Abstract
Brugada syndrome is a rare cardiac arrhythmia disorder, causally related to SCN5A mutations in around 20% of cases1–3. Through a genome-wide association study of 312 individuals with Brugada syndrome and 1,115 controls, we detected 2 significant association signals at the SCN10A locus (rs10428132) and near the HEY2 gene (rs9388451). Independent replication confirmed both signals (meta-analyses: rs10428132, P = 1.0 × 10−68; rs9388451, P = 5.1 × 10−17) and identified one additional signal in SCN5A (at 3p21; rs11708996, P = 1.0 × 10−14). The cumulative effect of the three loci on disease susceptibility was unexpectedly large (Ptrend = 6.1 × 10−81). The association signals at SCN5A-SCN10A demonstrate that genetic polymorphisms modulating cardiac conduction4–7 can also influence susceptibility to cardiac arrhythmia. The implication of association with HEY2, supported by new evidence that Hey2 regulates cardiac electrical activity, shows that Brugada syndrome may originate from altered transcriptional programming during cardiac development8. Altogether, our findings indicate that common genetic variation can have a strong impact on the predisposition to rare diseases.
Sudden cardiac death (SCD) is a leading cause of mortality in Western countries, with an incidence close to 1 per 1,000 individuals per year9. SCD results most frequently from ventricular fibrillation in the setting of coronary artery disease10. In 5–10% of cases, however, SCD occurs owing to rare inherited cardiac arrhythmias, which are typically associated with a distinctive electrocardiogram (ECG) pattern in the absence of identifiable structural heart disease10. One such disorder is Brugada syndrome, characterized by ST-segment elevation in right precordial ECG leads2. ST-segment elevation may be transient in nature and can be evoked by pharmacological sodium channel blockade. Loss-of-function mutations in SCN5A, encoding the pore-forming subunit of the cardiac sodium channel (Nav1.5) at 3p21, have been causally related to the disease in ~20% of cases3,11. However, whether arrhythmias arise as a result of abnormal conduction, repolarization or both is under debate12. Mutations in genes other than SCN5A have been found in a small subset of cases, but their involvement in Brugada syndrome remains unclear13.
Although Brugada syndrome is commonly considered a mendelian disorder with autosomal dominant transmission, studies in families harboring SCN5A mutations have demonstrated low disease penetrance14,15 and, in some instances, absence of the familial SCN5A mutation in some affected family members15. Also, many cases are sporadic16,17, and familial linkage analyses have largely been unsuccessful in uncovering new disease-causing genes. These observations suggest a more complex inheritance model. Identifying new genetic risk factors could assist in further diagnosis, provide new insights into underlying molecular mechanisms and yield new information relevant to the broader problem of SCD.
We conducted a genome-wide association study (GWAS) to explore the role of common genetic variants in susceptibility to Brugada syndrome. We established an international consortium enabling the recruitment of 1,114 unrelated, clinically well-defined cases from 13 centers in Europe, the United States and Japan (Supplementary Table 1). Every case had a Brugada syndrome type I ECG, as defined by consensus criteria, either at baseline or after drug challenge2, and SCN5A mutation status was systematically assessed.
We first conducted a GWAS on 383 cases of self-reported European ancestry and 898 control individuals from western France (cohort Données Epidémiologiques sur le Syndrome d’Insulino-Résistance (D.E.S.I.R.)18). Cases and controls were genotyped using Axiom Genome-Wide CEU 1 arrays (Affymetrix). After stringent quality control and the exclusion of rare SNPs, a total of 360,149 markers were available for further analysis (Online Methods). Multidimensional scaling on the combined cases and controls together with reference populations from the 1000 Genomes Project excluded 42 samples of non-European descent (Supplementary Fig. 1). Because controls were largely French individuals, whereas cases had a broader geographic origin, we supplemented the control set with individuals from four European populations from the 1000 Genomes Project that best matched the subsets of cases (Supplementary Fig. 2). After the exclusion of 29 cases for whom no matching controls were available, 2 homogeneous groups were defined. GWAS analysis was conducted separately on each group, and association data were combined in a meta-analysis, which included in total 312 cases and 1,115 controls (Online Methods).
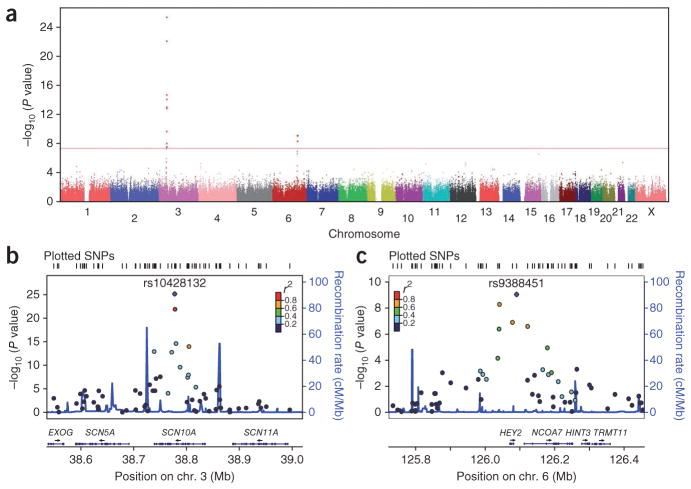
We found an excess of SNPs to be associated with Brugada syndrome compared to the number expected under the null hypothesis of no association (Supplementary Fig. 3). Two genomic regions showed association signals reaching genome-wide significance (Fig. 1a). The most significant association was obtained for rs10428132, a SNP located in the SCN10A gene at 3p22 (P = 6.79 × 10−26; Table 1). Nine other markers in linkage disequilibrium (LD) with rs10428132 (r2 = 0.20–0.76) also had associations that reached genome-wide significance (Fig. 1b and Supplementary Table 2). We detected another cluster of two SNPs in high LD (r2 = 0.81) at 6q22. The lead SNP at this locus was rs9388451 (P = 8.85 × 10−10), located downstream of the HEY2 gene (Fig. 1c and Supplementary Table 2). Neither conditional analysis at each associated locus nor GWAS following genome-wide imputation of non-genotyped SNPs uncovered any additional independent association signals at genome-wide significance (Online Methods and Supplementary Fig. 4). We confirmed both associations with similar effect sizes when the GWAS was restricted to 856 D.E.S.I.R. controls and 254 ancestry-matched cases by applying stringent exclusion criteria on the multidimensional scaling results (Supplementary Fig. 5).
