 60% of MTK1 expression was sufficient to prevent HRG-dependent cell migration. Perhaps indicating that a threshold of PI3K activity cannot be mobilized to lamellipodia or other cellular regions actively involved in cell migration. Similarly in the presence of insulin, PI3K is recruited to MTK1 (Fig. 3B, lane d), likely due to the presence of IRS-1 which is known to associate with PI3K [81]. The relationship between MTK1 and insulin in terms of cellular response remains to be explored, especially in regards to glucose homeostasis.
We also demonstrate that HER3/MTK1 association requires HER2 kinase activity, but HER2 does not remain in the complex. Lapatinib inhibits EGFR, HER2 and HER4 by binding to the ATP-binding pocket of these kinases, thereby inhibiting catalytic activity. Lapatinib shows a dose-dependent inhibition of EGFR with a Ki = 3 nM, HER2 with a Ki = 13 nM, and HER4 with a Ki = 347 nM [61]. In a dose response experiment, pre-treatment of MCF-7 cells with 50 nM lapatinib effectively inhibited HRG-induced HER3 phosphorylation and association with MTK1 (Fig. 4). Since HER4 has a Ki of 347 nM for lapatinib, EGFR and HER2 were the more likely candidates for lapatinib blockade of HER3 phosphorylation and subsequent MTK1/HER3 association. EGFR is expressed in MCF-7 and T-47D cells and can form heterodimers with HER3 [23,82]. However, we have performed experiments comparing EGF and HRG and found that the EGFR is not activated in response to HRG (data not shown). Together, our results suggest that lapatinib inhibited HER2 and effectively inhibited HER3 phosphorylation and formation of the MTK1/HER3 heterodimer. Additionally in HER2 negative and HER3 positive MDA-MB-231 cells, HRG does not induce formation of the MTK1/HER3 dimer (Fig. 4C). Therefore, these data support that HER2 protein expression and kinase activity is required for HER3 phosphorylation and MTK1/HER3 association.
Cell migration requires actin polymerization and intracellular coordination of actin binding proteins, which are regulated by signal transduction and subsequent second messenger signaling [42]. Migration is also an important part of cancer cell invasion and metastasis [83]. HER2 is linked to actin cytoskeletal reorganization in cancer [42] and heregulin stimulation of breast cancer cells enhances the conversion of g-actin to f-actin increasing cell migration [43,44], implicating HER3 with actin cytoskeletal reorganization. Additionally, our studies reveal that MEKK4 interacts with actin through a 40 amino acid region referred to as the actin interacting region (AIR), which is also found in yeast Ssk2p [45]. Deletion of the AIR within MEKK4 impairs the constitutive association between MEKK4 and actin (Fig. 5B). These results demonstrate a proximal localization of MEKK4 with the actin cytoskeleton. When MCF-7 cells were pre-treated with cytochalasin D to prevent actin polymerization, the interaction between MTK1 and HER3 was reduced by ~60%, indicating a cytoskeletal connection involving actin linking MTK1 and HER3. Future studies will be needed to determine whether the MTK1/HER3 dimer remains at the plasma membrane or mobilizes via actin filaments to other intracellular locations.
Although others have reported that HRG induces a proliferative response [68], we were unable to detect HRG-dependent proliferation in either T-47D (data not shown) or MCF-7 cells (Fig. 6A), although we detected an EGF-dependent proliferative response. As the cells were incubated with either HRG or EGF, we noticed that the cell media developed a more orange color in the presence of HRG. When the pH was recorded, we consistently observed that HRG caused an acidification of the extracellular medium while EGF had no effect. Given that HRG caused an association between MTK1 and HER3, we used siRNA to decrease the expression of MTK1 to determine whether MTK1 was involved in regulating extracellular pH. Although siRNA typically diminished MTK1 expression by only ~60% in MCF-7 cells (see Fig. 7D for a representative immunoblot), the change in extracellular pH was always diminished in the absence of MTK1 (Fig. 6B, lanes a & c). These results demonstrate that EGF and HRG induce different responses in MCF-7 cells and that the EGF-dependent proliferative response does not correlate with acidification of the extracellular medium. Since HRG did not induce proliferation of MCF-7 cells, we investigated whether HRG induced cell migration by using a scratch assay. After 24 h of HRG exposure in the presence of NS siRNA, 60% of the scratch area was filled in by cells migrating back into the scratched region (Fig. 7A). In contrast, only 20% of the scratch area was filled in with cells migrating back into the scratch region after 24 h in cells with MTK1 knockdown. Additionally, cells pre-treated with lapatinib migrated back into the scratch area by only 27%. These results demonstrate that HRG induces migration and not proliferation of MCF-7 cells, consistent with the results of Arsani et al. [63]. We also demonstrate that MTK1 is required to acidify the extracellular environment. The acidification of the extracellular environment likely helps optimize the enzymatic activity of matrix metalloproteinases or other enzymes needed for the migratory response.
