Abstract
Bioinformatic studies on a revised hierarchy of hematopoietic progenitors have provided a genome-wide view of lineage-affiliated transcriptional programs directing early hematopoiesis. Unexpectedly, lymphoid, myeloid and erythroid gene expression programs were primed with similar frequency at the multi-potent progenitor stage indicating a stochastic nature to this process. Multi-lineage transcriptional priming is quickly resolved upon erythroid lineage restriction with both lymphoid and myeloid transcriptional programs rapidly extinguished. However, expression of lymphoid and myeloid factors remains active past nominal lymphoid and myeloid lineage restrictions, revealing a common genetic network utilized by both pathways. Priming and resolution of multi-lineage potential is dependent on the activity of the DNA binding factor Ikaros. Ikaros primes the lymphoid transcriptional program in the HSC and represses the stem cell and other disparate transcriptional programs downstream of the HSC. Loss of Ikaros removes the lymphoid leg of the immune system and may confer aberrant self-renewing properties to myeloid progenitors.
Introduction
Differentiation into the three major hematopoietic lineages (erythroid, myeloid and lymphoid) has been extensively studied using both cellular and molecular approaches. These have delineated major steps in differentiation that demarcate lineage commitment and maturation. Nonetheless, the regulatory mechanisms that modulate the lineage differentiation properties of early progenitors have been elusive. One confounding issue is that the early hematopoietic progenitor hierarchy and their lineage affiliations are far more complex than initially perceived. Incomplete characterization of cellular intermediates in the multi-potent state and of early lineage restrictions, in addition to technical limitations of performing biochemical analysis on small number of progenitor cells have made it hard to delineate the mechanisms by which previously described early lineage-determining factors exert their function. It has been increasingly appreciated through genetic studies that widely-expressed epigenetic regulators play a key role in providing differentiation potential by altering the accessibility of gene expression programs in a lineage-specific manner, possibly by working in concert with lineage-specific transcription factors [1] [2,3{Yoshida, 2008 #715]] [4,5].
Here, we review new cellular and molecular advances that clarify the early transitions into the hemo-lymphoid pathways and that establish a molecular framework upon which this complex developmental process is executed. The role of Ikaros, an “early-acting” epigenetic regulator, the transcriptional networks through which it operates, and the molecular mechanisms that it uses to prime and ultimately establish lymphoid potential at the onset of hematopoiesis are discussed.
Lineage-affiliated transcriptional cascades identify both a lympho-myeloid and an erythroid pathway in differentiation
Lineage-affiliated transcriptional programs, referred to as signatures, were deduced from a hematopoietic stem cell (HSC) population (long- to short-term) and early lineage restricted progenitors by comparative computational analyses of their genome-wide expression profiles [6]. The multi-potent HSC, the lympho-myeloid bi-potent progenitor, LMPP (originally defined as lymphoid-primed multipotent progenitor) [7] [8] [9], the granulocyte-macrophage progenitor (GMP) and the megakaryocyte-erythrocyte progenitor (MEP) [10] (Fig.1) were isolated using an Ikaros-EGFP reporter that is differentially expressed within these progenitor populations [8]. Importantly, this reporter in combination with other stem cell markers provides a clear separation of HSC from LMPP not only in the wild type, but also in Ikaros null bone marrow where the absence of Flt3 expression, one of LMPP’s defining markers confounds analysis of Ikaros deficient progenitors [8].