Figure 1.
Genome-wide association analysis identifies two susceptibility loci for Brugada syndrome. (a) Manhattan plot showing the association of SNPs with Brugada syndrome in a GWAS of 312 cases and 1,115 controls. The red horizontal line marks the threshold for genome-wide significance (P = 5 × 10−8). Two loci reached genome-wide significance on chromosomes 3 and 6. (b) Association plots for 3q22 (left) and 6q22 (right). Each SNP is plotted with respect to its chromosomal location (x axis) and its association P value (left y axis). SNPs are colored according to their degree of LD (r2) with the leading variant represented by a purple diamond and labeled. The tall blue spikes indicate the recombination rate (right y axis) in that region of the chromosome. Coordinates are given according to NCBI Build 37.
Table 1.
Discovery and replication studies confirm three alleles on chromosomes 3 and 6 are associated with Brugada syndrome
| Marker | rs11708996 | rs10428132 | rs9388451 | |
|---|---|---|---|---|
| Genome position (Build 37) | Chr. 3: 38633923 | Chr. 3: 38777554 | Chr. 6: 126090377 | |
| Closest gene(s) | SCN5A | SCN10A | HEY2, NCOA7 | |
| Risk allele | C | T | C | |
| Protective allele | G | G | T | |
| GWAS (312 cases and 1,115 controls; European)a | RAFb | 0.23/0.15 | 0.69/0.41 | 0.65/0.50 |
| P | 2.70 × 10−5 | 6.79 × 10−26 | 8.85 × 10−10 | |
| OR (CI) | 1.64 (1.30–2.07) | 3.00 (2.45–3.69) | 1.83 (1.51–2.22) | |
| Replication 1 (594 cases and 806 controls; European)a | RAFb | 0.23/0.15 | 0.65/0.42 | 0.59/0.50 |
| P | 1.10 × 10−7 | 1.66 × 10−30 | 2.10 × 10−5 | |
| OR (CI) | 1.69 (1.39–2.04) | 2.35 (2.03–2.72) | 1.39 (1.19–1.61) | |
| Replication 2 (208 cases and 1,016 controls; Japanese)a | RAFb | 0.09/0.04 | 0.44/0.23 | 0.72/0.61 |
| P | 5.63 × 10−5 | 1.56 × 10−16 | 6.70 × 10−6 | |
| OR (CI) | 2.30 (1.53–3.45) | 2.56 (2.05–3.19) | 1.74 (1.37–2.21) | |
| Meta-analysis | P | 1.02 × 10−14 | 1.01 × 10−68 | 5.14 × 10−17 |
| OR (CI) | 1.73 (1.51–1.99) | 2.55 (2.30–2.84) | 1.58 (1.42–1.75) | |
| Heterogeneity | Q = 2.17 | Q = 3.68 | Q = 5.68 | |
| P = 0.338 | P = 0.159 | P = 0.058 | ||
| I2 = 4% | I2 = 45% | I2 = 65% | ||
| Cases with symptomsc (416 cases and 2,937 controls; meta-analysis)a | P | 6.88 × 10−8 | 1.15 × 10−39 | 5.01 × 10−8 |
| OR (CI) | 1.73 (1.42–2.12) | 2.84 (2.43–3.32) | 1.55 (1.32–1.81) |
Size of case and control sets; sample ancestry.
Risk allele frequency (RAF) in cases over controls.
Symptoms included ventricular tachycardia, ventricular fibrillation, syncope and near syncope.
We next considered candidate SNPs identified in previous GWAS on ECG traits4–7,19–23, using both genotype and imputation data (Supplementary Table 3). After the removal of redundant SNPs from the same haplotype (r2 > 0.2) and the exclusion of the haplotype containing rs10428132, we observed a significant enrichment in association for the remaining 54 markers (P = 5.0 × 10−8; Supplementary Fig. 6). The SNP with the lowest association P value, rs11708996 (Supplementary Table 3), was located in SCN5A and has previously been associated with variability in PR interval and QRS duration6,7, two ECG parameters that reflect cardiac conduction, a process possibly affected in Brugada syndrome12. Although residing next to the strongest association signal at the 3p22 locus, rs11708996 showed no LD with rs10428132 (r2 = 0.06), and its association P value in GWAS data remained unchanged in analysis conditioning on rs10428132, thus suggesting that it represents an independent association (data not shown). Considering the established involvement of SCN5A in Brugada syndrome, we carried this SNP forward to the validation step of the study. We also considered SNPs at loci harboring the 11 other susceptibility genes13 but found no enrichment in association at statistically significant levels (genomic inflation factor (λGC) = 0.98; Online Methods and Supplementary Fig. 7).
We sought to replicate the two genome-wide significant signals (rs10428132 and rs9388451) as well as rs11708996 in an independent case-control set of 594 individuals of European descent with Brugada syndrome and 806 previously genotyped D.E.S.I.R. individuals24 (Online Methods). All three SNPs showed robust association, with a similar direction of effect as seen in the discovery set, and the signal at rs10428132 reached genome-wide statistical significance in the replication set alone (Table 1). To seek additional replication and assess associations in other ancestry groups, we tested a third independent case-control set comprising 208 Japanese cases and 1,016 ancestry-matched controls. We again replicated the three associations, with rs10428132 exceeding the genome-wide significance threshold (Table 1). Meta-analysis of the discovery and replication sets yielded highly significant association P values for all three loci (rs10428132, P = 1.01 × 10−68; rs11708996, P = 1.02 × 10−14; rs9388451, P = 5.14 × 10−17), with the corresponding effect sizes ranging from 1.58 to 2.55 (Table 1). Effect sizes were similar when the meta-analysis was restricted to symptomatic individuals, with the association for rs10428132 reaching genome-wide statistical significance (Table 1).