MTK1 is not tyrosine phosphorylated in response to HRG stimulation of T-47D cells (Fig. 2A, lane a) or MCF-7 cells (Fig. 2A, lane g). However, the association between MTK1 and HER3 is dependent on HRG. Since MTK1 is not tyrosine phosphorylated, it is unlikely that MTK1 is a substrate for HER2. Conversely, we have not investigated whether the catalytic activity of MTK1 is required for the association between MTK1 and HER3. Our data demonstrate phosphorylation of serine 686 of HER3 (LERGEpSIEP) in response to HRG by sequencing using LC–MS/MS (Fig. 2B). Since we were unable to sequence any HER3 peptides due to lack of HER3 association with MTK1 in the absence of HRG stimulation, we cannot state whether serine 686 is phosphorylated under basal conditions. Phosphorylation of HER3 at serine 686 has been previously reported, but the functional significance of this modification is not known [84]. Earlier studies have shown MEKK4 functions as a serine/threonine kinase regulating MKK6, which is upstream of p38 [2,3,11,13]. Our current results suggest that MTK1 functions as a scaffolding protein for phosphoHER3. However, since MTK1 is a serine/threonine kinase, it is possible that the catalytic activity of MTK1 is required to regulate changes in extracellular pH and cell migration.
In summary, HER2/HER3 heterodimerization has been well characterized, but we show for the first time that HRG stimulation results in the formation of a MTK1/HER3 heterodimer, which is sustained for up to 150 min in breast cancer cells. We also demonstrate that HER2 kinase activity is needed for association of MTK1 with phosphoHER3. Furthermore, disruption of actin polymerization inhibits HRG induced MTK1/HER3 association, which suggests that f-actin is required to bring MTK1 and phosphoHER3 together linking MTK1 to the actin cytoskeleton. Additionally, we report that MTK1 is required for HRG induced extracellular acidification and cell migration. Lapatinib also inhibited HRG induced extracellular acidification and cell migration, showing that HER2 is required in this signaling process. In conclusion, over-expression of HER2 in breast cancer has been targeted with drugs like trastuzumab and lapatinib, however drug resistance or unresponsiveness persists among many patients [85,86]. The link between HER3 up-regulation to HER2 drug resistance makes HER3 a desirable molecular target for anti-cancer drug development. However, the lack of HER3 catalytic activity has made HER3 somewhat less tractable as a drug target. By identifying the recruitment of MTK1 to phosphoHER3, we now shed new light onto the HER3 branch of the HER2/HER3 signaling tree.
Supplementary Material
Acknowledgments
We would like to express our thanks to Dr. George Tsaprailis, Dr. Linda Breci and Andrea Hunt of the Southwest Environmental Health Sciences Center (University of Arizona) for the help with LC/MS. This work was supported by NIEHS Training grant ES007091, ES006694, ES012007 and ES04940.
Abbreviations
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
- MAP3K4
mitogen-activated protein kinase kinase kinase 4
- MEKK4
mitogen-activated, extracellular signal-regulated kinase kinase
- MTK1
mitogen activated protein three kinase
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- HER
human epidermal growth factor receptor
- Ssk2
suppressor of sensor kinase
- HRG
heregulin
- LC–MS/MS
liquid chromatography and tandem mass spectrometry
- f-actin
filamentous actin
- g-actin
globular actin
- cyto D
cytochalasin D
- AIR
actin interacting region
- pTyr
phosphotyrosines
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.cellsig.2013.08.043.