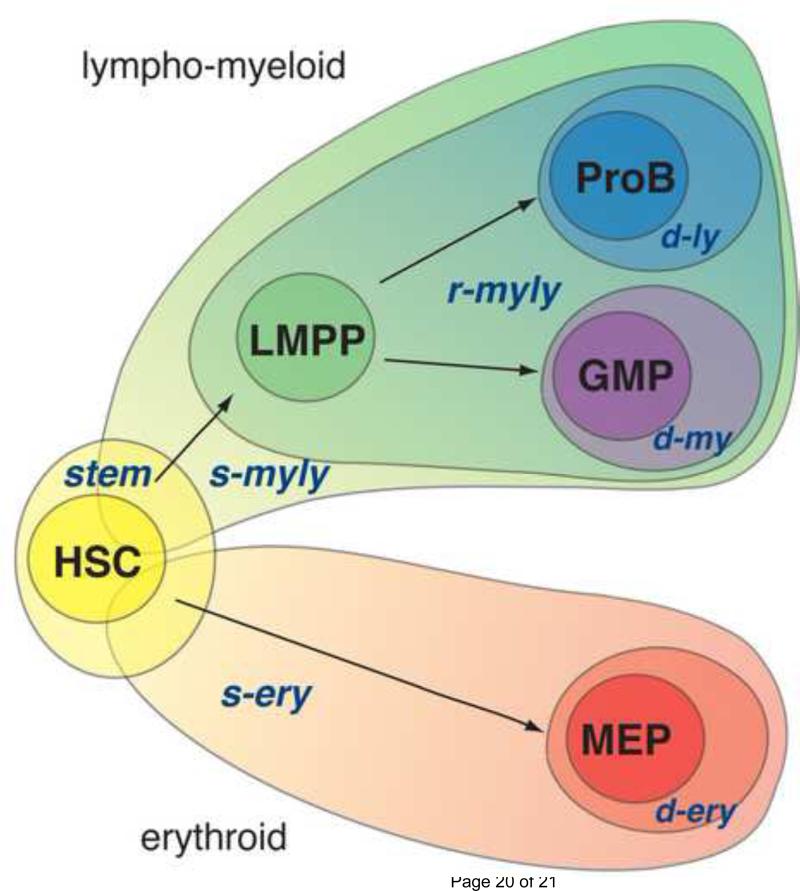
Figure 1. Cascades of lineage-affiliated signatures directing early hematopoiesis.
S-myly is the first in a series of lymphoid-myeloid lineage specific signatures that is primed in the HSC and propagated along the lymphoid and myeloid pathways. The rmyly is primed in the LMPP and marks restriction into a bi-potent lympho-myeloid state. The d-ly and d-my, are indicative of further restriction into the lymphoid or myeloid pathways. S-ery is the first of the erythoid lineage specific signatures that is primed in the HSC. The d-ery is primed after s-ery in committed megakaro-erythrocyte progenitors (MEPs) and demarcates commitment to the erythroid lineage. The stem signature is only expressed in the HSC and demarcates its properties such as self-renewal and multi-potency. Within the HSC, it is widely co-expressed with the s-myly and s-ery signatures. The stem signature is rapidly repressed in lineage-restricted progenitors that lack self-renewing potential.
Lineage-affiliated signatures fall within two transcriptional cascades that are primed in the HSC (denoted by the s(tem) prefix) and differentially propagated in the erythroid or in the lympho-myeloid pathways (denoted by the r(restricted) or the d(ifferentiated) prefix) (Fig.1). The first signature in the erythroid transcriptional cascade is representative of the potential for erythroid differentiation. This is primed in the HSC, established in erythroid progenitors (MEP) and extinguished in lympho-myeloid progenitors (LMPP, GMP and proB). The second signature in this cascade is d-ery and represents restriction into the erythroid lineage. Erythroid differentiation factors are present in both signatures indicating their early priming at the multi-potent state and rapid establishment in early lineage restricted progenitors [6].