We next assessed the cumulative effect of the three loci on susceptibility to Brugada syndrome (Fig. 2a and Supplementary Table 4). We found that disease risk increased consistently with increasing numbers of carried risk alleles (Ptrend = 6.1 × 10−81), with the estimated odds ratio (OR) reaching 21.5 in the presence of more than four risk alleles versus less than 2 (Fig. 2b). Under a multiplicative model, assuming a prevalence of 0.05% for Brugada syndrome2 and using the ORs and allele frequencies reported in Table 1, the sibling relative risk attributable to the three lead SNPs was estimated at λS = 1.4, with the three loci accounting for 7% of the variance in disease susceptibility (Online Methods). The proportion of the phenotypic variance explained by only these three loci, as well as their cumulative effect, are unexpectedly large in comparison to previously reported effects for common genetic variation on susceptibility to common disease25. However, as 1.5% of the European population is expected to carry more than four risk alleles (Supplementary Table 4), these three polymorphisms are unlikely to by themselves explain the occurrence of Brugada syndrome and are only associated with a low absolute risk for this rare condition. Furthermore, the ORs reported here were calculated using data from case-control collections and thus may overestimate relative risks.
Figure 2.
Cumulative effect of alleles at the three associated loci on susceptibility to Brugada syndrome. (a) Distribution of risk alleles among individuals with Brugada syndrome (white bars) and among control individuals (black bars) from Europe (top) and Japan (bottom). (b) ORs calculated according to the number of risk alleles carried. A meta-analysis was performed as described in the Online Methods, using individuals carrying no or one risk allele as the reference. Each black bar represents the log(OR) value (horizontal bar) and the 95% confidence interval (on the log scale; vertical bar).
In subsequent analyses comparing SNP allele dosages in subsets of cases, we could not detect any consistent association of risk alleles with symptoms, SCN5A mutation status or Brugada syndrome type I ECG at baseline compared to after drug challenge (Supplementary Table 5).
Our strongest association (rs10428132) resides in intron 14 of the SCN10A gene, located adjacent to SCN5A on chromosome 3p21–22. The particular haplotype tagged by this SNP, which also contains a nonsynonymous variant affecting SCN10A (rs6795970, r2 = 0.97 with rs10428132), has previously been associated with variability in PR interval and QRS duration in the general population4–7. SCN10A, which encodes the sodium channel isoform Nav1.8, was originally reported to be highly expressed in nociceptive sensory neurons of the dorsal root ganglia and cranial sensory ganglia26. Besides being expressed in cardiac neurons27, SCN10A was also shown in recent studies to be expressed in the working myocardium4,28 and the specialized conduction system7,29, indicating a possible role for Nav1.8 in cardiac electrical function. An independent report implicated the rs6801957 SNP (r2 = 0.97 with rs10428132) as a probable causal variant on the associated haplotype30. This SNP, which alters a highly conserved nucleotide within a consensus T-box–binding site, affects TBX5- or TBX3-mediated enhancer activity30 and is thus expected to affect TBX5/TBX3-regulated expression of SCN5A and SCN10A in vivo31. Further studies are required to determine whether the effects of this SNP on conduction and Brugada syndrome are mediated through regulation of SCN5A, SCN10A or both. The other independent association at 3p21–22 (rs11708996) involved another haplotype previously associated with cardiac conduction6,7. For both haplotypes, the PR- and QRS-prolonging allele was associated with disease risk, providing support for the concept that the cardiomyocyte depolarization process has an important role in the pathogenesis of Brugada syndrome12.
In contrast to both signals at the SCN5A-SCN10A locus, to our knowledge, the signal at 6q22 has never been detected by previous GWAS on ECG traits. The corresponding SNP tagged an LD block that entirely encompasses the HEY2 gene and extends into the first intron of NCOA7 (also called ERAP140), which encodes a nuclear receptor coactivator that interacts with estrogen receptor α to modulate its activity32. NCOA7 is predominantly expressed in the brain32 and has not been implicated in cardiovascular function. In contrast, HEY2 (also called HESR2, HRT2 and CHF1) encodes a basic helix-loop-helix (bHLH) transcriptional repressor that is expressed in the cardiovascular system8.
Studies we have conducted in Hey2-targeted mice provide strong support for the role of this gene in Brugada syndrome. In the developing mouse heart, Hey2 expression is confined to the (subepicardial) compact myocardium of the ventricle33. Hey2-null mice exhibit a spectrum of developmental anomalies, including ventricular wall thinning, abnormal right ventricular morphology and postnatal cardiomyopathic changes34–37. The expression of Gja5 (encoding Cx40), Nppa and Tbx5, normally enriched in the (subendocardial) trabecular component of the ventricle, is expanded into the compact myocardium in Hey2-deficient embryos34,38,39. Because such transmural heterogeneity in expression is similarly well established for Nav1.5 (high expression in subendocardium, low expression in subepicardium)40, loss of Hey2 might also affect the transmural expression gradient of this ion channel implicated in Brugada syndrome2,3. Indeed, in hearts from homozygous Hey2-null embryos, we observed higher Nav1.5 expression in the compact layer than in wild-type hearts, flattening the expression gradient of this channel (Supplementary Fig. 8a).
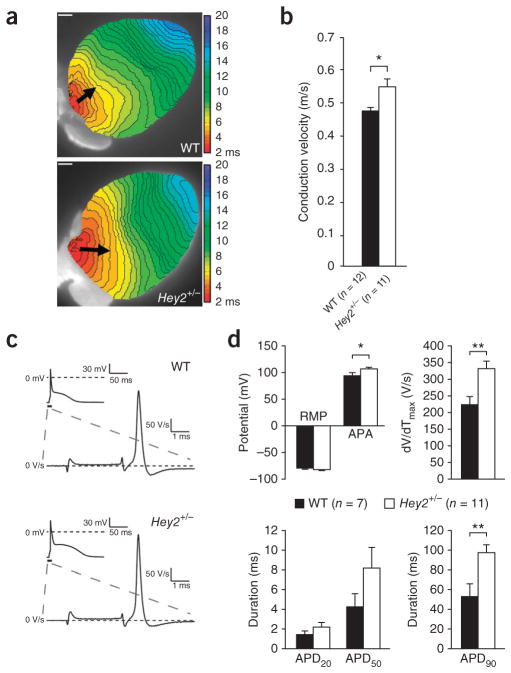
The functional consequences of Hey2 loss were investigated in adult heterozygous Hey2 mice (Hey2+/−), which have structurally normal hearts (Supplementary Fig. 9). In vivo surface ECG parameters were unchanged in Hey2+/− mice (Supplementary Fig. 10). However, conduction velocity was significantly increased in the right ventricular outflow tract (RVOT) of isolated Hey2+/− hearts (Fig. 3a, b), whereas conduction velocity was unaffected in the right and left ventricular free wall (Supplementary Fig. 11). Action potential upstroke velocity was increased in Hey2+/− myocytes isolated from the RVOT region (Fig. 3c, d), pointing to increased sodium channel function, despite undetectable changes in Nav1.5 expression in adult hearts from Hey2+/− mice in immunohistochemistry analysis (Supplementary Fig. 8b). Furthermore, the prolonged repolarization parameters observed in these cells suggest an additional regulatory role for Hey2 in repolarizing currents (Fig. 3d). Future work should address whether the observed alterations in action potential characteristics and conduction are mediated through ion channel correlates, subtle structural heart alterations or both. Nevertheless, the preferential involvement of the RVOT is in line with ECG manifestations in right precordial leads and concurs with the observation that the RVOT is a common site of origin of ventricular arrhythmias in individuals with Brugada syndrome41.