References
- 1.Lange-Carter CA, Pleiman CM, Gardner AM, Blumer KJ, Johnson GL. Science. 1993;260:315–319. doi: 10.1126/science.8385802. [DOI] [PubMed] [Google Scholar]
- 2.Gerwins P, Blank JL, Johnson GL. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- 3.Takekawa M, Posas F, Saito H. EMBO J. 1997;16:4973–4982. doi: 10.1093/emboj/16.16.4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda T, Takekawa M, Saito H. Science. 1995;269:554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- 5.Bettinger BT, Amberg DC. J Cell Biochem. 2007;101:34–43. doi: 10.1002/jcb.21289. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman JI, Kaplan S. J Am Coll Cardiol. 2002;39:1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 7.Stevens MV, Parker P, Vaillancourt RR, Camenisch TD. Dev Dyn. 2006;235:2761–2770. doi: 10.1002/dvdy.20922. [DOI] [PubMed] [Google Scholar]
- 8.Abell AN, Rivera-Perez JA, Cuevas BD, Uhlik MT, Sather S, Johnson NL, Minton SK, Lauder JM, Winter-Vann AM, Nakamura K, Magnuson T, Vaillancourt RR, Heasley LE, Johnson GL. Mol Cell Biol. 2005;25:8948–8959. doi: 10.1128/MCB.25.20.8948-8959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chi H, Sarkisian MR, Rakic P, Flavell RA. Proc Natl Acad Sci U S A. 2005;102:3846–3851. doi: 10.1073/pnas.0500026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkisian MR, Bartley CM, Chi H, Nakamura F, Hashimoto-Torii K, Torii M, Flavell RA, Rakic P. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takekawa M, Saito H. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 12.Miyake Z, Takekawa M, Ge Q, Saito H. Mol Cell Biol. 2007;27:2765–2776. doi: 10.1128/MCB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mita H, Tsutsui J, Takekawa M, Witten EA, Saito H. Mol Cell Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abell AN, Granger DA, Johnson GL. J Biol Chem. 2007;282:30476–30484. doi: 10.1074/jbc.M705783200. [DOI] [PubMed] [Google Scholar]
- 15.Kraus MH, Issing W, Miki T, Popescu NC, Aaronson SA. Proc Natl Acad Sci U S A. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemmon MA. Breast Dis. 2003;18:33–43. doi: 10.3233/bd-2003-18105. [DOI] [PubMed] [Google Scholar]
- 17.Muller WJ, Sinn E, Pattengale PK, Wallace R, Leder P. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- 18.Yarden Y, Sliwkowski MX. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 19.Fiddes RJ, Janes PW, Sivertsen SP, Sutherland RL, Musgrove EA, Daly RJ. Oncogene. 1998;16:2803–2813. doi: 10.1038/sj.onc.1201815. [DOI] [PubMed] [Google Scholar]
- 20.Krueger JS, Keshamouni VG, Atanaskova N, Reddy KB. Oncogene. 2001;20:4209–4218. doi: 10.1038/sj.onc.1204541. [DOI] [PubMed] [Google Scholar]
- 21.Wu WS, Wu JR, Hu CT. Cancer Metastasis Rev. 2008;27:303–314. doi: 10.1007/s10555-008-9112-4. [DOI] [PubMed] [Google Scholar]
- 22.Darcy KM, Zangani D, Wohlhueter AL, Huang RY, Vaughan MM, Russell JA, Ip MM. J Histochem Cytochem. 2000;48:63–80. doi: 10.1177/002215540004800107. [DOI] [PubMed] [Google Scholar]
- 23.Eccles SA. Int J Dev Biol. 2011;55:685–696. doi: 10.1387/ijdb.113396se. [DOI] [PubMed] [Google Scholar]