The second transcriptional cascade demarcates differentiation into the lymphoid and myeloid pathways (Fig.1) [6]. The s-myly (s-myeloid-lymphoid) signature is the first in this cascade primed in the HSC and augmented in the LMPP, GMP and proB, but extinguished in the MEP. The s-myly is the largest of the stem cell primed lineage-affiliated signatures and provides a transcriptional backbone utilized during development of both the adaptive and innate immune systems. Many of the s-myly genes are involved in molecular pathways that support a variety of immune cell functions. Subsequent lineage-affiliated signatures present in this cascade are activated downstream of the HSC and represent progressive lineage restriction into the lymphoid and myeloid pathways. The r-myly signature is representative of the transition from HSC to LMPP and contains a second layer of lympho-myeloid genes first primed in the LMPP. Subsets of this signature are differentially maintained in the GMP and proB (Fig.1). Finally, the d-my and d-ly are representative of further lineage restrictions into the myeloid and lymphoid lineages, respectively. The relatively small size of the d-ly signature deduced by comparison of the LMPP, HSC, GMP and MEP reflects the fact that LMPP is only partially restricted to lymphoid development. It retains bi-potency for both lymphoid and myeloid differentiation. Further comparison between the LMPP and early proB transcriptomes have revealed a more extensive lymphoid signature that is primed downstream of the LMPP [11,12].
Atop of both lineage-affiliated transcriptional cascades lies an HSC-specific (stem) transcriptional signature (Fig. 1). This is highly expressed within a multi-potent HSC population that ranges in self-renewing potential and is extinguished in the non-self renewing progenitors, the LMPP, GMP and MEP. Several of the genes in this signature are implicated in mediating the HSC's functional properties such as interactions with a specialized niche, quiescence and self-renewing divisions [6].
Taken together these global gene expression studies have revealed two lineage-affiliated transcriptional cascades primed in the multi-potent HSC and propagated in a differential manner in lineage-restricted progenitors. Restriction into the erythroid differentiation pathway involves the rapid elimination of lymphoid and myeloid and a rapid amplification of erythroid transcriptional programs. In contrast, early lymphoid and myeloid programs are primed concomitantly in the HSC and remain associated through lymphoid and myeloid lineage restrictions. Priming of additional layers of lymphoid and myeloid gene expression during lineage restrictions coincide with the slow emergence of discrete immune cell fates built upon an early-primed common transcriptional program.
Co-priming of lymphoid, myeloid and erythoid transcription
Transcriptional studies performed in progenitor populations were further validated at the single progenitor level by multiplex RT-PCR analysis with candidates deduced from the s-series of lineage affiliated signatures (i.e. s-ery and s-myly) [6]. These studies showed lymphoid, myeloid, and erythroid lineage gene expression and co-expression at comparable frequency in the HSC population, consistent with a stochastic lineage differentiation potential. In the MEP, solely erythroid genes were expressed, consistent with erythroid lineage commitment, while in the bi-potent LMPP, myeloid and lymphoid gene expression was augmented, and the majority of these progenitors co-expressed genes affiliated with both lineages. Taken together, these studies have demonstrated that lymphoid-, myeloid- and erythoid-affiliated transcriptional programs are all accessible at the earliest point of hematopoiesis. This was unexpected based on previous studies that concluded that lymphoid transcriptional priming was initiated in lineage-restricted progenitors such as the CLP and more recently in the LMPP [13][14]. The difference between these studies lies in the choice of genes utilized for single progenitor transcriptional analyses. Whereas the genes employed by Ng et. al. were selected from the s-class of lineage affiliated transcriptional signatures such as the s-myly and s-ery, which were deduced based on expression in the HSC compartment, earlier studies utilized genes that are expressed at later stages of lymphoid lineage restriction, and which are representative of the d-ly or r-myly signatures.
The comparable frequency of expression and co-expression of lineage-affiliated transcripts within the HSC population indicate that lineage transcriptional priming in multipotent progenitors occurs in a stochastic manner. Stem cell-specific and lineage-affiliated transcripts were found to be widely co-expressed in the HSC, indicating that the transcriptional mechanisms supporting self-renewal and lineage differentiation are not mutually exclusive. However, upon lineage restriction the stem cell transcriptional program is rapidly extinguished whereas the s-class lineage transcriptional programs are augmented. This likely prohibits lineage-restricted progenitors from acquiring aberrant self-renewing properties.