Figure 3.
Increased conduction velocity and sodium channel availability in the RVOT of adult Hey2+/− mice. (a) Representative optical activation maps from isolated wild-type (WT; n = 12) and Hey2+/− (n = 11) hearts stimulated at the RVOT at a basic cycle length of 120 ms (scale bars, 1 mm). Arrows indicate conduction along the RVOT epicardium through the connection of similar isochrones. Isochrones (0.5-ms intervals) in the RVOT of Hey2+/− hearts are less crowded compared to wild-type isochromes, indicating faster conduction in the RVOT of Hey2+/− hearts (as indicated by increased arrow length). (b) On average, conduction velocity in the RVOT of Hey2+/− hearts was significantly increased compared to that in wild-type hears (*P < 0.05; Student’s t test). Error bars, s.e.m. (c) Representative action potentials measured in adult wild-type (WT; n = 7) and Hey2+/− (n = 11) cardiomyocytes isolated from RVOT. (d) Average action potential characteristics measured at 4 Hz. RVOT cardiomyocytes from Hey2+/− mice showed a significant increase in maximal upstroke velocity (dV/dTmax, a measure of sodium channel availability and a determinant of conduction velocity) and action potential amplitude (APA), indicating increased sodium channel availability (*P < 0.05; **P < 0.01; Student’s t test). Resting membrane potential (RMP), action potential duration at 20% and 50% repolarization (APD20 and APD50, respectively) were not significantly different in wild-type and Hey2+/− mice; action potential duration at 90% repolarization (APD90) was significantly increased in RVOT cardiomyocytes from Hey2+/− mice. Results are expressed as mean ± s.e.m.
In conclusion, we have identified Hey2 as a transcriptional regulator of cardiac electrical function involved in the pathogenesis of Brugada syndrome. Furthermore, we provide new evidence that common variants, previously shown to modulate ECG conduction indices, also modulate susceptibility to a rare arrhythmia disorder. Most notably, this study demonstrates that the GWAS paradigm can be successfully applied to a rare disorder, previously considered monogenic, to identify common genetic variants with unexpectedly strong modifier effects.
URLs
Affymetrix Power Tools, http://www.affymetrix.com/partners_programs/programs/developer/tools/powertools.affx; GTOOL, http://www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html; R statistical package, http://www.r-project.org/; 1000 Genomes Project, http://www.1000genomes.org/; 1000 Genomes phase I integrated variant set release, http://mathgen.stats.ox.ac.uk/impute/data_download_1000G_phase1_integrated.html.
ONLINE METHODS
Case and control samples
Individuals with Brugada syndrome, defined by the presence of a type 1 ECG2, were recruited from 13 centers in Europe (Nantes, Paris, Amsterdam, Pavia, Copenhagen, Munich, Münster and London), the United States (Utica and Nashville) and Japan (Nagasaki, Shiga and Osaka). Only index cases were included from extended pedigrees. Appropriate medical ethical committee approval was obtained at each participating clinical center. Informed consent was available from all subjects. Clinical data (age at diagnostic ECG, SCN5A mutation status, symptoms and family history of sudden cardiac death) and ECGs were collected centrally and reviewed. A Brugada syndrome type I ECG pattern was defined on the basis of the criteria drawn out at the Second Consensus Conference on Brugada Syndrome2, namely, a coved type ST elevation at baseline or after a drug challenge test, in one or more leads in the right precordial leads, including the third and fourth intercostal space. Drug challenge tests were performed according to consensus criteria2. Control subjects were drawn from the D.E.S.I.R. cohort18 for the GWAS and the European replication set and were drawn from the Sado study42 for the Japanese replication set. No statistical method was used to predetermine sample size.
GWAS genotyping
SNP genotyping was performed on population-optimized Affymetrix Axiom Genome-Wide CEU 1 array plates following the standard manufacturer’s protocol. Each array contains 567,097 SNPs. Fluorescence intensities were quantified using the Affymetrix GeneTitan Multi-Channel Instrument, and primary analysis was conducted with Affymetrix Power Tools following the manufacturer’s recommendations (see URLs). Genotype calling, a two-dimensional clustering analysis, was performed using the ‘apt’ program. Individuals with genotype call rate of lower than 97% were removed, as were those with fewer than 10,000 markers reporting a heterozygous state (the threshold was determined after visual inspection). Monomorphic SNPs were excluded, as were those with minor allele frequency (MAF) of <10% (n = 175,153), a call rate of <95% (n = 19,986) or Hardy-Weinberg disequilibrium in controls (n = 2,054 with P < 1 × 10−4 when testing for Hardy-Weinberg equilibrium). Note that Hardy-Weinberg disequilibrium was also tested in demographically homogenous cases to identify very large deviations (P < 1 × 10−7). Additional SNPs were excluded for batch effect: such SNPs were defined as those with significant differences in allele frequency in one plate versus all others within cases and within controls only (n = 68) or with unexplained large differences observed in controls versus the 1000 Genomes Project European (non-Finnish) population (n = 9,686).