- 24.Menard S, Tagliabue E, Campiglio M, Pupa SM. J Cell Physiol. 2000;182:150–162. doi: 10.1002/(SICI)1097-4652(200002)182:2<150::AID-JCP3>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs IB, Siemer I, Buhler H, Schmider A, Henrich W, Lichtenegger W, Schaller G, Kuemmel S. Anticancer Res. 2006;26:4397–4401. [PubMed] [Google Scholar]
- 26.Carraway KL, III, Sliwkowski MX, Akita R, Platko JV, Guy PM, Nuijens A, Diamonti AJ, Vandlen RL, Cantley LC, Cerione RA. J Biol Chem. 1994;269:14303–14306. [PubMed] [Google Scholar]
- 27.Lyne JC, Melhem MF, Finley GG, Wen D, Liu N, Deng DH, Salup R. Cancer J Sci Am. 1997;3:21–30. [PubMed] [Google Scholar]
- 28.Stern DF. J Mammary Gland Biol Neoplasia. 2008;13:215–223. doi: 10.1007/s10911-008-9083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alroy I, Yarden Y. FEBS Lett. 1997;410:83–86. doi: 10.1016/s0014-5793(97)00412-2. [DOI] [PubMed] [Google Scholar]
- 30.Amin DN, Sergina N, Lim L, Goga A, Moasser MM. Biochem J. 2012;447:417–425. doi: 10.1042/BJ20120724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Way TD, Lin JK. Future Oncol. 2005;1:841–849. doi: 10.2217/14796694.1.6.841. [DOI] [PubMed] [Google Scholar]
- 32.Kurokawa H, Arteaga CL. Clin Cancer Res. 2003;9:511S–515S. [PubMed] [Google Scholar]
- 33.Kurokawa H, Lenferink AE, Simpson JF, Pisacane PI, Sliwkowski MX, Forbes JT, Arteaga CL. Cancer Res. 2000;60:5887–5894. [PubMed] [Google Scholar]
- 34.Shou J, Massarweh S, Osborne CK, Wakeling AE, Ali S, Weiss H, Schiff R. J Natl Cancer Inst. 2004;96:926–935. doi: 10.1093/jnci/djh166. [DOI] [PubMed] [Google Scholar]
- 35.Bedard PL, de Azambuja E, Cardoso F. Curr Cancer Drug Targets. 2009;9:148–162. doi: 10.2174/156800909787581024. [DOI] [PubMed] [Google Scholar]
- 36.Engel RH, Kaklamani VG. Drugs. 2007;67:1329–1341. doi: 10.2165/00003495-200767090-00006. [DOI] [PubMed] [Google Scholar]
- 37.Esteva FJ, Pusztai L. Oncology. 2005;19:5–16. [PubMed] [Google Scholar]
- 38.Scaltriti M, Verma C, Guzman M, Jimenez J, Parra JL, Pedersen K, Smith DJ, Landolfi S, Ramon y Cajal S, Arribas J, Baselga J. Oncogene. 2009;28:803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 39.Stern HM. Sci Transl Med. 2012;4:127rv122. doi: 10.1126/scitranslmed.3001539. [DOI] [PubMed] [Google Scholar]
- 40.Garrett JT, Olivares MG, Rinehart C, Granja-Ingram ND, Sanchez V, Chakrabarty A, Dave B, Cook RS, Pao W, McKinely E, Manning HC, Chang J, Arteaga CL. Proc Natl Acad Sci U S A. 2011;108:5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hellyer NJ, Cheng K, Koland JG. Biochem J. 1998;333(Pt 3):757–763. doi: 10.1042/bj3330757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldner JC, Brandt BH. Exp Cell Res. 2002;272:93–108. doi: 10.1006/excr.2001.5385. [DOI] [PubMed] [Google Scholar]
- 43.Adam L, Vadlamudi R, Kondapaka SB, Chernoff J, Mendelsohn J, Kumar R. J Biol Chem. 1998;273:28238–28246. doi: 10.1074/jbc.273.43.28238. [DOI] [PubMed] [Google Scholar]
- 44.Hijazi MM, Thompson EW, Tang C, Coopman P, Torri JA, Yang D, Mueller SC, Lupu R. Int J Oncol. 2000;17:629–641. doi: 10.3892/ijo.17.4.629. [DOI] [PubMed] [Google Scholar]