Reconciliation of cellular differentiation with multi-lineage transcriptional programs
The lineage-affiliated transcriptional cascades faithfully predict the cellular properties of a revised progenitor hierarchy. Identification of the LMPP, a major bi-potent lymphomyeloid progenitor downstream of the HSC supports an early separation of the immune cell fates from the megakaryo-erythroid fates in the adult hematopoietic system [7] [8] [9]. A similar separation of lympho-myeloid from erythroid cell fates has also been revealed by studies on the fetal hematopoietic system [14][15].
The early thymic progenitor (ETP), a possible counterpart of the LMPP [16][17], was recently shown to have in addition to T cell potential significant myeloid potential retained through the DN2 stage of T cell differentiation (Fig. 2, ETP) [18][19][20] [21]. The CLP [22], the LMPP's downstream progeny in the bone marrow have comparable B and T cell developmental potential but reduced myeloid potential [11][12][23]. An unexpected T cell potential was also recently revealed in the GMP when this progenitor was exposed to Notch1 signaling in vitro [6], indicating that a latent lymphoid program detected in the GMP is functional under the right differentiation conditions. With B cell differentiation potential greatly reduced in this progenitor, this result suggests that a common mechanism guiding restrictions into the myeloid and T cell pathways exists.
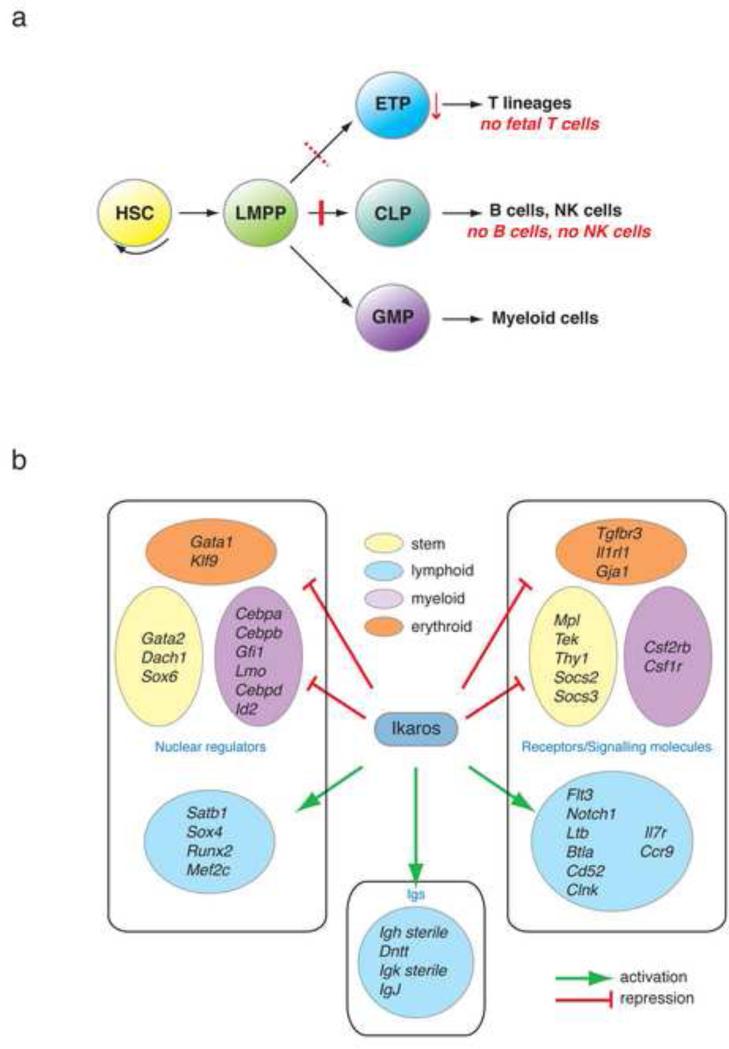
Figure 2. Ikaros effects on priming of lymphoid lineage potential.