Demographic analyses
The ancestry of individuals was assessed using a multidimensional scaling technique, as implemented in PLINK43. SNPs were selected for short-range LD independence. Pruning was performed using a two-step procedure to accommodate longer range LD (this is particularly important, as the Axiom Human array is enriched in SNPs in the human leukocyte antigen (HLA) region). In a first step, we applied the threshold r2 < 0.2 within a 20-kb LD block or within 50 SNPs. In a second step, we applied the same threshold within a 10-Mb distance or within 100 SNPs on the pruned data set. We then created an identity-by-state (IBS) matrix including all individuals and applied the multidimensional scaling method (–mds option in PLINK) to retrieve the first five components. Three matrices were estimated using our cases and controls together with all 1000 Genomes Project populations (IC) and all European (except Finnish) populations (E). At each level, we excluded outliers on the first two components using an expectation-maximization-fitted Gaussian mixture clustering method44 implemented in the R package M-CLUST, assuming either three (for IC) or two (for E) clusters and noise. Outlier position was assigned using nearest-neighbor-based classification45 (NNclust in R package PrabClus). Outliers were excluded from the analysis, as previously done in GWAS46.
GWAS
Using the clustering algorithm described above, we defined two homogenous groups (A and B). To carry out the genome-wide analysis, each SNP was tested within groups A and B separately, using logistic regression and assuming an additive genetic model with adjustment for the first five components retrieved. No additional covariates were added, as advised47. Instead, the results from groups A and B were combined into a meta-analysis using an inverse normal strategy48, whereby the summary P values for each test (and effect direction) are combined into a signed z score that, properly weighted, yields N (μ = 0, σ2 = 1). Because the number of controls exceeded by far the number of cases in all studies, we used the effective sample size (weighting studies A and B) using METAL software as advised49. In addition, we performed a second genome-wide analysis on a homogenous sample of 254 cases and 806 controls of apparent French origin (largest geographically homogenous sample; Supplementary Fig. 5).
Concordance rate between Axiom and 1000 Genomes Project data
We genotyped 95 HapMap individuals on Affymetrix Axiom Genome-Wide CEU 1 arrays using the same process as described above. We could retrieve the genotypes of 58 of these 95 individuals from the 1000 Genomes Project database. The concordance rate was tested using PLINK (merging mode 7, which compares the common non-missing genotypes). The concordance rate was 99.4% over a total of 20,853,552 genotypes and 100% over the 174 genotypes corresponding to the 3 associated SNPs.
Genome-wide imputation analysis
Genotyped SNPs in cases and controls were phased using the SHAPE-IT (v.1) program50. Imputation of 6.1 million frequent SNPs (MAF > 0.05 in Europeans) was carried out using IMPUTE v2 (ref. 51). Chromosome regions were split in chunks of approximately 7 Mb. The reference panel was Phase I integrated variant set release (v3) in NCBI Build 37 (hg19) coordinates (see URLs). For each chromosomal chunk, a set of genetically matched panel individuals was selected, according to the last strategy used by IMPUTE52. Imputed SNPs were combined with SNPs extracted from the 1000 Genomes Project data set under the IMPUTE format. We merged both data sets using GTOOL. We applied a logistic regression (additive model) as implemented in SNPTEST45 (options -frequentist 1 and -score), using the first five components as covariates. Individuals from the 1000 Genomes Project data set were added for all SNPs either genotyped or imputed.
Post-analysis quality control
For each significantly associated SNP in a region (called here lead SNP), we visually inspected the cluster graph (Supplementary Fig. 12a) and the association results of SNPs in LD (locus plot).
Enrichment analysis
We tested enrichment in moderately associated SNPs grouped by distinctive properties. The first group included SNPs that have been reported to be associated with ECG traits in previous GWAS. We used the list published in ref. 53 after removal of redundant SNPs (one SNP was selected randomly for each group of SNPs with r2 > 0.2) and exclusion of SNPs located in the same haplotype block as rs10428132. The second list comprised every SNP located within 1-Mb intervals (500 kb upstream and 500 kb downstream) centered on the susceptibility genes for Brugada syndrome listed in ff 13. We removed the SCN5A gene, as it would have disproportionately inflated the overall statistics. Quantile-quantile plots were first visually inspected to check for enrichment. The significance of enrichment suggested by the quantile-quantile plot for the first group (ECG-associated SNPs) was formally tested by a simple Fisher combination test.
Replication genotyping
The three significantly associated SNPs from the GWAS stage (rs10428132, rs9388451 and rs11708996) were typed for the replication steps by TaqMan SNP Genotyping assays (Applied Biosystems) according to the manufacturer’s protocol on a LightCycler 480 Real-Time PCR System (Roche) or an ABI Prism 7900HT Sequence Detection System (Applied Biosystems). Assays C__26860683_10, C___2824356_10 and C__44417794_10 were functionally tested by Applied Biosystems for the SNPs rs10428132, rs9388451 and rs11708996, respectively. Reaction conditions included an initial denaturation step at 95 °C for 10 min, followed by 50 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 30 s. Data were analyzed with LightCycler 480 software (release 1.5.0 SP4; Supplementary Fig. 12b) or by ABI 7900HT Sequence Detection Systems software (Supplementary Fig. 12c). Outliers were excluded from the analysis. After the first round of genotyping, two samples per SNP genotype cluster were sequenced to verify genotype (Supplementary Fig. 12b). Furthermore, 15 samples that were previously genotyped on Affymetrix Axiom Genome-Wide CEU 1 arrays were used as control samples: the genotypes obtained by TaqMan assays were perfectly concordant with those generated with Axiom array technology.
SNP imputation for the control population used in the European replication
The genotypes of the three SNPs tested in replication were imputed using IMPUTE software for 806 D.E.S.I.R. individuals on the basis of genotype data sets previously generated on Illumina Human CNV370-Duo arrays24,54. We selected a threshold of 0.9 for genotype calling, meaning that the genotypes whose posterior probability exceeded this threshold were called, whereas those where this probability was below the threshold were set as unknown. We genotyped 49 individuals on both Affymetrix and Illumina arrays. Once hard calls were obtained using GTOOL (see URLs) with -Gen mode and –threshold option set to 0.9, concordance rates were calculated using PLINK (merging mode 7). Perfect concordance was observed between genotyped and imputed SNPs (100% concordance rate for 144 genotypes).