- 45.Yuzyuk T, Foehr M, Amberg DC. Mol Biol Cell. 2002;13:2869–2880. doi: 10.1091/mbc.02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock C, Gassner B, Hauck CR, Arnold H, Mally S, Eble JA, Dieterich P, Schwab A. J Physiol. 2005;567:225–238. doi: 10.1113/jphysiol.2005.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan SD, Antoniucci DM, Fok KS, Alajoki ML, Harkins RN, Thompson SA, Wada HG. J Biol Chem. 1995;270:22608–22613. doi: 10.1074/jbc.270.38.22608. [DOI] [PubMed] [Google Scholar]
- 48.Kim JM, Min SK, Kim H, Kang HK, Jung SY, Lee SH, Choi Y, Roh S, Jeong D, Min BM. Int J Mol Med. 2007;19:393–400. [PubMed] [Google Scholar]
- 49.Sennoune SR, Luo D, Martinez-Zaguilan R. Cell Biochem Biophys. 2004;40:185–206. doi: 10.1385/CBB:40:2:185. [DOI] [PubMed] [Google Scholar]
- 50.Rofstad EK, Mathiesen B, Kindem K, Galappathi K. Cancer Res. 2006;66:6699–6707. doi: 10.1158/0008-5472.CAN-06-0983. [DOI] [PubMed] [Google Scholar]
- 51.Halfter UM, Derbyshire ZE, Vaillancourt RR. Biochem J. 2005;388:17–28. doi: 10.1042/BJ20041236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Derbyshire ZE, Halfter UM, Heimark RL, Sy TH, Vaillancourt RR. Mol Cell Biochem. 2005;271:77–90. doi: 10.1007/s11010-005-5386-9. [DOI] [PubMed] [Google Scholar]
- 53.Stock C, Schwab A. Pflugers Arch. 2009;458:981–992. doi: 10.1007/s00424-009-0677-8. [DOI] [PubMed] [Google Scholar]
- 54.Beerli RR, Hynes NE. J Biol Chem. 1996;271:6071–6076. doi: 10.1074/jbc.271.11.6071. [DOI] [PubMed] [Google Scholar]
- 55.Aguilar Z, Akita RW, Finn RS, Ramos BL, Pegram MD, Kabbinavar FF, Pietras RJ, Pisacane P, Sliwkowski MX, Slamon DJ. Oncogene. 1999;18:6050–6062. doi: 10.1038/sj.onc.1202993. [DOI] [PubMed] [Google Scholar]
- 56.Carraway KL, III, Cantley LC. Cell. 1994;78:5–8. doi: 10.1016/0092-8674(94)90564-9. [DOI] [PubMed] [Google Scholar]
- 57.Neve RM, Holbro T, Hynes NE. Oncogene. 2002;21:4567–4576. doi: 10.1038/sj.onc.1205555. [DOI] [PubMed] [Google Scholar]
- 58.Aissouni Y, Zapart G, Iovanna JL, Dikic I, Soubeyran P. Biochem Biophys Res Commun. 2005;338:808–814. doi: 10.1016/j.bbrc.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 59.Britsch S. Adv Anat Embryol Cell Biol. 2007;190:1–65. [PubMed] [Google Scholar]
- 60.Plowman GD, Green JM, Culouscou JM, Carlton GW, Rothwell VM, Buckley S. Nature. 1993;366:473–475. doi: 10.1038/366473a0. [DOI] [PubMed] [Google Scholar]
- 61.Wood ER, Truesdale AT, McDonald OB, Yuan D, Hassell A, Dickerson SH, Ellis B, Pennisi C, Horne E, Lackey K, Alligood KJ, Rusnak DW, Gilmer TM, Shewchuk L. Cancer Res. 2004;64:6652–6659. doi: 10.1158/0008-5472.CAN-04-1168. [DOI] [PubMed] [Google Scholar]
- 62.Teixido C, Arguelaguet E, Pons B, Aracil M, Jimeno J, Somoza R, Mares R, Ramon YCS, Hernandez-Losa J. Int J Oncol. 2012;41:317–324. doi: 10.3892/ijo.2012.1425. [DOI] [PubMed] [Google Scholar]
- 63.Asrani K, Keri RA, Galisteo R, Brown SA, Morgan SJ, Ghosh A, Tran NL, Winkles JA. Mol Cancer Res. 2013;11:393–404. doi: 10.1158/1541-7786.MCR-12-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atlas E, Cardillo M, Mehmi I, Zahedkargaran H, Tang C, Lupu R. Mol Cancer Res. 2003;1:165–175. [PubMed] [Google Scholar]
- 65.Fiddes RJ, Janes PW, Sanderson GM, Sivertsen SP, Sutherland RL, Daly RJ. Cell Growth Differ. 1995;6:1567–1577. [PubMed] [Google Scholar]