A. Ikaros effects on the lymphoid and myeloid differentiation pathways. Ikaros is required for the generation of the CLP and ETP by priming lymphoid lineage-specific signatures in the HSC and LMPP. B. Ikaros activates an extensive lymphoid transcriptional program while it represses smaller cohorts of myeloid-, stem cell-and erythroid-specific genes in the HSC and LMPP. Key downstream targets of Ikaros that are positively (green arrow) or negatively (red block) regulated are shown in the diagram. Signature affiliation, gene function and sub-cellular localization of these genes also are indicated.
The clear bi-potency of the LMPP and the latent bi-potency of its early progeny support a sharing of genetic resources through the GMP, ETP-DN1-2 and CLP-proB stages of differentiation. The subsequent focus on one cell fate coincides with the activation of lineage-affiliated transcriptional programs at later stages of development. These appear to harness the common transcriptional backbone provided by the shared genetic resources and empower one immune cell fate over another.
Ikaros priming of lymphoid potential
The zinc finger DNA binding protein Ikaros (encoded on Ikzf1) has emerged as a key regulator in early lymphocyte development and homeostasis (Fig. 2A) [24] [25] [26] [8] [27]. In Ikaros null mice, B cell development is arrested prior to the pro-B and CLP stage of differentiation [28] [17,26]. Although T cell development is present, early T cell progenitors (ETP/DN1) are greatly reduced in number [26]. More recent studies have shown that although the first step in lympho-myeloid lineage differentiation is made in the absence of Ikaros, its product is a disabled LMPP that is unable to differentiate into the lymphoid pathways but can differentiate into the myeloid pathways [8]. In contrast, another key transcription factor E2A may be required for LMPP generation from the HSC [29]. Further evaluation of Ikaros’ transcriptional effects in early progenitors has revealed a dual role for this factor in the activation and repression of cell fate-affiliated transcriptional programs. Ikaros’ early control of lymphocyte development relies on its ability to activate a cascade of lymphoid lineage transcriptional programs and to repress transcriptional programs that interfere with lymphocyte differentiation (Fig. 2).
Reciprocal regulation of lympho-myeloid priming by Ikaros
Ikaros’ first role, manifested in the multi-potent HSC, is to reciprocally regulate the expression of lymphoid and myeloid promoting factors (Fig. 2B), thus providing an appropriate transcriptional platform (the s-myly signature) upon which further restrictions into the lymphoid and myeloid pathways can be made in a regulated fashion. The cytokine receptors Ltb, Flt3, Il7r, the signaling molecules Notch1, Clnk, Btla and the nuclear regulators Mef2c, Sox4 and Satb1 are examples of s-myly genes that support the adaptive immune cell fate and that have diminished expression upon loss of Ikaros in the LMPP. In contrast, the cytokine receptors Csf1r and Csf2rb and the nuclear factors Cebpa, Cebpb, Sfpi1, Gfi1 are examples of myeloid promoting factors whose expression is augmented upon loss of Ikaros (Fig. 2B). Nonetheless, the s-myly signature in its majority remains unaffected and Ikaros’ specific effects on a subset of its components provides clues by which to delineate the early transcriptional networks required for lymphoid lineage restriction [6].
An Ikaros-based transcriptional network has been recently proposed to regulate early lympho-myeloid lineage decisions [30]. In this model, Ikaros activates Gfi1, which then down-regulates the myeloid promoting Sfpi1 (PU.1) to achieve B cell development. However, our studies provide the opportunity to examine gene expression in discrete cell types in which these fate decisions are made. We find that expression of both Gfi1 and Sfpi1 is increased in HSC, LMPP and GMP in the absence of Ikaros, an observation that is inconsistent with this model.
The loss of lymphoid potential observed in the Ikaros null LMPP could be a consequence of Ikaros’ role as a direct positive and negative regulator of an extensive group of lymphoid- or myeloid-affiliated genes within the s-myly signature. Additionally, discrete changes in the activity of lineage-affiliated transcriptional regulators that lie downstream of Ikaros can perturb the balanced expression of their affiliated transcriptional programs and generate an LMPP with a pre-determined fate in myeloid differentiation.