Replications and meta-analyses
The case-control replication study was performed using a logistic regression method that accounts for genotype calling uncertainty. This method, based on missing data theory, allows the unbiased estimation of ORs and confidence intervals and is implemented in SNPTEST (options–method ml). Pooled ORs were obtained by averaging the ORs from all stages (GWAS and European and Japanese replications) and weighted by the inverse of the variance. Heterogeneity was tested using the Cochran’s Q test and was also measured using Higgins’ index55. We generated genetic scores for individuals on the basis of an allelic scoring system involving our three SNPs. These scores were created either through the number of at-risk alleles for the European discovery and Japanese replication populations or the risk allele dosage in the European replication population. Risk allele dosages from the European replication population were collapsed using dosage. The distribution of imputed dosage is shown in Supplementary Figure 13a. Results were similar between imputed dosage (β = 0.62921, σ = 0.05427, P = 4.43 × 10−31) and collapsed imputed dosage (β = 0.61778, σ = 0.05386, P = 1.88 × 10−30). Moreover, we compared the genetic scores obtained with genotyped versus imputed SNPs for 49 individuals who were genotyped on Axiom Genome-Wide CEU 1 arrays and imputed with the European replication population and observed high correlation between methods (Supplementary Fig. 13b). Finally, we tested whether a non-additive model (recessive or dominant) might be a better fit for each genome-wide significant SNP. A heterozygote effect was added to the logistic regression analysis along with the linear effect (effect of the number of alternative alleles) in each study and in a meta-analysis. We did not detect any consistent deviation from the additive model (Supplementary Table 6).
Estimation of the genetic score effect by multiple imputation
Despite this high concordance, we chose to estimate genotype score risk within the Multiple Imputation framework56. Ten data sets were created where each uncertain genotype was replaced by a value simulated under the probability distribution obtained through genetic imputation (IMPUTE output consisting of P(AA), P(AB) and P(BB)). The β value and variance were obtained using standard procedures57. We let m be the number of simulations (called replicates). For each simulation, we carried out a logistic regression (either on score as an ordinal value or on each score versus baseline). The Multiple Imputation effect estimation was calculated as follows:
The variance was calculated as the sum of within-replicate and between-replicate variances
Confidence intervals were retrieved using the 95% quantile of a Student distribution with a number of degrees of freedom, which is a function of the two components of the variance. We used the ‘glm’ function of the R statistical package (see URLs) to perform logistic regression. R was used to create complete data sets from IMPUTE output.
Calculation of sibling relative risk and liability-scale variance
Assuming a multiplicative model (within and between variants), the contribution to the sibling relative risk of a set of N SNPs was calculated using the following formula
where pj and ORj denote the RAF and corresponding allelic OR at the jth SNP58. Assuming disease prevalence K, the liability-scale variance59 explained by these SNPs was calculated as follows:
In the expression, T = ϕ−1 (1 − K), T1 = ϕ−1 (1 − λSK) and ω = z/K, where z is the height of the standard Gaussian density at T.
Surface ECG analysis
Mice (male and female, aged 3–5 months) were anesthetized using isoflurane inhalation (0.8–1.0 volume percentage in oxygen), and surface ECGs were recorded from subcutaneous 23-gauge needle electrodes attached to each limb using the Powerlab acquisition system (ADInstruments). ECG traces were signal averaged and analyzed for heart rate (RR interval) and for PR-interval, QRS-interval and QT-interval duration using LabChart7Pro software (ADInstruments). QT intervals were corrected for heart rate using the formula (RR in ms).
Optical mapping in isolated hearts
Mice were anesthetized by an intraperitoneal injection of pentobarbital, after which the heart was excised, cannulated, incubated in 10 ml Tyrode’s solution containing 15 μM Di-4 ANEPPS and subsequently placed in an optical mapping setup. Hearts were perfused at 37 °C with Tyrode’s solution (128 mM NaCl, 4.7 mM KCl, 1.45 mM CaCl2, 0.6 mM MgCl2, 27 mM NaHCO3, 0.4 mM NaH2PO4 and 11 mM glucose (pH maintained at 7.4 by equilibration with a mixture of 95% O2 and 5% CO2)) containing blebbistatin to prevent movement artifacts. Excitation light was provided by a 5-W power LED (filtered 510 ± 20 nm). Fluorescence (filtered for >610 nm) was transmitted through a tandem lens system on a CMOS sensor (100 × 100 elements; MICAM Ultima). Hearts were paced at a basic cycle length (BCL) of 120 ms (twice the diastolic stimulation threshold) from the center of the right or left ventricle epicardial surface or from the RVOT epicardial surface. Optical action potentials were analyzed with custom software. Local activation was defined as the maximum positive slope of the action potential, and local activation times were used to construct ventricular activation maps. To calculate conduction velocity in longitudinal and transversal directions in the right and left ventricles, the difference in activation time between at least three consecutive electrode terminals separated by a known distance located parallel (longitudinal) or perpendicular (transversal) to the direction of propagation was measured. The direction of propagation was determined using isochronal maps. For c velocity measurements in the RVOT, transverse fiber direction was defined as the slowest conduction velocity.
Action potential measurements in isolated cardiomyocytes
Cardiomyocytes were isolated by enzymatic dissociation as previously described60. After perfusion of the heart with enzyme solution, the RVOT area at the base of the right ventricle just below the truncus pulmonalis was excised and used during the subsequent isolation process43. Quiescent rod-shaped cross-striated cells with a smooth surface were selected for measurements. Action potentials were recorded with the amphotericin-B-perforated patch-clamp using an Axopatch 200B Clamp amplifier (Molecular Devices Corporation). Signals were filtered (low pass, 10 kHz) and digitized (40 kHz), and action potentials were corrected for the estimated change in liquid junction potential. Action potentials were measured at 36 ± 0.2 °C using a modified Tyrode’s solution containing 140 mM NaCl, 5.4 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5.5 mM glucose and 5.0 mM HEPES, pH 7.4 (NaOH). Pipettes (1.5–2.5 MΩ) were filled with solution containing 125 mM potassium gluconate, 20 mM KCl, 10 mM NaCl, 0.22 mM amphotericin-B and 10 mM HEPES, pH 7.2 (KOH). Action potentials were elicited at 4 Hz by 3 ms, 1.2× threshold current pulses through the patch pipette. We analyzed resting membrane potential (RMP), action potential amplitude (APA), maximal upstroke velocity (dV/dtmax) and action potential duration at 20, 50 and 90% repolarization (APD20, APD50 and APD90, respectively). Data from ten consecutive action potentials were averaged. Results are expressed as mean ± s.e.m. Two sets of data were considered significantly different if the P value of the unpaired Student’s t test with Bonferroni correction was <0.05.