- 66.Hernandez L, Smirnova T, Kedrin D, Wyckoff J, Zhu L, Stanley ER, Cox D, Muller WJ, Pollard JW, Van Rooijen N, Segall JE. Cancer Res. 2009;69:3221–3227. doi: 10.1158/0008-5472.CAN-08-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xue C, Liang F, Mahmood R, Vuolo M, Wyckoff J, Qian H, Tsai KL, Kim M, Locker J, Zhang ZY, Segall JE. Cancer Res. 2006;66:1418–1426. doi: 10.1158/0008-5472.CAN-05-0550. [DOI] [PubMed] [Google Scholar]
- 68.Yang C, Klein EA, Assoian RK, Kazanietz MG. Biochem J. 2008;410:167–175. doi: 10.1042/BJ20070781. [DOI] [PubMed] [Google Scholar]
- 69.Kanungo J, Potapova I, Malbon CC, Wang H. J Biol Chem. 2000;275:24032–24039. doi: 10.1074/jbc.M002747200. [DOI] [PubMed] [Google Scholar]
- 70.Sun BK, Kim JH, Nguyen HN, Oh S, Kim SY, Choi S, Choi HJ, Lee YJ, Song JJ. Oncol Rep. 2011;25:537–544. doi: 10.3892/or.2010.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones RB, Gordus A, Krall JA, MacBeath G. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 72.Holbro T, Beerli RR, Maurer F, Koziczak M, Barbas CF, III, Hynes NE. Proc Natl Acad Sci U S A. 2003;100:8933–8938. doi: 10.1073/pnas.1537685100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaught DB, Stanford JC, Young C, Hicks DJ, Wheeler F, Rinehart C, Sanchez V, Koland J, Muller WJ, Arteaga CL, Cook RS. Cancer Res. 2012;72:2672–2682. doi: 10.1158/0008-5472.CAN-11-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baulida J, Kraus MH, Alimandi M, Di Fiore PP, Carpenter G. J Biol Chem. 1996;271:5251–5257. doi: 10.1074/jbc.271.9.5251. [DOI] [PubMed] [Google Scholar]
- 75.Hynes NE, Lane HA. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 76.Khazaie K, Schirrmacher V, Lichtner RB. Cancer Metastasis Rev. 1993;12:255–274. doi: 10.1007/BF00665957. [DOI] [PubMed] [Google Scholar]
- 77.Schmitz-Peiffer C, Whitehead JP. IUBMB Life. 2003;55:367–374. doi: 10.1080/1521654031000138569. [DOI] [PubMed] [Google Scholar]
- 78.Chan BT, Lee AV. J Mammary Gland Biol Neoplasia. 2008;13:415–422. doi: 10.1007/s10911-008-9101-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Soltoff SP, Carraway KL, III, Prigent SA, Gullick WG, Cantley LC. Mol Cell Biol. 1994;14:3550–3558. doi: 10.1128/mcb.14.6.3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smirnova T, Zhou ZN, Flinn RJ, Wyckoff J, Boimel PJ, Pozzuto M, Coniglio SJ, Backer JM, Bresnick AR, Condeelis JS, Hynes NE, Segall JE. Oncogene. 2012;31:706–715. doi: 10.1038/onc.2011.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kooijman R, Lauf JJ, Kappers AC, Rijkers GT. J Exp Med. 1995;182:593–597. doi: 10.1084/jem.182.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yarden RI, Lauber AH, El-Ashry D, Chrysogelos SA. Endocrinology. 1996;137:2739–2747. doi: 10.1210/endo.137.7.8770893. [DOI] [PubMed] [Google Scholar]
- 83.Yamazaki D, Kurisu S, Takenawa T. Cancer Sci. 2005;96:379–386. doi: 10.1111/j.1349-7006.2005.00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 85.Garrett JT, Arteaga CL. Cancer Biol Ther. 2011;11:793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nahta R, Esteva FJ. Breast Cancer Res. 2006;8:215. doi: 10.1186/bcr1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.