Repression of stem cell transcriptional programs by Ikaros
Ikaros’ second task is to extinguish the genetic programs that support stemness and multi-potency within the LMPP (Fig. 2B). Loss of Ikaros in the LMPP results in an increase in the expression of a subset of stem cell genes. These are normally extinguished in this lineage-restricted progenitor that has limited self-renewing potential. The cytokine receptors Mpl and Tek, both implicated in self-renewal are significantly up-regulated in the mutant LMPP [6]. Signaling molecules such as Thy1, Socs2 and Socs3 and the nuclear regulators Gata2, Dach1 and Sox6 are other examples of stem-cell-specific genes up-regulated upon loss of Ikaros (Fig. 2B). Genetic components of the early erythroid signature (s-ery) were also aberrantly increased in Ikaros deficient LMPP, albeit to a lesser degree. The nuclear regulators Gata1 and Klf9, the cytokine receptors, Il1rl1, Tgfbr3 and the adherens junction molecule Gja1 are such examples (Fig. 2B). The functional consequences of the persisting expression of these genes in Ikaros null LMPP beg further investigation.
Concluding remarks
Genome-wide transcriptional studies performed at the progenitor population level combined with single cell progenitor analyses have provided the molecular framework upon which early lineage transitions are conducted. Both differentiation and transcriptional studies support an early separation of the erythroid from the lymphomyeloid lineages early in development. In contrast, the lymphoid and myeloid cell fates and their supporting genetic programs appear to be intertwined through nominal lineage restrictions. This close relationship may reflect the evolutionary mechanism that gave rise to the immune system with its adaptive leg arising late and relying on adaptation of available resources already utilized for innate immunity [31][32]. The latent lymphoid or myeloid cell fates still retained in myeloid or lymphoid progenitors may also serve as a backup mechanism for alternative immune cell production when the main pathway is blocked by stress or genetic insult.
A key regulator in the development of the adaptive immune system is the zinc finger DNA binding factor Ikaros whose stable association with chromatin regulators implies a role in modulating lineage-specific transcriptional programs through chromatin-based mechanisms [33][34][35]. Indeed Ikaros’ major role is to reciprocally regulate the priming of lymphoid and myeloid transcriptional programs within the HSC and LMPP. Transcriptional priming precedes the stable establishment of lineage-specific transcriptional programs and is associated with the activity of chromatin remodeling complexes in many developmental systems. A further understanding of the mechanisms that carefully balance lineage decisions during development of the immune system can provide us with the means to manipulate immune cell fates for both academic and therapeutic purposes.
Acknowledgments
We would like to thank Drs; Bruce Morgan, Idit Hazan and John Seavitt for critically reading the manuscript. The authors were supported by the NIH-R37-AI33062 and NIHR01-AI42254 grants to K. Georgopoulos.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1••.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [This paper shows that a combination of permissive and repressive histone modifications are associated on the promoter regions of transcription factors that are important for differentiation of the ES cells. This bivalent histone code is defined by presence of H3K27 and H3K4 indicating concomitant recruitment of dispsrate chromatin remodelers (PRC1/2 and Mll) Authors propose that bivalent histone domains repress developmental genes in ES cells while keeping them poised for activation.] [DOI] [PubMed] [Google Scholar]