Other methods
Procedures for immunohistochemistry hybridization on mouse heart sections were performed as described previously40.
Supplementary Material
Acknowledgments
We thank L. Beekman, C. de Gier-de Vries, B. de Jonge and the Genomic Platform of Nantes (Biogenouest Genomics) for technical support. We are also grateful to the French Clinical Network against Inherited Cardiac Arrhythmias, which includes the University Hospitals of Nantes, Bordeaux, Rennes, Tours, Brest, Strasbourg, La Réunion, Angers and Montpellier. This study was funded by research grants from the Leducq Foundation (CVD-05; Alliance Against Sudden Cardiac Death), the Ministry of Education, Culture, Sports, Science and Technology of Japan (grant-in-aid for the Project in Sado for Total Health, PROST), the French Ministry of Health (PHRC AOR04070, P040411 and PHRCI DGS2001/0248), INSERM (ATIP-Avenir program to R.R.) and the French Regional Council of Pays-de-la-Loire. This research was also supported by the Netherlands Heart Institute (grant 061.02 to C.A.R. and C.R.B.) and the Division for Earth and Life Sciences (ALW; project 836.09.003 to C.A.R.) with financial aid from the Netherlands Organization for Scientific Research (NWO). C.R.B. acknowledges support from the Netherlands Heart Foundation (NHS 2007B202 and 2009B066). J.B. was supported by a research grant from the European Society of Cardiology and the Netherlands Heart Institute (ICIN) and by the French-Dutch Academy through the Van Gogh program. E.S.-B. was supported by the Interdisziplinären Zentrums für Klinische Forschung (IZKF) of the University of Münster and the Collaborative Research Center SFB656. M.G. acknowledges support from the German Research Foundation (DFG-SFB 688 and TP A16). This manuscript is dedicated to the memory of Denis Escande, who founded the Leducq Foundation Network Alliance Against Sudden Cardiac Death.
Footnotes
AUTHOR CONTRIBUTIONS
C.R.B., J.-J.S. and R.R. designed the study. Y.M. and J.-B.G. evaluated all ECGs. C.D. coordinated the statistical analyses, which C.D., F.S., P.L. and E.C. carried out. F.G., A.D., S.L. and E.C. performed genotyping for the GWAS. J.B., J.V., V. Portero and K.H. carried out genotyping in the validation sets. A.A.W., H.L.T., H.L.M., V. Probst, F.K., S. Bézieau, S.C., S.K., B.M.B., E.S.-B., S.Z., L.C., P.J.S., F.D., M.T., C.A., S. Bartkowiak, P.G., V.F., A.L., D.M.R., P.W., E.R.B., R.B., J.T.-H., M.S.O., N.M., A.N., M.H., S.O., K.H., W.S. and T.A. recruited subjects and participated in clinical and molecular diagnostics. P.F., B.B., O.L., H.W., T.M. and N.E. provided controls. M.G., D.W. and C.W. provided the mice. C.A.R., A.O.V., B.J.B. and R.W. acquired and analyzed electrophysiological data. V.M.C., C.A.R. and R.W. acquired and analyzed protein expression data. C.R.B., J.B., C.A.R., C.D., J.-J.S., V.M.C., R.C. and R.R. interpreted the data. C.R.B., J.-J.S., V. Probst, D.M.R., A.A.W., S.K., E.S.-B., A.L. and R.R. obtained funding. C.R.B., J.B., C.D. and R.R. drafted the manuscript. All coauthors critically revised the manuscript for intellectual content. C.R.B. and R.R. led the study together.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 2.Antzelevitch C, et al. Brugada syndrome: Report of the Second Consensus Conference Endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–670. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 3.Kapplinger JD, et al. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chambers JC, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 5.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 6.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sotoodehnia N, et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat Genet. 2010;42:1068–1076. doi: 10.1038/ng.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 9.Straus SMJM, et al. The incidence of sudden cardiac death in the general population. J Clin Epidemiol. 2004;57:98–102. doi: 10.1016/S0895-4356(03)00210-5. [DOI] [PubMed] [Google Scholar]
- 10.Priori SG, et al. Task Force on Sudden Cardiac Death, European Society of Cardiology, Summary of Recommendations. Europace. 2002;4:3–18. doi: 10.1053/eupc.2001.0214. [DOI] [PubMed] [Google Scholar]
- 11.Chen Q, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 12.Wilde AAM, et al. The pathophysiological mechanism underlying Brugada syndrome: depolarization versus repolarization. J Mol Cell Cardiol. 2010;49:543–553. doi: 10.1016/j.yjmcc.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crotti L, et al. Spectrum and prevalence of mutations involving BrS1-through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Priori SG, et al. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: a prospective evaluation of 52 families. Circulation. 2000;102:2509–2515. doi: 10.1161/01.cir.102.20.2509. [DOI] [PubMed] [Google Scholar]
- 15.Probst V, et al. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome (clinical perspective) Circ Cardiovasc Genet. 2009;2:552–557. doi: 10.1161/CIRCGENETICS.109.853374. [DOI] [PubMed] [Google Scholar]
- 16.Schulze-Bahr E, et al. Sodium channel gene (SCN5A) mutations in 44 index patients with Brugada syndrome: different incidences in familial and sporadic disease. Hum Mutat. 2003;21:651–652. doi: 10.1002/humu.9144. [DOI] [PubMed] [Google Scholar]
- 17.Hermida JS, et al. Prospective evaluation of the familial prevalence of the Brugada syndrome. Am J Cardiol. 2010;106:1758–1762. doi: 10.1016/j.amjcard.2010.07.049. [DOI] [PubMed] [Google Scholar]
- 18.Balkau B. An epidemiologic survey from a network of French Health Examination Centres, (D.E.S.I.R.): epidemiologic data on the insulin resistance syndrome. Rev Epidemiol Sante Publique. 1996;44:373–375. [PubMed] [Google Scholar]
- 19.Arking DE, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38:644–651. doi: 10.1038/ng1790. [DOI] [PubMed] [Google Scholar]
- 20.Newton-Cheh C, et al. Common variants at ten loci influence QT interval duration in the QTGEN Study. Nat Genet. 2009;41:399–406. doi: 10.1038/ng.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfeufer A, et al. Common variants at ten loci modulate the QT interval duration in the QTSCD Study. Nat Genet. 2009;41:407–414. doi: 10.1038/ng.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eijgelsheim M, et al. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marroni F, et al. A genome-wide association scan of RR and QT interval duration in 3 European genetically isolated populations: the EUROSPAN project. Circ Cardiovasc Genet. 2009;2:322–328. doi: 10.1161/CIRCGENETICS.108.833806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 25.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akopian AN, Sivilotti L, Wood JN. A tetrodotoxin-resistant voltage-gated sodium channel expressed by sensory neurons. Nature. 1996;379:257–262. doi: 10.1038/379257a0. [DOI] [PubMed] [Google Scholar]
- 27.Verkerk AO, et al. Functional Nav1.8 channels in intracardiac neurons: the link between SCN10A and cardiac electrophysiology. Circ Res. 2012;111:333–343. doi: 10.1161/CIRCRESAHA.112.274035. [DOI] [PubMed] [Google Scholar]
- 28.Yang T, et al. Blocking Scn10a channels in heart reduces late sodium current and is antiarrhythmic. Circ Res. 2012;111:322–332. doi: 10.1161/CIRCRESAHA.112.265173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pallante BA, et al. Contactin-2 expression in the cardiac Purkinje fiber network. Circ Arrhythm Electrophysiol. 2010;3:186–194. doi: 10.1161/CIRCEP.109.928820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Boogaard M, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnolds DE, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122:2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shao W, Halachmi S, Brown M. ERAP140, a conserved tissue-specific nuclear receptor coactivator. Mol Cell Biol. 2002;22:3358–3372. doi: 10.1128/MCB.22.10.3358-3372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fischer A, Gessler M. Hey genes in cardiovascular development. Trends Cardiovasc Med. 2003;13:221–226. doi: 10.1016/s1050-1738(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 34.Xin M, et al. Essential roles of the bHLH transcription factor Hrt2 in repression of atrial gene expression and maintenance of postnatal cardiac function. Proc Natl Acad Sci USA. 2007;104:7975–7980. doi: 10.1073/pnas.0702447104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kokubo H, et al. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res. 2004;95:540–547. doi: 10.1161/01.RES.0000141136.85194.f0. [DOI] [PubMed] [Google Scholar]
- 36.Gessler M, et al. Mouse gridlock: no aortic coarctation or deficiency, but fatal cardiac defects in Hey2−/− mice. Curr Biol. 2002;12:1601–1604. doi: 10.1016/s0960-9822(02)01150-8. [DOI] [PubMed] [Google Scholar]
- 37.Sakata Y, et al. Ventricular septal defect and cardiomyopathy in mice lacking the transcription factor CHF1/Hey2. Proc Natl Acad Sci USA. 2002;99:16197–16202. doi: 10.1073/pnas.252648999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koibuchi N, Chin MT. CHF1/Hey2 plays a pivotal role in left ventricular maturation through suppression of ectopic atrial gene expression. Circ Res. 2007;100:850–855. doi: 10.1161/01.RES.0000261693.13269.bf. [DOI] [PubMed] [Google Scholar]
- 39.Fischer A, et al. Hey basic helix-loop-helix transcription factors are repressors of GATA4 and GATA6 and restrict expression of the GATA target gene ANF in fetal hearts. Mol Cell Biol. 2005;25:8960–8970. doi: 10.1128/MCB.25.20.8960-8970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remme CA, et al. The cardiac sodium channel displays differential distribution in the conduction system and transmural heterogeneity in the murine ventricular myocardium. Basic Res Cardiol. 2009;104:511–522. doi: 10.1007/s00395-009-0012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nademanee K, et al. Prevention of ventricular fibrillation episodes in Brugada syndrome by catheter ablation over the anterior right ventricular outflow tract epicardium. Circulation. 2011;123:1270–1279. doi: 10.1161/CIRCULATIONAHA.110.972612. [DOI] [PubMed] [Google Scholar]
- 42.Sakuma M, et al. Incidence and outcome of osteoporotic fractures in 2004 in Sado City, Niigata Prefecture, Japan. J Bone Miner Metab. 2008;26:373–378. doi: 10.1007/s00774-007-0841-1. [DOI] [PubMed] [Google Scholar]
- 43.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frayley C, Raftery AE. MCLUST Version 3 for R: Normal Mixture Modeling and Model-Based Clustering. Department of Statistics, University of Washington; Seattle: 2006. [Google Scholar]
- 45.Byers S, Raftery AE. Nearest-neighbor clutter removal for estimating features in spatial point processes. J Am Stat Assoc. 1998;93:577–584. [Google Scholar]
- 46.Postel-Vinay S, et al. Common variants near TARDBP and EGR2 are associated with susceptibility to Ewing sarcoma. Nat Genet. 2012;44:323–327. doi: 10.1038/ng.1085. [DOI] [PubMed] [Google Scholar]
- 47.Pirinen M, Donnelly P, Spencer CCA. Including known covariates can reduce power to detect genetic effects in case-control studies. Nat Genet. 2012;44:848–851. doi: 10.1038/ng.2346. [DOI] [PubMed] [Google Scholar]
- 48.Stouffer SA, Suchman EA, Devinney LC, Star SA, Williams RM., Jr . Studies in Social Psychology in World War II. Vol. 1. Princeton University Press; Princeton, NJ: 1949. The American Soldier: Adjustment During Army Life. [Google Scholar]
- 49.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 51.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 52.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3. 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kolder ICRM, Tanck MWT, Bezzina CR. Common genetic variation modulating cardiac ECG parameters and susceptibility to sudden cardiac death. J Mol Cell Cardiol. 2012;52:620–629. doi: 10.1016/j.yjmcc.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 54.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 55.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 56.Little RJA, Rubin DB. Statistical Analysis with Missing Data. Wiley and Sons; New York: 1987. [Google Scholar]
- 57.Rubin DB. Multiple Imputation for Nonresponse in Surveys. 2008 http://onlinelibrary.wiley.com/book/10.1002/9780470316696.
- 58.Lin S, Chakravarti A, Cutler DJ. Exhaustive allelic transmission disequilibrium tests as a new approach to genome-wide association studies. Nat Genet. 2004;36:1181–1188. doi: 10.1038/ng1457. [DOI] [PubMed] [Google Scholar]
- 59.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Remme CA, et al. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.