- 2.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 3.McMahon KA, Hiew SY, Hadjur S, Veiga-Fernandes H, Menzel U, Price AJ, Kioussis D, Williams O, Brady HJ. Mll has a critical role in fetal and adult hematopoietic stem cell self-renewal. Cell Stem Cell. 2007;1:338–345. doi: 10.1016/j.stem.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Yoshida T, Zhang J, Hazan I, Naito T, Ng SY, Snippert HJ, Heller EJ, Lawton L, Williams CJ, Georgopoulos K. The role of the chromatin remodeler Mi-2beta in hematopoietic stem cell self-renewal and multi-lineage differentiation. Genes Dev. 2008;22:1174–1189. doi: 10.1101/gad.1642808. [This study demonstrates that the ATP-dependent chromatin remodeler Mi-2β, an Ikaros partner regulates two important properties of the HSC, self-renewal and multi-potency. Loss of Mi-2β, causes normally quiescent HSC to undergo aberrant cell divisions that result in their exhaustion. Loss of self-renewal correlates with loss of a significant fraction of stem cell specific genes. Mi-2β deficient HSC readily differentiate into the erythroid lineage but their differentiation in their myeloid is blocked downstream of the LMPP. Priming of the myeloid lineage program is severely affected in the HSC-LMPP compartment with dire consequences for LMPP's further differentiation into the myeloid pathway. This provides a diagramatically opposing scenario to that described for Ikaros mutant progenitors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 6••.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide Lineage-Specific Transcriptional Networks Underscore Ikaros-Dependent Lymphoid Priming in Hematopoietic Stem Cells. Immunity. 2009 doi: 10.1016/j.immuni.2009.01.014. [Using whole-genome transcriptional profiles of HSC and early progenitors, this study deduced a set of molecular signatures that represent and guide early hematopoietic differentiation. The developmental potential of early progenitors is revealed by these molecular signatures. These signatures are applied to analysis of Ikaros–null progenitors and reveal that Ikaros reciprocally modulates lymphoid and myeloid gene expression while also extinguishing stem and erythroid gene expression in lympho-myeloid restricted progenitors.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 8••.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lai AY, Kondo M. Asymmetrical lymphoid and myeloid lineage commitment in multipotent hematopoietic progenitors. J Exp Med. 2006203:1867–1873. doi: 10.1084/jem.20060697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 11••.Rumfelt LL, Zhou Y, Rowley BM, Shinton SA, Hardy RR. Lineage specification and plasticity in CD19- early B cell precursors. J Exp Med. 2006;203:675–687. doi: 10.1084/jem.20052444. [Using 12 color flow cytometry and functional analyses, authors have dissected the CD19-B cell precursor populations and identified MPP, CLP and Fr.A, Pre-ProB subsets. Unexpected myeloid activity is revealed in the CLP and residual T cell activity in the Fr.A pre-proB. Expression profiling indicates the up-regulation of ‘late’ lymphoid genes in the CLP or in Fr.A.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [A subdivision of the Flt3+CLP population is performed here using a λ5-promoter driven hCD25 reporter. The reporter negative population consitutes 95% of Flt3+CLP and has B, T and myeloid differentiation potential, while the reporter positive subset ( 5% of the Flt3+CLP) is restricted to the B cell fate and displays expression of B cell genes indicating that initiation of B cell commitment within the CLP.] [DOI] [PubMed] [Google Scholar]
- 13.Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- 14••.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 200726:407–419. doi: 10.1016/j.immuni.2007.02.013. [This study provides first evidence for LMPP activity in the fetal hematopoietic system. A hierarchical order of l lineage transcriptiona priming is described from the HSC to the LMPP . It is shown that during transition from HSC to LMPP, priming of megakaryocyte and erythroid genes is down-regulated whereas expression of early primed myleoid genes is augmented. Priming of lymphoid lineage affiliated genes is also shown to occur within a fraction of the LMPP and downstream of the HSC suggesting a model by which lymphocyte development occurs on an already establsihed myeloid transcriptioonal platform. The difference in lymphoid priming reported by this study and by Ng et al which places lymphoid transcriptional priing within the HSC compartment lies in the choice of “early” vs “late”lymphoid lineage genes used in the single progenitor transcript analysis. Whereas the Manson study uses llymphoid genes first activated upon lymphoid lineage restriction the Ng study uses lymphoid genes primed in the HSC and revealed by comparative analysis of HSC and progenitor transcriptomes.] [DOI] [PubMed] [Google Scholar]
- 15.Kawamoto H, Katsura Y. A new paradigm for hematopoietic cell lineages: revision of the classical concept of the myeloid-lymphoid dichotomy. Trends Immunol. 2009;30:193–200. doi: 10.1016/j.it.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Shortman K, Wu L. Early T lymphocyte progenitors. Annu Rev Immunol. 1996;14:29–47. doi: 10.1146/annurev.immunol.14.1.29. [DOI] [PubMed] [Google Scholar]
- 17.Allman D, Sambandam A, Kim S, Miller JP, Pagan A, Well D, Meraz A, Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 18.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J Exp Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19••.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [Using in vitro assays and a transgenic reporter of RAG activity, the authors demonstrated that ETPs retain myeloid potential and produce macrophage and granulocyte progeny in vivo.] [DOI] [PubMed] [Google Scholar]
- 20••.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [Using in vitro assays and adoptive transfer of thymic progenitors expanded in fetal thymic organ cultures, the authors demonstrated that early stage thymocyte progenitors retain myeloid potential, and that this potential is not extinguished until differentiation beyond the DN2 stage.] [DOI] [PubMed] [Google Scholar]
- 21.Rothenberg EV, Moore JE, Yui MA. Launching the T-cell-lineage developmental programme. Nat Rev Immunol. 2008;8:9–21. doi: 10.1038/nri2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 23.Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes Dev. 2009;23:2376–2381. doi: 10.1101/gad.1836009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 25.Avitahl N, Winandy S, Friedrich C, Jones B, Ge Y, Georgopoulos K. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999;10:333–343. doi: 10.1016/s1074-7613(00)80033-3. [DOI] [PubMed] [Google Scholar]
- 26.Winandy S, Wu L, Wang JH, Georgopoulos K. Pre-T cell receptor (TCR) and TCR-controlled checkpoints in T cell differentiation are set by Ikaros. J Exp Med. 1999;190:1039–1048. doi: 10.1084/jem.190.8.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mullighan C, Downing J. Ikaros and acute leukemia. Leuk Lymphoma. 2008;49:847–849. doi: 10.1080/10428190801947500. [DOI] [PubMed] [Google Scholar]
- 28.Wang JH, Nichogiannopoulou A, Wu L, Sun L, Sharpe AH, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 29•.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A Proteins Promote Development of Lymphoid-Primed Multipotent Progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [In addition to providing the transcriptome of Tcfe2a in early hematopoietic progenitors, this study demonstrates that E2a activity is required for differentiation of LMPPs from HSCs and to antagonize GMP differentiation from LMPPs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Spooner CJ, Cheng JX, Pujadas E, Laslo P, Singh H. A recurrent network involving the transcription factors PU.1 and Gfi1 orchestrates innate and adaptive immune cell fates. Immunity. 2009;31:576–586. doi: 10.1016/j.immuni.2009.07.011. [Using data from knockout mice and shRNA gene knockdown experiments, this study concludes that Ikaros promotes B cell differentiation through down-regulation of Gfi-1, an inhibitor of PU.1 activity. However, this study should be regarded with the caveat that Ng et al have shown that Gfi-1 is up-regulated in Ikaros null HSC, LMPP and GMP, and the data for Ikaros’ negative regulation of Gfi-1 consisted of a measurement of fluorescence of a Gfi-1 reporter in a heterogenous progenitor population (consisting of LT-, ST-HSC and LMPP). The bi-modal changes in report expression detected in the mutant (both higher and lower relative to wild) likely reflects changes in progenitor subsets caused by the loss of Gfi-1.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothenberg EV, Pant R. Origins of lymphocyte developmental programs: transcription factor evidence. Semin Immunol. 2004;16:227–238. doi: 10.1016/j.smim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 32.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 34.O'Neill DW, Schoetz SS, Lopez RA, Castle M, Rabinowitz L, Shor E, Krawchuk D, Goll MG, Renz M, Seelig HP, et al. An ikaros-containing chromatin- remodeling complex in adult-type erythroid cells. Mol Cell Biol. 2000;20:7572–7582. doi: 10.1128/mcb.20.20.7572-7582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sridharan R, Smale ST. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J Biol Chem. 2007;282:30227–30238. doi: 10.1074/jbc.M702541200. [DOI] [PubMed] [Google Scholar]